Abstract
Background
Although molecular tools are increasingly employed to decipher invertebrate systematics, earthworm (Annelida: Clitellata: ‘Oligochaeta’) taxonomy is still largely based on conventional dissection, resulting in data that are mostly unsuitable for dissemination through online databases. In order to evaluate if micro-computed tomography (μCT) in combination with soft tissue staining techniques could be used to expand the existing set of tools available for studying internal and external structures of earthworms, μCT scans of freshly fixed and museum specimens were gathered.
Findings
Scout images revealed full penetration of tissues by the staining agent. The attained isotropic voxel resolutions permit identification of internal and external structures conventionally used in earthworm taxonomy. The μCT projection and reconstruction images have been deposited in the online data repository GigaDB and are publicly available for download.
Conclusions
The dataset presented here shows that earthworms constitute suitable candidates for μCT scanning in combination with soft tissue staining. Not only are the data comparable to results derived from traditional dissection techniques, but due to their digital nature the data also permit computer-based interactive exploration of earthworm morphology and anatomy. The approach pursued here can be applied to freshly fixed as well as museum specimens, which is of particular importance when considering the use of rare or valuable material. Finally, a number of aspects related to the deposition of digital morphological data are briefly discussed.
Similar content being viewed by others
Data description
Purpose of data acquisition
The present dataset constitutes the first attempt at comparative micro-computed tomography (μCT) scanning of earthworm (Annelida: Clitellata: ‘Oligochaeta’) specimens. When used in combination with staining techniques that permit enhancing soft tissue contrast [1], μCT could become a promising technique for resolving pervasive issues in earthworm taxonomy and systematics. To this end, the application of μCT to freshly fixed and museum specimens was evaluated, and results were compared with data derived from traditional dissection techniques. The main methodological and taxonomical results of the study are presented in an accompanying publication [2].
The aim of the present report is to provide the earthworm research community with a reference dataset for future analyses of soft-bodied organisms based on non-destructive imaging techniques. In addition, uninhibited data access and enforced data deposition, as practiced here, are briefly discussed.
Scanned specimens
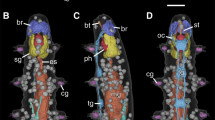
Scans of four lumbricid (‘Oligochaeta’: Lumbricidae) earthworm specimens are part of the present dataset. One freshly fixed and one museum specimen (stored in ethanol for several decades) were scanned for each of the two different species employed in the study, i.e. Aporrectodea caliginosa (Savigny, 1826) and Aporrectodea trapezoides (Dugès, 1828). All four specimens were stained using an ethanol-based phosphotungstic acid (PTA) solution, which was adapted from protocols described previously [3]. In order to increase the isotropic voxel resolution of the three-dimensional (3D) image stack, only the first ca. 35 segments of each specimen were scanned. These segments harbor all internal and external structures commonly used in earthworm taxonomy. Specific specimen data and supplementary image files have been deposited in the publicly accessible database of the Museum of Comparative Zoology, MCZbase (http://mczbase.mcz.harvard.edu/). In addition, hyperlinks to each specimen entry in MCZbase are provided on the dataset website in the GigaScience Database (GigaDB) online repository [4].
Data acquisition and processing
The four scans were produced using a μCT system equipped with a cone-beam tungsten X-ray source (SkyScan 1173, Bruker microCT, Kontich, Belgium). The specific scanning parameters are provided in the accompanying publication [2], and can also be found in the log file (.log) of each dataset folder available for download at GigaDB[4].
Each scan resulted in a set of 960 projection images in tagged image file format (TIFF, .tif). No binning protocols were employed during data acquisition. The projection images covered 2240 × 2240 pixels at 16-bit dynamic range. Reconstruction of the two-dimensional (2D) projection images into a 3D volumetric image stack was performed using the software NRecon 1.6.6.0 (Bruker microCT, Kontich, Belgium). This program runs under the reconstruction engine NReconServer 1.6.6, which employs a Feldkamp algorithm for volumetric reconstruction [5]. The two reconstruction parameters with significant effect on the quality of the final data were ring artifact and beam hardening correction. The output format for the 3D volumetric image stacks was bitmap image file (BMP, .bmp) at 8-bit dynamic range and 2240 × 2240 pixel size. In order to reduce final file size, the volume of interest (VOI) function, a 3D cropping tool, was used to remove all uninformative parts of the data following reconstruction. This resulted in changes to the pixel dimensions of each reconstructed image stack, but did not lead to spatial distortions in any of the three dimensions. Further information on the contents and size of both the projection and the reconstruction data folders is provided in Table 1.
Data quality
The quality of the data was ascertained through visual inspection of the scout projection and reconstruction images. Primary criteria were i) the full penetration of the sample by the staining agent and ii) the absence of artifacts. Although a total of eight scans were obtained in the course of the study, four of these scans were either trial scans or showed significant artifacts [2]. Therefore, only the four most representative scans have been deposited in GigaDB. Nonetheless, these scans represent the full taxonomic and morphological breadth of species and sample types included in the study. The imagery allows for an identification of numerous internal and external structures. No significant difference in the approach was observed when employing freshly fixed or museum specimens, nor between the two species analyzed.
Potential uses
The potential uses of the dataset presented here include morphometric or volumetric analyses of internal organs, studies of ingested sediment particles, the possibility of online collaborative dataset annotation, or interactive data exploration using digital 2D and 3D visualization tools.
The methodological approach itself is suitable for high-throughput scanning of hundreds or even thousands of earthworm specimens as well as other soft-bodied organisms [2]. This would result in large morphological taxon sampling, one of the prerequisites for broad taxonomic and systematic studies. Furthermore, non-invasive imaging techniques such as μCT leave specimens intact and generate digital data suitable for online dissemination, an important condition for effective data mining.
Availability and requirements
Data availability
The dataset is available at GigaDB and has a citable digital object identifier (DOI) [4]. Each of the eight folders has been packed using tape archiver (tar, .tar), before being compressed using GNU zip (gzip, .gz). The folders can be individually downloaded using a set of tools, e.g. File Transfer Protocol (FTP).
Dataset name: MicroCT scans of freshly fixed and museum earthworm specimens
Operating system: Platform-independent
License: Creative Commons 0 (CC0) public domain dedication (https://creativecommons.org/publicdomain/zero/1.0/)
Data requirements
Following download, the reconstructed images can, for example, be rapidly visualized using the ‘File:Import:Image Sequence’ command chain in the Java-based imaging software ImageJ (http://imagej.nih.gov/ij/). In addition, numerous other 2D and 3D visualization tools are available for free [6]. Given the size of the reconstructed image folders, a computer system with about 4 GB main random access memory (RAM) and 1 GB video RAM should be used.
Discussion
The dataset presented here permits full open access both to μCT-derived raw data (here: the projection images) as well as derivative data (here: the reconstructed image stacks). The availability of μCT raw data files has been deemed important, primarily due to the rapid increase in the performance of reconstruction algorithms, which in the future could lead to improved data reconstruction [7]. Furthermore, one reviewer as well as the editor of the accompanying publication [2] requested data deposition for purposes of data transparency, which was achieved here through storage and archiving of the dataset in GigaDB[4]. Despite these advances, a lack of coherent policy for data archiving and enforced data deposition in digital morphology remains [8], and metadata standards for data gathered using non-invasive imaging techniques are still not available [7].
Availability of supporting data
The dataset supporting the results of this article is available in the GigaScience Database online repository [4].
Abbreviations
- 2D:
-
Two-dimensional
- 3D:
-
Three-dimensional
- BMP:
-
Bitmap image file
- CC0:
-
Creative Commons 0 1.0 public domain dedication
- DOI:
-
Digital object identifier
- FTP:
-
File Transfer Protocol
- GigaDB:
-
GigaScience Database
- GNU:
-
GNU’s not unix
- gzip:
-
GNU zip
- MCZ IZ:
-
Museum of Comparative Zoology Invertebrate Zoology
- μCT:
-
Micro-computed tomography
- PTA:
-
Phosphotungstic acid
- RAM:
-
Random access memory
- ROI:
-
region of interest
- TIFF:
-
Tagged image file format
- VOI:
-
Volume of interest.
References
Metscher BD: MicroCT for comparative morphology: simple staining methods allow high-contrast 3D imaging of diverse non-mineralized animal tissues. BMC Physiol. 2009, 9: 11-10.1186/1472-6793-9-11. doi:10.1186/1472-6793-9-11
Fernández R, Kvist S, Lenihan J, Giribet G, Ziegler A: Sine systemate chaos? A versatile tool for earthworm taxonomy: non-destructive imaging of freshly fixed and museum specimens using micro-computed tomography. PLoS ONE. 2014, 9 (5): e96617-doi:10.1371/journal.pone.0096617
Metscher BD: X-ray microtomographic imaging of intact vertebrate embryos. Cold Spring Harb Protoc. 2011, 12: 1462-1471. doi:10.1101/pdb.prot067033
Lenihan J, Kvist S, Fernández R, Giribet G, Ziegler A: MicroCT scans of freshly fixed and museum earthworm specimens. GigaSci Database. 2014,http://dx.doi.org/10.5524/100092,
Xiao S, Bresler Y, Munson DC: Proceedings of the 2003 International Conference on Image Processing (ICP 2003): 14–18 September 2003; Barcelona, Spain. 2003, IEEE, 819-822. doi:10.1109/ICIP.2003.1246806
Elicieri KW, Berthold MR, Goldberg IG, Ibánez L, Manjunath BS, Martone ME, Murphy RF, Peng H, Plant AL, Roysam BR, Stuurman N, Swedlow JR, Tamancak P, Carpenter AE: Biological imaging software tools. Nature Meth. 2012, 9: 697-710. 10.1038/nmeth.2084. doi:10.1038/nmeth.2084
Rowe T, Frank LR: The disappearing third dimension. Science. 2011, 331: 712-714. 10.1126/science.1202828. doi:10.1126/science.1202828
Ziegler A, Ogurreck M, Steinke T, Beckmann F, Prohaska S, Ziegler A: Opportunities and challenges for digital morphology. Biol Direct. 2010, 5: 45-10.1186/1745-6150-5-45. doi:10.1186/1745-6150-5-45
Acknowledgements
We are grateful to Christopher Laumer for help with reagent preparation. We thank Christopher Hunter for assistance with data upload and management. Scott Edmunds, Sarah Faulwetter, and Brian D. Metscher provided comments that helped to improve the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: JL SK RF AZ. Performed the experiments: JL SK RF. Analyzed the data: JL SK RF AZ. Contributed reagents/materials/analysis tools: JL SK RF GG AZ. Wrote the paper: JL SK RF GG AZ. All authors read and approved the final manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Lenihan, J., Kvist, S., Fernández, R. et al. A dataset comprising four micro-computed tomography scans of freshly fixed and museum earthworm specimens. GigaSci 3, 6 (2014). https://doi.org/10.1186/2047-217X-3-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2047-217X-3-6