Abstract
Filamin-A (FLNA), also called actin-binding protein 280 (ABP-280), was originally identified as a non-muscle actin binding protein, which organizes filamentous actin into orthogonal networks and stress fibers. Filamin-A also anchors various transmembrane proteins to the actin cytoskeleton and provides a scaffold for a wide range of cytoplasmic and nuclear signaling proteins. Intriguingly, several studies have revealed that filamin-A associates with multiple non-cytoskeletal proteins of diverse function and is involved in several unrelated pathways. Mutations and aberrant expression of filamin-A have been reported in human genetic diseases and several types of cancer. In this review, we discuss the implications of filamin-A in cancer progression, including metastasis and DNA damage response.
Similar content being viewed by others
Introduction
The cytoskeleton, a complex network of protein fibers in eukaryotic cells, provides a dynamic structural framework that is crucial for maintaining normal cell activity, including cell shape, cellular motion, division, and intracellular transport among other processes [1–3]. Eukaryotic cells contain three main types of cytoskeletal filaments: microfilaments, intermediate filaments, and microtubules [1, 3]. Microfilaments (also called actin filaments or F-actin) are composed of linear polymers of actin subunits that form the thinnest filaments of the cytoskeleton. The actin filament is a polar macromolecule characterized by the elongation of one filament end coupled with shrinkage at the other. This dynamic interplay generates force and causes net movement of the intervening strand [4]. Actin filaments also act as tracks for the movement of myosin molecules that attach to the microfilament and "walk" along them [5]. In addition, actin filaments cross-link into bundles to form the dynamic actin cytoskeletal network, which is in turn finely tuned by multiple families of cytoskeletal proteins [6], called actin binding proteins. These proteins typically share a conserved, α-actinin-like F-actin binding domain (ABD) [7–9].
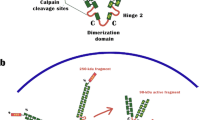
One actin binding protein that contains the actinin-like F-actin binding domain is the filamin family. This family is composed of three homologous proteins (FLNA, FLNB, and FLNC) that are products of different genes and their mRNA splice variants [10]. The three filamin genes are highly conserved and filamin proteins exhibit 60-80% overall amino acid identity, with the greatest divergence observed at the two hinge regions, sharing 45% identity [11]. Filamin-A (FLNA), also known as human actin-binding protein 280 (ABP-280) or filamin-1, is encoded by the X-linked gene FLNA[12, 13]. As shown in Figure 1, the human filamin-A is a homodimer with large subunits of 280KD, forming a V-shaped structure [11, 14–16]. At the NH2 terminus of the monomer, there is an actin-binding domain (ABD), followed by 24 tandem repeats of ~96 amino acids in length. Between repeats 15 and 16, there is a hinge domain, and repeat 24 is separated from repeat 23 by a second hinge domain. The last 65 amino acids of repeat 24 mediates the dimerization of filamin-A subunits [14].
Structure of human filamin-A protein. Filamin-A is a V-shaped homodimer. Each monomer is a protein of 2647 amino acids that contains 24 tandem repeats. Each repeat contains ~96 amino acids. Filamin-A can be divided into 3 major domains: F-Actin-Binding Domain (ABD), the filamin-A Rod regions: Rod-1 containing repeat 1–15 and Rod-2 containing repeats 16–23; C-Terminal Domain (FCTD) of repeats 16–24, containing major partner interaction domains. In the FCTD, a defined C-terminal filamin-A dimerization domain of ~65 amino acids is located in repeat 24, and a filamin-A hinge regions of about 34 amino acids is located between repeat 23 and 24, and between repeat 15–16
Filamin-A binds and cross-links cortical actin filaments into a dynamic three-dimensional structure through its N-terminal actin-binding domain [17, 18]. In addition to filamentous actin, filamin-A interacts with more than 60 functionally diverse cellular proteins, including trans-membrane receptors, signaling molecules, DNA damage repair proteins. These diverse interactions suggest that filamin-A is a key component of a versatile signaling scaffold complex [15, 17–19]. In this review, we discuss the role of filamin-A in human diseases, with an emphasis on cancer.
Filamin-A mutations and human genetic diseases
Due to its versatile function in regulating cell motility and signaling, defects in the FLNA gene have been demonstrated as the cause for a wide range of developmental malformations involving the brain, bone, limbs, and heart [10, 19]. Periventricular nodular heterotopia (PNH) is a well characterized neuron development disorder, mainly caused by loss-of-function mutations in FLNA gene [20–25], while other possible genetic contributions cannot be excluded [26–29]. In patients affected by PNH, clusters of grey matter along the ventricles consisting of neurons fail to migrate to the cortex during prenatal development. X-linked PNH is mainly confined to females, indicating that FLNA null mutations in males are predominantly associated with prenatal death. In addition, PNH patients can co-present abnormal cerebral migration with other brain, skeletal, or visceral abnormalities [30–35], while higher chance of FLNA mutation has been implicated as the causes for these cases. Furthermore, clustered missense mutations in FLNA have been identified in a diverse spectrum of congenital malformations in humans [19, 36], including otopalatodigital syndrome (OPD), frontometaphyseal dysplasia (FMD) and Melnick–Needles syndrome (MNS). Moreover, using a familial and genealogical survey, it was discovered that missense mutations in FLNA were the likely genetic cause of familial cardiac valvular dystrophy [33, 37, 38].
The importance of filamin-A controlled cell adhesion and migration in human development has been confirmed using mouse models and established cell lines of filamin-A deficiency [39–43]. These approaches have been successful in reconstructing the phenotypes observed in patients carrying FLNA defects, indicating that impaired filamin-A is likely responsible for several human genetic diseases. The underlying pathological mechanisms of these diseases are complex and might be attributed to the loss of FLNA partner bindings or through aberrant FLNA protein interactions. Nevertheless, the effect of most of patient mutations on FLNA partner interactions remains to be elucidated at the molecular level.
Filamin-A in cytoskeleton reorganization and cell shape determination
The cytoskeleton provides rigid structural support that is responsible for maintaining cell shape. Somewhat paradoxically, this rigid network is also highly active and dynamic, which endows the cells with plasticity and adaptability to respond to stimuli from the surrounding environment. As an actin binding protein, filamin-A plays an essential role in these processes. Filamin-A is responsible for cross-linking actin filaments into orthogonal networks. These Filamin-A-actin networks possess active and reversible organizational properties, which protect cell from various shear stresses [44–46]. As shown in Figure 1, the unique structure of filamin-A, homodimer and multiple actin binding sites (ABD and Rod 1 domain), forges its high avidity binding to F-actin [47]. FLNA hinges alternatively confer flexibility to the filamin-A molecule [44]. The conformation of Rod 2 region also modulates the interaction of FLNA with multiple binding partners (Table 1), which allow the binding affinity of FLNA to actin filaments to be finely tuned to the arrangement of F-actin which varies throughout the cytoplasm [48–51]. Furthermore, filamin-A has been shown to regulate intermediate filament systems via the interaction with vimentin, a component of intermediate filaments [52, 53]. These conserved structural functions allow filamin-A to be a highly dynamic mediator of cytoskeleton reorganization.
Filamin-A also connects actin networks to plasma membrane. The distinctive shape of a cell depends on the organization of the actin network together with the proteins that connect actin filaments to the plasma membrane. When attached to a planner network of actin filaments, the membrane maintains as a flat surface. However, when the underlying actin network is organized into parallel bundles of actin filaments, the membrane acquires the finger-like shape of cell protrusion, such as lamellipodia, pseudopodia and filopodia. In addition to cross-linking actin filaments into 3D networks, filamin-A interacts with a wide variety of proteins, including transmembrane proteins and signaling molecules (Table 1). Additionally, filamin-A acts an adaptor that mediates the complex connections between the integral membrane proteins and actin filaments, which enables filamin-A to modulate cell shape at specific areas. Moreover, actin filaments are enriched in the cortex of many cells, a narrow zone just beneath the plasma membrane, where the protrusive structures are formed. At both the leading edge and the rear of polarized motile cells, filamin-A can bind actin to influence the nature of cytoplasmic protrusions and retractions [94, 95]. Interestingly, it has been shown recently that filamin-A organizes perinuclear actin networks and regulates the nuclear shape via RefilinB (FAM101B) [91].
In addition, Filamin-A acts as a scaffold for signaling molecules that mediate intracellular protein trafficking. Membrane receptors sense and transmit extracellular biochemical signals across cell membranes, which is dependent on the association with the F-actin cytoskeleton. Filamin-A regulates matrix–cytoskeleton signaling by virtue of binding to both actin networks and the cytoplasmic tails of membrane receptors (Table 1). One of the most well-studied interactions is with the integrin family of proteins which have recently been described at the atomic level [48, 49]. In response to intergrin binding, the filamin-A molecule is mechanically stretched, which alters partner binding affinity while opening new partner binding sites. It is hypothesized that this phenomenon underlies FLNA’s mechanism for regulating the cytoskeleton. Following integrin binding to extracellular matrix ligands, filamin-A coordinates the actin-remodeling activities of GTPase signaling factors as shown in Table 1, and anchors them in proximity to the cell membrane, leading the formation of lamellipodia and filopodia [10]. Upon stimulation, membrane receptors are continually endocytosed and recycled back to the cell membrane by trafficking [96, 97]. While the biochemical pathways of protein trafficking are not completely defined, increasing evidence demonstrates that filamin-A is pivotal for the intracellular trafficking and for deployment of cell β1 integrins on the cell membrane, which are essential for maintaining directed cell migration[53, 98, 99].
Filmin-A and cell-matrix interaction
The extracellular matrix (ECM) not only provides essential physical support to cells but also initiates crucial biochemical and biomechanical cues that are required for tissue morphogenesis, tumor invasion, and wound healing [100, 101]. However, the mechanisms by which cells recognize and respond to changes in ECM properties are not clear. It is believed that adhesion receptors such as integrins, discoidin domain receptors and syndecans mediate cell to cell communication [102–104]. Upon stimulation by extracellular matrix ligands, intergrins are activated and recruit filamin-A to actin filaments, which facilitate cell spread through matrix-to-cell (outside-in) signals. As a result, filamin-A may enhance integrin–ligand interactions by binding to the cytoplasmic domain of the β1-intergrin subunit through inside-out signaling [98].
Although interactions with adhesion receptors facilitate the interplay between the cell and the ECM, the mechanisms by which filamin-A regulates cell migration are not simply unidirectional. For example, the strong association between integrin and filamin-A impairs migration [105], and the competition binding by talin to replace filamin-A can resume signaling and movement, which altogether underscores the importance of a balance of multiple interactions during migration [66, 67, 105]. Xu et al. reported that filamin-A regulates focal adhesion disassembly, and that down-regulation of filamin-A enhances the cleavage of focal adhesion proteins [106]. Furthermore, filamin-A has been shown to play a role in ECM degradation and it has been demonstrated that knockdown of filamin-A increases the expression of matrix metalloproteinase-9 (MMP-9) [107] which induces MMP2 activation [108].
Filamina A and cancer metastasis
It is known than filamin-A interacts with many proteins related to cancer metastasis [17, 41, 52, 83, 109, 110] (Table 1). Although a large volume of studies associate filamin-A with cancer metastasis, the specific roles of filamin-A during metastatic invasion remain elusive. This enigma is a reflection of the complex nature of metastasis, a multifactorial process which involves tumor cell detachment from the primary sites, followed by tumor cell invasion of the EMC and migration, and finally intravasation, survival in the vasculature, extravasation, and colonization at the secondary sites. Metastasis requires the cancer cells to be able to adapt to different cells shapes, to resist to mechanical stress, and to be highly motile. In addition, it is highly important that the cell is able to re-attach to ECM at secon dary sites. It should come as no surprise that all of these processes are related to the filamin-A cytoskeleton network. Thus, it is conceivable that lack of filamin-A would render the tumor cells less mobile and less invasive, more sensitive to mechanical stress, and result in inefficiency in attachment to ECM at secondary sites to form metastasis colonies. It is also possible that cancer cells with appropriate filamin-A levels would likely have an advantage during metastasis. Alternatively, during the early stages of tumorigenesis, in situ cancer cells need to be detached from the original sites to initiate metastasis. It stands to reason that filamin-A may be required for the cancer cells to remain attached to the original site, and that mis-regulation or loss of filamin-A may increase the risk of initiation of cancer metastasis. Due to the dual roles of filamin-A in the regulation of cancer cell mobility and cell interaction with the extracellular matrix, it is unsurprising that inconsistent results were reported on filamin-A’s role in cancer metastasis (see a summary in Table 2).
In support of the hypothesis that filamin-A promotes cancer metastasis, a comparative proteomic study has identified that high levels of filamin-A were correlated with increased metastatic potential of hepatocellular carcinoma [112]. Using a three-dimensional migration approach, Quite et al. found that filamin-A is required for podosome stabilization, podosome rosette formation, extracellular matrix degradation, and three-dimensional mesenchymal migration [121]. Li et al. identified the interaction of pro-PrP with filamin-A and the binding enhances association between FLNA and integrin β1, which promotes cell spreading and migration and further contributes to melanomagenesis [62]. Recently, we have shown that knockdown of filamin-A in melanoma C8161 cells reduced metastasis in xenograft mouse models, and that filamin-A inhibition reduced the mobility and invasiveness of breast cancer cell lines that do not over-express ErbB2 in vitro[116]. These studies support the model that filamin-A expression is likely responsible for remodeling cancer cell shape and mobility which is integral for initiating metastasis.
Filamin-A readily undergoes proteolysis at its two hinge regions to generate 170 kDa, 150 kDa, 120 kDa, 110 kDa and 90 kDa of major fragments. Based on IHC on human prostate tissue microarray, Bedolla et al. found that filamin-A proteolysis is associated with a reduction of metastatic potential of prostate cancer. The prostate cancer metastasis correlates with cytoplasmic localization of full-length filamin-A but not nuclear filamin-A fragments [114]. It was further suggested that metastasis may be prevented by cleavage and subsequent nuclear translocation of this protein [114]. Castoria et al. recently reported that the interaction of filamin-A with androgen receptor (AR) at the cytoskeleton can be rapidly induced upon stimulation by androgen. Filamin-A/AR complex further recruits integrin beta 1, activating a cascade that drives cell migration [79]. Cells expressing mutant AR that lack a filamin-A interacting domain fail migrate in response to androgen stimulation [79].
At the same time, a few reports suggest that reduction of the full-length filamin-A promotes metastasis by a different mechanism. Baldassarre et al. showed that knockdown of filamin-A in fibrosarcoma increased matrix metalloprotease 2 activity and activation, enhanced the ability of cells to remodel the ECM, and increased cellular invasive potential without significantly altering two-dimensional random cell migration [108]. In addition, using breast cancer cells that over-express ErbB2, Xu et al. reported that knockdown of filamin-A promoted cleavage of focal adhesion in cancer cells and stimulated cancer cell migration, invasion, and metastasis. The same authors also reported that local breast cancers during early development have a higher levels of filamin-A than late counterparts [106]. Although the question of whether the filamin-A levels witnessed in these cancer tissues were inclusive of surrounding ECM among the in situ carcinoma remains elusive, this study suggests that loss of filamin-A enhances FAK turnover, which results in the disrupting cancer cell attachment to the EMC. Furthermore, O’Connell et al. demonstrated that Wnt5a-activated calpain 1 is able of cleaving filamin-A, which causes cytoskeleton remodeling and enhances melanoma cell motility [120].
The apparent discrepancies among the literature strongly indicate that levels of filamin-A in the cancer cells may not be the sole indicator of predicting whether a cancer is more or less metastatic. The type of cancer cells (such as carcinoma verses fibrosarcoma), concurrent expression of other relevant genes in the cancer cells (such as ErbB2), their interactions with the extracellular environment, and the proteolysis of filamin-A also appear to be major collaborating factors for filamin-A mediated metastatic invasion.
Filamin-A in cell signaling and cancer progression
In addition to the regulation of metastasis, filamin-A is also involved in other aspects of tumor progression. The interaction between filamin-A and R-RAS has been suggested to be responsible for maintaining the endothelial barrier function [56]. Knockout of mouse FlnA significantly reduces the oncogenic properties of K-RAS, including lung tumor formation, proliferation of K-RAS expressing fibroblasts, and the activation of the downstream signaling molecules such as ERK and AKT [76]. Found in most cases of infant acute lymphoblastic leukaemia (ALL) and acute myeloblastic leukaemia (AML), the fusion of MLL and its partner gene leads to a gain of function of the MLL gene, which affects the differentiation of the hematopoietic pluripotent stem cells or lymphoid and myeloid committed stem cells [122]. FLNA was also identified as a new partner gene fused to MLL [123]. Recently, Muscolini et al. identified the interaction between filamin-A and NF-κB inducing kinase (NIK), and found that filamin-A is essential for mediating the activation of the IKKα/NF-κB cascade through CD28 signaling [77].
Interaction of FLNA with membrane transforming receptors has also been reported. Najib et al. found that filamin-A can compete with PI3K-p85 binding to the G protein-coupled receptor (GPCR) sst2 to negatively regulate PI3K signaling [78]. It has also been demonstrated that filamin-A modulates signaling events and cellular responses induced by external stimuli, including EGF receptor [124] and tumor necrosis factor (TNF) receptor-associated factor 2 (TRAF2) [82].
Although FLNA mutations can be the cause of human genetic disease, increased cancer incidence has not been observed in these patients. Furthermore, filamin-A is dispensable for cell-autonomous survival and loss of filamin-A expression or depletion of filamin-A with RNAi does not cause cell death or not impede growth [88, 125–127]. Nevertheless, disruption of filamin-A function may contribute to the biology of cancers and provide the tumor cells with a growth advantage. Therefore, further studies are required to address whether anomaly in FLNA can initiate tumorigenesis.
DNA damage response and nuclear functions of Filamin-A
In the past two decades, a striking discovery related to filamin-A is its nuclear functions, especially in DNA damage response. In 2001, Yuan and Shen first reported the interaction between filamin-A and BRCA2, a critical protein involved in DNA damage repair [89]. Coincidentally, Velkova et al. reported interaction of filamin-A with BRCA1, another DNA repair protein, in 2010 [87]. It has been demonstrated that inhibition of filamin-A moderately inhibited homologous recombinational DNA repair, sensitized cells to ionizing radiation and cisplatin, and prolonged the recovery of G2 arrest following irradiation [90]. Velkova first suggested a role of filamin-A in non-homologous end joining repair of the DNA double strand breaks [87] and has since fundamentally established a role of filamin-A in DNA damage response through the interactions with BRCA1 and BRCA2 [87, 88, 90, 126, 127]. Because DNA damage response signaling and DNA repair play a critical role in the maintenance of genomic integrity [128], these studies also raised a possibility that mis-regulation of filamin-A function may contribute to the initiation of tumorigenesis.
It is likely that filamin-A acts in DNA damage as auxiliary factor to provide nuclear matrix support for the repair machinery instead of serving as an enzyme directly involved in processing DNA damage. Perhaps due to this indirect role in DNA damage response, the specific consequence of filamin-A inhibition in cell sensitivities to DNA damage depends on the type of damage or how the DNA damage is induced. For example, although inhibition of filamin-A sensitizes cells to radiation, cisplatin, and bleomycin, it surprisingly also confers a resistance to topo-isomerse poisons [126].
It has also been demonstrated that filamin-A interacts with RefilinB (FAM101B) to organize perinuclear actin networks and regulate nuclear shape, which is essential for normal nuclear function maintenance [91]. Deng et al. also demonstrated that filamin-A is a nucleolar protein that suppresses ribosomal RNA gene transcription [92]. In addition, Oiu et al. identified two nuclear binding partners of filamin-A, TAF1B and mKIAA1093, from mouse embryonic cDNA libraries, which further endorses the proposed nuclear function of filamin-A [93].
Filamin-A as a biomarker and potential target for cancer therapy
Due to its functions in the control of cell mobility, cell-ECM interactions, cell signaling, and DNA damage response, it is conceivable that filamin-A may be developed as a biomarker for cancer diagnosis and outcome prediction. When compared with normal tissues, increased expression of filamin-A was observed in various types of cancers. In some cases, different stages of cancers have varied level of filamin-A expression. Thus, when compared with normal tissues, filamin-A expression may be considered as an alternative diagnostic marker. In addition, variable expression levels of filamin-A protein among different cancers may serve as a useful tool for individualized therapy and outcome prediction.
The correlation of filamin-A expression patterns in different stages of cancer tissues and cancer outcomes has been reported. Uhlen et al. employed antibody-based proteomics to evaluate the expression and localization of FLNA profiles in 48 normal human tissues and 20 different cancers [129]. They found that ~50 % of the colorectal cancer had moderate to high levels of filamin-A, whereas filamin-A was undetectable in normal colon glandular cells. Similarly, the expression of filamin-A was increased in pancreatic cancer, while in normal pancreas, exocrine ductal cells had low to moderate levels of filamin-A [129]. A Gene profiling approach was also employed to assess filamin-A expression in human cancer. Up-regulation of filamin-A was detected in salivary gland adenoid cystic carcinoma [130], peripheral cholangiocarcinomas [131], human glioblastomas [132] and in pancreatic cancer [133]. On the contrary, under-expression of filamin-A was also observed in some types of cancers, including human bladder cancer [134, 135] and colon adenocarcinoma [136]. These reports suggest that, tumor tissues may have different expression levels of filamin-A from the surrounding normal tissue, and thus FLNA may serve as a useful diagnostic marker.
As discussed in early sections, various reports have implicated that the levels of filamin-A expression in the cancer cells may be correlated with the cancer metastatic potential and sensitivities to therapeutic DNA damage agents. It is conceivable that the level of filamin-A in cancer tissue can be developed as an alternative marker for outcome prediction and individualized therapy. As we previously discussed, the role of filamin-A in DNA damage sensitivity is dependent on the type of DNA damage [126]. A further implication of these findings is that expression level of filamin-A in cancer cells may be exploited as a biomarker to predict the effectiveness of different types of therapeutic DNA damage agents to the cancer subtype [126].
The role of filamin-A in DNA damage response presents an intriguing possibility that targeting filamin-A may be useful for cancer therapy. In a panel of human melanoma cell lines established from cancer patients, a correlation between filamin-A expression and drug sensitivity was observed, i.e. the lower filamin-A level the cell line had, the more sensitive the cell was to bleomycin and cisplatin treatment [127]. In addition to chemotherapeutic drugs, filamin-A deficiency sensitizes melanoma and breast cancer cells to ionizing radiation [88, 89]. These data was further confirmed by xenograft animal models, which demonstrated that tumors generated from filamin-A deficient cells displayed a better response to chemotherapy [127]. In addition, Nallapalli et al. found that filamin-A played an important role during lung tumor growth [76]. These studies predict that inhibition of filamin-A sensitizes cancer cells to specific therapeutic DNA damage agents. Considering the metastatic role of filamin-A in cancer cell mobility and re-attachment to secondary sites, it is also conceivable to predict that inhibition of filamin-A would also reduce distant cancer metastasis. Thus, filamin-A may also be an ideal therapeutic target for metastasis control.
Small molecules to block filamin-A function have been attempted. PTI-125 is novel compound binding to filamin-A with 200 femtomolar affinity, which disrupts the toxic signaling of amyloid-β42 (Aβ42), reducing amyloid-related Alzheimer’s disease pathogenesis [137]. The same research group also designed filamin-A peptide fragments containing the Ultra-low-dose naloxone (NLX) binding site, which blocked the protective effect of NLX on both the MOR–Gs coupling and downstream cAMP accumulation induced by chronic morphine, presumably by interfering with NLX’s binding to filamin-A [138]. Considering that the dimerization of filamin-A at the C-terminus is a common feature for filamin-A function, we propose that disruption of the filamin-A dimer formation could be a valuable approach.
In addition to simply inhibiting filamin-A expression, alternative strategies to disrupt the filamin-A functions have been suggested. Because filamin-A cleavage has been shown to be associated with reduced metastasis, Bedolla et al. suggest that strategies to induce filamin-A cleavage may be developed to reduce cancer metastasis [114]. This strategy may be feasible because it is known that filamin-A proteolysis by calpains is regulated by AKT-dependent phosphorylation at serine 2152 [139], and treatment with PI3K inhibitor that abrogates AKT activity induces filamin-A cleavage and localization of the cleavage product into nucleus [140].
In summary, due to its multiple functions as a scaffolding protein, filamin-A integrates intracellular signaling and mediates a variety of cellular processes (Figure 2). Its functions in cancer, especially in metastasis and sensitivity to DNA damage, may be exploited for cancer therapy and outcome prediction. However, the specific consequence of filamin-A inhibition in metastasis and DNA damage sensitivity is likely dependent on the cancer stages and types of DNA damage agents. Extensive studies are required to fully develop filamin-A as a valid cancer marker and therapeutic target.
Schematic presentation of filamin-A functions. Through the interactions with its binding partners, filamin-A is endowed with versatile cellular functions, including maintenance of dynamic F-actin networks and regulating cell shape; mediating the communication between cytoskeleton and ECM; acting as a scaffold for cell signaling to regulate cell motility; facilitating intracellular trafficking and promoting membrane protein recycling; regulating RNA transcription through interactions with transcriptional factors and RNA polymerase machinery; modulating nuclear receptor signaling through the binding with androgen receptor; and mediating DNA damage response through interactions with BRCA1, BRCA2
References
Wickstead B, Gull K: The evolution of the cytoskeleton. J Cell Biol. 2012, 194 (4): 513-525.
Doherty GJ, McMahon HT: Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Annu Rev Biophys. 2008, 37: 65-95. 10.1146/annurev.biophys.37.032807.125912
Boggiano JC, Fehon RG: Growth control by committee: intercellular junctions, cell polarity, and the cytoskeleton regulate Hippo signaling. Dev Cell. 2012, 22 (4): 695-702. 10.1016/j.devcel.2012.03.013
Galkin VE, Orlova A, Egelman EH: Actin filaments as tension sensors. Curr Biol. 2012, 22 (3): R96-R101. 10.1016/j.cub.2011.12.010
Dominguez R, Holmes KC: Actin structure and function. Annu Rev Biophys. 2011, 40: 169-186. 10.1146/annurev-biophys-042910-155359
Otto JJ: Actin-bundling proteins. Curr Opin Cell Biol. 1994, 6 (1): 105-109. 10.1016/0955-0674(94)90123-6
Van Troys M, Vandekerckhove J, Ampe C: Structural modules in actin-binding proteins: towards a new classification. Biochim Biophys Acta. 1999, 1448 (3): 323-348. 10.1016/S0167-4889(98)00152-9
Matsudaira P: Modular organization of actin crosslinking proteins. Trends Biochem Sci. 1991, 16 (3): 87-92.
Low SH, Mukhina S, Srinivas V, Ng CZ, Murata-Hori M: Domain analysis of alpha-actinin reveals new aspects of its association with F-actin during cytokinesis. Exp Cell Res. 2010, 316 (12): 1925-1934. 10.1016/j.yexcr.2010.03.004
Zhou AX, Hartwig JH, Akyurek LM: Filamins in cell signaling, transcription and organ development. Trends Cell Biol. 2010, 20 (2): 113-123. 10.1016/j.tcb.2009.12.001
van der Flier A, Sonnenberg A: Structural and functional aspects of filamins. Biochim Biophys Acta. 2001, 1538 (2–3): 99-117.
Wang K: Filamin, a new high-molecular-weight protein found in smooth muscle and nonmuscle cells. Purification and properties of chicken gizzard filamin. Biochemistry. 1977, 16 (9): 1857-1865. 10.1021/bi00628a015
Wang K, Ash JF, Singer SJ: Filamin, a new high-molecular-weight protein found in smooth muscle and non-muscle cells. Proc Natl Acad Sci USA. 1975, 72 (11): 4483-4486. 10.1073/pnas.72.11.4483
Gorlin JB, Yamin R, Egan S, Stewart M, Stossel TP, Kwiatkowski DJ, Hartwig JH: Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J Cell Biol. 1990, 111 (3): 1089-1105. 10.1083/jcb.111.3.1089
Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS: Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001, 2 (2): 138-145. 10.1038/35052082
Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers HR, Stossel TP: Actin-binding protein requirement for cortical stability and efficient locomotion. Science. 1992, 255 (5042): 325-327. 10.1126/science.1549777
Ohta Y, Hartwig JH, Stossel TP: FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat Cell Biol. 2006, 8 (8): 803-814. 10.1038/ncb1437
Popowicz GM, Schleicher M, Noegel AA, Holak TA: Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem Sci. 2006, 31 (7): 411-419. 10.1016/j.tibs.2006.05.006
Feng Y, Walsh CA: The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004, 6 (11): 1034-1038. 10.1038/ncb1104-1034
Sheen VL, Dixon PH, Fox JW, Hong SE, Kinton L, Sisodiya SM, Duncan JS, Dubeau F, Scheffer IE, Schachter SC, et al: Mutations in the X-linked filamin 1 gene cause periventricular nodular heterotopia in males as well as in females. Hum Mol Genet. 2001, 10 (17): 1775-1783. 10.1093/hmg/10.17.1775
Fox JW, Lamperti ED, Eksioglu YZ, Hong SE, Feng Y, Graham DA, Scheffer IE, Dobyns WB, Hirsch BA, Radtke RA, et al: Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998, 21 (6): 1315-1325. 10.1016/S0896-6273(00)80651-0
Chardon JW, Mignot C, Aradhya S, Keren B, Afenjar A, Kaminska A, Beldjord C, Heron D, Boycott KM: Deletion of filamin A in two female patients with periventricular nodular heterotopia. Am J Med Genet A. 2012, 158A (6): 1512-1516. 10.1002/ajmg.a.35409
Moro F, Carrozzo R, Veggiotti P, Tortorella G, Toniolo D, Volzone A, Guerrini R: Familial periventricular heterotopia: missense and distal truncating mutations of the FLN1 gene. Neurology. 2002, 58 (6): 916-921. 10.1212/WNL.58.6.916
Dobyns WB, Andermann E, Andermann F, Czapansky-Beilman D, Dubeau F, Dulac O, Guerrini R, Hirsch B, Ledbetter DH, Lee NS, et al: X-linked malformations of neuronal migration. Neurology. 1996, 47 (2): 331-339. 10.1212/WNL.47.2.331
Ferland RJ, Batiz LF, Neal J, Lian G, Bundock E, Lu J, Hsiao YC, Diamond R, Mei D, Banham AH, et al: Disruption of neural progenitors along the ventricular and subventricular zones in periventricular heterotopia. Hum Mol Genet. 2009, 18 (3): 497-516.
Cardoso C, Boys A, Parrini E, Mignon-Ravix C, McMahon JM, Khantane S, Bertini E, Pallesi E, Missirian C, Zuffardi O, et al: Periventricular heterotopia, mental retardation, and epilepsy associated with 5q14.3-q15 deletion. Neurology. 2009, 72 (9): 784-792. 10.1212/01.wnl.0000336339.08878.2d
Ferland RJ, Gaitanis JN, Apse K, Tantravahi U, Walsh CA, Sheen VL: Periventricular nodular heterotopia and Williams syndrome. Am J Med Genet A. 2006, 140 (12): 1305-1311.
Sheen VL, Ganesh VS, Topcu M, Sebire G, Bodell A, Hill RS, Grant PE, Shugart YY, Imitola J, Khoury SJ, et al: Mutations in ARFGEF2 implicate vesicle trafficking in neural progenitor proliferation and migration in the human cerebral cortex. Nat Genet. 2004, 36 (1): 69-76. 10.1038/ng1276
Sheen VL, Wheless JW, Bodell A, Braverman E, Cotter PD, Rauen KA, Glenn O, Weisiger K, Packman S, Walsh CA, et al: Periventricular heterotopia associated with chromosome 5p anomalies. Neurology. 2003, 60 (6): 1033-1036. 10.1212/01.WNL.0000052689.03214.61
Parrini E, Ramazzotti A, Dobyns WB, Mei D, Moro F, Veggiotti P, Marini C, Brilstra EH, Dalla Bernardina B, Goodwin L, et al: Periventricular heterotopia: phenotypic heterogeneity and correlation with Filamin A mutations. Brain. 2006, 129 (Pt 7): 1892-1906.
Jefferies JL, Taylor MD, Rossano J, Belmont JW, Craigen WJ: Novel cardiac findings in periventricular nodular heterotopia. Am J Med Genet A. 2010, 152A (1): 165-168. 10.1002/ajmg.a.33110
Sheen VL, Jansen A, Chen MH, Parrini E, Morgan T, Ravenscroft R, Ganesh V, Underwood T, Wiley J, Leventer R, et al: Filamin A mutations cause periventricular heterotopia with Ehlers-Danlos syndrome. Neurology. 2005, 64 (2): 254-262. 10.1212/01.WNL.0000149512.79621.DF
de Wit MC, de Coo IF, Lequin MH, Halley DJ, Roos-Hesselink JW, Mancini GM: Combined cardiological and neurological abnormalities due to filamin A gene mutation. Clin Res Cardiol. 2011, 100 (1): 45-50. 10.1007/s00392-010-0206-y
Kakita A, Hayashi S, Moro F, Guerrini R, Ozawa T, Ono K, Kameyama S, Walsh CA, Takahashi H: Bilateral periventricular nodular heterotopia due to filamin 1 gene mutation: widespread glomeruloid microvascular anomaly and dysplastic cytoarchitecture in the cerebral cortex. Acta Neuropathol. 2002, 104 (6): 649-657.
Sheen VL, Walsh CA: Periventricular heterotopia: new insights into Ehlers-Danlos syndrome. Clin Med Res. 2005, 3 (4): 229-233. 10.3121/cmr.3.4.229
Robertson SP, Twigg SR, Sutherland-Smith AJ, Biancalana V, Gorlin RJ, Horn D, Kenwrick SJ, Kim CA, Morava E, Newbury-Ecob R, et al: Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet. 2003, 33 (4): 487-491. 10.1038/ng1119
Kyndt F, Gueffet JP, Probst V, Jaafar P, Legendre A, Le Bouffant F, Toquet C, Roy E, McGregor L, Lynch SA, et al: Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation. 2007, 115 (1): 40-49.
Bernstein JA, Bernstein D, Hehr U, Hudgins L: Familial cardiac valvulopathy due to filamin A mutation. Am J Med Genet A. 2011, 155A (9): 2236-2241.
Feng Y, Chen MH, Moskowitz IP, Mendonza AM, Vidali L, Nakamura F, Kwiatkowski DJ, Walsh CA: Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc Natl Acad Sci USA. 2006, 103 (52): 19836-19841. 10.1073/pnas.0609628104
Hart AW, Morgan JE, Schneider J, West K, McKie L, Bhattacharya S, Jackson IJ, Cross SH: Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet. 2006, 15 (16): 2457-2467. 10.1093/hmg/ddl168
Leung R, Wang Y, Cuddy K, Sun C, Magalhaes J, Grynpas M, Glogauer M: Filamin A regulates monocyte migration through Rho small GTPases during osteoclastogenesis. J Bone Miner Res. 2010, 25 (5): 1077-1091.
Lian G, Lu J, Hu J, Zhang J, Cross SH, Ferland RJ, Sheen VL: Filamin a regulates neural progenitor proliferation and cortical size through Wee1-dependent Cdk1 phosphorylation. J Neurosci. 2012, 32 (22): 7672-7684. 10.1523/JNEUROSCI.0894-12.2012
Lynch CD, Gauthier NC, Biais N, Lazar AM, Roca-Cusachs P, Yu CH, Sheetz MP: Filamin depletion blocks endoplasmic spreading and destabilizes force-bearing adhesions. Mol Biol Cell. 2011, 22 (8): 1263-1273. 10.1091/mbc.E10-08-0661
Gardel ML, Nakamura F, Hartwig JH, Crocker JC, Stossel TP, Weitz DA: Prestressed F-actin networks cross-linked by hinged filamins replicate mechanical properties of cells. Proc Natl Acad Sci USA. 2006, 103 (6): 1762-1767. 10.1073/pnas.0504777103
Kasza KE, Nakamura F, Hu S, Kollmannsberger P, Bonakdar N, Fabry B, Stossel TP, Wang N, Weitz DA: Filamin A is essential for active cell stiffening but not passive stiffening under external force. Biophys J. 2009, 96 (10): 4326-4335. 10.1016/j.bpj.2009.02.035
Glogauer M, Arora P, Chou D, Janmey PA, Downey GP, McCulloch CA: The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J Biol Chem. 1998, 273 (3): 1689-1698. 10.1074/jbc.273.3.1689
Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP: Structural basis of filamin A functions. J Cell Biol. 2007, 179 (5): 1011-1025. 10.1083/jcb.200707073
Ithychanda SS, Qin J: Evidence for multisite ligand binding and stretching of filamin by integrin and migfilin. Biochemistry. 2011, 50 (20): 4229-4231. 10.1021/bi2003229
Pentikainen U, Ylanne J: The regulation mechanism for the auto-inhibition of binding of human filamin A to integrin. J Mol Biol. 2009, 393 (3): 644-657. 10.1016/j.jmb.2009.08.035
Lad Y, Kiema T, Jiang P, Pentikainen OT, Coles CH, Campbell ID, Calderwood DA, Ylanne J: Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. EMBO J. 2007, 26 (17): 3993-4004. 10.1038/sj.emboj.7601827
Samuelsson SJ, Luther PW, Pumplin DW, Bloch RJ: Structures linking microfilament bundles to the membrane at focal contacts. J Cell Biol. 1993, 122 (2): 485-496. 10.1083/jcb.122.2.485
Kim H, Nakamura F, Lee W, Hong C, Perez-Sala D, McCulloch CA: Regulation of cell adhesion to collagen via beta1 integrins is dependent on interactions of filamin A with vimentin and protein kinase C epsilon. Exp Cell Res. 2010, 316 (11): 1829-1844. 10.1016/j.yexcr.2010.02.007
Kim H, Nakamura F, Lee W, Shifrin Y, Arora P, McCulloch CA: Filamin A is required for vimentin-mediated cell adhesion and spreading. Am J Physiol Cell Physiol. 2010, 298 (2): C221-C236. 10.1152/ajpcell.00323.2009
Hartwig JH, Stossel TP: Isolation and properties of actin, myosin, and a new actinbinding protein in rabbit alveolar macrophages. J Biol Chem. 1975, 250 (14): 5696-5705.
Nakamura F, Hartwig JH, Stossel TP, Szymanski PT: Ca2+ and calmodulin regulate the binding of filamin A to actin filaments. J Biol Chem. 2005, 280 (37): 32426-32433. 10.1074/jbc.M502203200
Griffiths GS, Grundl M, Allen JS 3rd, Matter ML: R-Ras interacts with filamin a to maintain endothelial barrier function. J Cell Physiol. 2011, 226 (9): 2287-2296. 10.1002/jcp.22565
Gawecka JE, Griffiths GS, Ek-Rylander B, Ramos JW, Matter ML: R-Ras regulates migration through an interaction with filamin A in melanoma cells. PLoS One. 2010, 5 (6): e11269. 10.1371/journal.pone.0011269
Falet H, Pollitt AY, Begonja AJ, Weber SE, Duerschmied D, Wagner DD, Watson SP, Hartwig JH: A novel interaction between FlnA and Syk regulates platelet ITAM-mediated receptor signaling and function. J Exp Med. 2010, 207 (9): 1967-1979. 10.1084/jem.20100222
Smith TC, Fang Z, Luna EJ: Novel interactors and a role for supervillin in early cytokinesis. Cytoskeleton (Hoboken). 2010, 67 (6): 346-364.
Lin R, Karpa K, Kabbani N, Goldman-Rakic P, Levenson R: Dopamine D2 and D3 receptors are linked to the actin cytoskeleton via interaction with filamin A. Proc Natl Acad Sci USA. 2001, 98 (9): 5258-5263. 10.1073/pnas.011538198
Kim KM, Gainetdinov RR, Laporte SA, Caron MG, Barak LS: G protein-coupled receptor kinase regulates dopamine D3 receptor signaling by modulating the stability of a receptor-filamin-beta-arrestin complex. A case of autoreceptor regulation. J Biol Chem. 2005, 280 (13): 12774-12780.
Li C, Yu S, Nakamura F, Pentikainen OT, Singh N, Yin S, Xin W, Sy MS: Pro-prion binds filamin A, facilitating its interaction with integrin beta1, and contributes to melanomagenesis. J Biol Chem. 2010, 285 (39): 30328-30339. 10.1074/jbc.M110.147413
Li C, Yu S, Nakamura F, Yin S, Xu J, Petrolla AA, Singh N, Tartakoff A, Abbott DW, Xin W, et al: Binding of pro-prion to filamin A disrupts cytoskeleton and correlates with poor prognosis in pancreatic cancer. J Clin Invest. 2009, 119 (9): 2725-2736. 10.1172/JCI39542
Nakamura F, Pudas R, Heikkinen O, Permi P, Kilpelainen I, Munday AD, Hartwig JH, Stossel TP, Ylanne J: The structure of the GPIb-filamin A complex. Blood. 2006, 107 (5): 1925-1932. 10.1182/blood-2005-10-3964
Kanaji T, Ware J, Okamura T, Newman PJ: GPIbalpha regulates platelet size by controlling the subcellular localization of filamin. Blood. 2012, 119 (12): 2906-2913. 10.1182/blood-2011-08-376566
Kiema T, Lad Y, Jiang P, Oxley CL, Baldassarre M, Wegener KL, Campbell ID, Ylanne J, Calderwood DA: The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006, 21 (3): 337-347. 10.1016/j.molcel.2006.01.011
Sharma CP, Ezzell RM, Arnaout MA: Direct interaction of filamin (ABP-280) with the beta 2-integrin subunit CD18. J Immunol. 1995, 154 (7): 3461-3470.
Ott I, Fischer EG, Miyagi Y, Mueller BM, Ruf W: A role for tissue factor in cell adhesion and migration mediated by interaction with actin-binding protein 280. J Cell Biol. 1998, 140 (5): 1241-1253. 10.1083/jcb.140.5.1241
Klaile E, Muller MM, Kannicht C, Singer BB, Lucka L: CEACAM1 functionally interacts with filamin A and exerts a dual role in the regulation of cell migration. J Cell Sci. 2005, 118 (Pt 23): 5513-5524.
Lad Y, Jiang P, Ruskamo S, Harburger DS, Ylanne J, Campbell ID, Calderwood DA: Structural basis of the migfilin-filamin interaction and competition with integrin beta tails. J Biol Chem. 2008, 283 (50): 35154-35163. 10.1074/jbc.M802592200
Ithychanda SS, Das M, Ma YQ, Ding K, Wang X, Gupta S, Wu C, Plow EF, Qin J: Migfilin, a molecular switch in regulation of integrin activation. J Biol Chem. 2009, 284 (7): 4713-4722.
Tu Y, Wu S, Shi X, Chen K, Wu C: Migfilin and Mig-2 link focal adhesions to filamin and the actin cytoskeleton and function in cell shape modulation. Cell. 2003, 113 (1): 37-47. 10.1016/S0092-8674(03)00163-6
Stahlhut M, van Deurs B: Identification of filamin as a novel ligand for caveolin-1: evidence for the organization of caveolin-1-associated membrane domains by the actin cytoskeleton. Mol Biol Cell. 2000, 11 (1): 325-337.
Muriel O, Echarri A, Hellriegel C, Pavon DM, Beccari L, Del Pozo MA: Phosphorylated filamin A regulates actin-linked caveolae dynamics. J Cell Sci. 2011, 124 (Pt 16): 2763-2776.
Scott MG, Pierotti V, Storez H, Lindberg E, Thuret A, Muntaner O, Labbe-Jullie C, Pitcher JA, Marullo S: Cooperative regulation of extracellular signal-regulated kinase activation and cell shape change by filamin A and beta-arrestins. Mol Cell Biol. 2006, 26 (9): 3432-3445. 10.1128/MCB.26.9.3432-3445.2006
Nallapalli RK, Ibrahim MX, Zhou AX, Bandaru S, Naresh S, Redfors B, Pazooki D, Zhang Y, Boren J, Cao Y, et al: Targeting filamin A reduces K-RAS-induced lung adenocarcinomas and endothelial response to tumor growth in mice. Mol Cancer. 2012, 11 (1): 50. 10.1186/1476-4598-11-50
Muscolini M, Sajeva A, Caristi S, Tuosto L: A novel association between filamin A and NF-kappaB inducing kinase couples CD28 to inhibitor of NF-kappaB kinase alpha and NF-kappaB activation. Immunol Lett. 2011, 136 (2): 203-212. 10.1016/j.imlet.2011.01.011
Najib S, Saint-Laurent N, Esteve JP, Schulz S, Boutet-Robinet E, Fourmy D, Lattig J, Mollereau C, Pyronnet S, Susini C, et al: A switch of G protein-coupled receptor binding preference from phosphoinositide 3-kinase (PI3K)-p85 to filamin A negatively controls the PI3K pathway. Mol Cell Biol. 2012, 32 (5): 1004-1016. 10.1128/MCB.06252-11
Castoria G, D'Amato L, Ciociola A, Giovannelli P, Giraldi T, Sepe L, Paolella G, Barone MV, Migliaccio A, Auricchio F: Androgen-induced cell migration: role of androgen receptor/filamin A association. PLoS One. 2011, 6 (2): e17218. 10.1371/journal.pone.0017218
Ozanne DM, Brady ME, Cook S, Gaughan L, Neal DE, Robson CN: Androgen receptor nuclear translocation is facilitated by the f-actin cross-linking protein filamin. Mol Endocrinol. 2000, 14 (10): 1618-1626. 10.1210/me.14.10.1618
Marti A, Luo Z, Cunningham C, Ohta Y, Hartwig J, Stossel TP, Kyriakis JM, Avruch J: Actin-binding protein-280 binds the stress-activated protein kinase (SAPK) activator SEK-1 and is required for tumor necrosis factor-alpha activation of SAPK in melanoma cells. J Biol Chem. 1997, 272 (5): 2620-2628. 10.1074/jbc.272.5.2620
Leonardi A, Ellinger-Ziegelbauer H, Franzoso G, Brown K, Siebenlist U: Physical and functional interaction of filamin (actin-binding protein-280) and tumor necrosis factor receptor-associated factor 2. J Biol Chem. 2000, 275 (1): 271-278. 10.1074/jbc.275.1.271
Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP: The small GTPase RalA targets filamin to induce filopodia. Proc Natl Acad Sci USA. 1999, 96 (5): 2122-2128. 10.1073/pnas.96.5.2122
Ueda K, Ohta Y, Hosoya H: The carboxy-terminal pleckstrin homology domain of ROCK interacts with filamin-A. Biochem Biophys Res Commun. 2003, 301 (4): 886-890. 10.1016/S0006-291X(03)00048-2
Nakamura F, Heikkinen O, Pentikainen OT, Osborn TM, Kasza KE, Weitz DA, Kupiainen O, Permi P, Kilpelainen I, Ylanne J, et al: Molecular basis of filamin A-FilGAP interaction and its impairment in congenital disorders associated with filamin A mutations. PLoS One. 2009, 4 (3): e4928. 10.1371/journal.pone.0004928
Bellanger JM, Astier C, Sardet C, Ohta Y, Stossel TP, Debant A: The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat Cell Biol. 2000, 2 (12): 888-892. 10.1038/35046533
Velkova A, Carvalho MA, Johnson JO, Tavtigian SV, Monteiro AN: Identification of Filamin A as a BRCA1-interacting protein required for efficient DNA repair. Cell Cycle. 2010, 9 (7): 1421-1433.
Yue J, Wang Q, Lu H, Brenneman M, Fan F, Shen Z: The cytoskeleton protein filamin-A is required for an efficient recombinational DNA double strand break repair. Cancer Res. 2009, 69 (20): 7978-7985. 10.1158/0008-5472.CAN-09-2177
Yuan Y, Shen Z: Interaction with BRCA2 suggests a role for filamin-1 (hsFLNa) in DNA damage response. J Biol Chem. 2001, 276 (51): 48318-48324.
Meng X, Yuan Y, Maestas A, Shen Z: Recovery from DNA damage-induced G2 arrest requires actin-binding protein filamin-A/actin-binding protein 280. J Biol Chem. 2004, 279 (7): 6098-6105.
Gay O, Gilquin B, Nakamura F, Jenkins ZA, McCartney R, Krakow D, Deshiere A, Assard N, Hartwig JH, Robertson SP, et al: RefilinB (FAM101B) targets filamin A to organize perinuclear actin networks and regulates nuclear shape. Proc Natl Acad Sci USA. 2011, 108 (28): 11464-11469. 10.1073/pnas.1104211108
Deng W, Lopez-Camacho C, Tang JY, Mendoza-Villanueva D, Maya-Mendoza A, Jackson DA, Shore P: Cytoskeletal protein filamin A is a nucleolar protein that suppresses ribosomal RNA gene transcription. Proc Natl Acad Sci USA. 2012, 109 (5): 1524-1529. 10.1073/pnas.1107879109
Qiu H, Nomiyama R, Moriguchi K, Fukada T, Sugimoto K: Identification of novel nuclear protein interactions with the N-terminal part of filamin A. Biosci Biotechnol Biochem. 2011, 75 (1): 145-147. 10.1271/bbb.100567
Flanagan LA, Chou J, Falet H, Neujahr R, Hartwig JH, Stossel TP: Filamin A, the Arp2/3 complex, and the morphology and function of cortical actin filaments in human melanoma cells. J Cell Biol. 2001, 155 (4): 511-517. 10.1083/jcb.200105148
Nakamura F, Osborn E, Janmey PA, Stossel TP: Comparison of filamin A-induced cross-linking and Arp2/3 complex-mediated branching on the mechanics of actin filaments. J Biol Chem. 2002, 277 (11): 9148-9154. 10.1074/jbc.M111297200
Ulrich F, Heisenberg CP: Trafficking and cell migration. Traffic. 2009, 10 (7): 811-818. 10.1111/j.1600-0854.2009.00929.x
Pellinen T, Ivaska J: Integrin traffic. J Cell Sci. 2006, 119 (Pt 18): 3723-3731.
D'Addario M, Arora PD, Fan J, Ganss B, Ellen RP, McCulloch CA: Cytoprotection against mechanical forces delivered through beta 1 integrins requires induction of filamin A. J Biol Chem. 2001, 276 (34): 31969-31977. 10.1074/jbc.M102715200
Meyer SC, Sanan DA, Fox JE: Role of actin-binding protein in insertion of adhesion receptors into the membrane. J Biol Chem. 1998, 273 (5): 3013-3020. 10.1074/jbc.273.5.3013
Frantz C, Stewart KM, Weaver VM: The extracellular matrix at a glance. J Cell Sci. 2010, 123 (Pt 24): 4195-4200.
Larsen M, Artym VV, Green JA, Yamada KM: The matrix reorganized: extracellular matrix remodeling and integrin signaling. Curr Opin Cell Biol. 2006, 18 (5): 463-471. 10.1016/j.ceb.2006.08.009
Xian X, Gopal S, Couchman JR: Syndecans as receptors and organizers of the extracellular matrix. Cell Tissue Res. 2010, 339 (1): 31-46. 10.1007/s00441-009-0829-3
Harburger DS, Calderwood DA: Integrin signalling at a glance. J Cell Sci. 2009, 122 (Pt 2): 159-163.
Leitinger B, Hohenester E: Mammalian collagen receptors. Matrix Biol. 2007, 26 (3): 146-155. 10.1016/j.matbio.2006.10.007
Calderwood DA, Huttenlocher A, Kiosses WB, Rose DM, Woodside DG, Schwartz MA, Ginsberg MH: Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat Cell Biol. 2001, 3 (12): 1060-1068. 10.1038/ncb1201-1060
Xu Y, Bismar TA, Su J, Xu B, Kristiansen G, Varga Z, Teng L, Ingber DE, Mammoto A, Kumar R, et al: Filamin A regulates focal adhesion disassembly and suppresses breast cancer cell migration and invasion. J Exp Med. 2010, 207 (11): 2421-2437. 10.1084/jem.20100433
Zhu TN, He HJ, Kole S, D'Souza T, Agarwal R, Morin PJ, Bernier M: Filamin A-mediated down-regulation of the exchange factor Ras-GRF1 correlates with decreased matrix metalloproteinase-9 expression in human melanoma cells. J Biol Chem. 2007, 282 (20): 14816-14826. 10.1074/jbc.M611430200
Baldassarre M, Razinia Z, Brahme N, Buccione R, Calderwood DA: Filamin A controls matrix metalloprotease activity and regulates cell invasion in human fibrosarcoma cells. J Cell Sci. 2012, 2012: 17.
Byfield FJ, Wen Q, Levental I, Nordstrom K, Arratia PE, Miller RT, Janmey PA: Absence of filamin A prevents cells from responding to stiffness gradients on gels coated with collagen but not fibronectin. Biophys J. 2009, 96 (12): 5095-5102. 10.1016/j.bpj.2009.03.046
Kim H, Sengupta A, Glogauer M, McCulloch CA: Filamin A regulates cell spreading and survival via beta1 integrins. Exp Cell Res. 2008, 314 (4): 834-846. 10.1016/j.yexcr.2007.11.022
Adams M, Simms RJ, Abdelhamed Z, Dawe HR, Szymanska K, Logan CV, Wheway G, Pitt E, Gull K, Knowles MA, et al: A meckelin-filamin A interaction mediates ciliogenesis. Hum Mol Genet. 2012, 21 (6): 1272-1286. 10.1093/hmg/ddr557
Ai J, Huang H, Lv X, Tang Z, Chen M, Chen T, Duan W, Sun H, Li Q, Tan R, et al: FLNA and PGK1 are two potential markers for progression in hepatocellular carcinoma. Cell Physiol Biochem. 2011, 27 (3–4): 207-216.
Alper O, Stetler-Stevenson WG, Harris LN, Leitner WW, Ozdemirli M, Hartmann D, Raffeld M, Abu-Asab M, Byers S, Zhuang Z, et al: Novel anti-filamin-A antibody detects a secreted variant of filamin-A in plasma from patients with breast carcinoma and high-grade astrocytoma. Cancer Sci. 2009, 100 (9): 1748-1756. 10.1111/j.1349-7006.2009.01244.x
Bedolla RG, Wang Y, Asuncion A, Chamie K, Siddiqui S, Mudryj MM, Prihoda TJ, Siddiqui J, Chinnaiyan AM, Mehra R, et al: Nuclear versus cytoplasmic localization of filamin A in prostate cancer: immunohistochemical correlation with metastases. Clin Cancer Res. 2009, 15 (3): 788-796. 10.1158/1078-0432.CCR-08-1402
Carabalona A, Beguin S, Pallesi-Pocachard E, Buhler E, Pellegrino C, Arnaud K, Hubert P, Oualha M, Siffroi JP, Khantane S, et al: A glial origin for periventricular nodular heterotopia caused by impaired expression of Filamin-A. Hum Mol Genet. 2012, 21 (5): 1004-1017. 10.1093/hmg/ddr531
Xi J, Yue J, Lu H, Campbell N, Yang Q, Lan S, Haffty BG, Yuan C, Shen Z: Inhibition of Filamin-A Reduces Cancer Metastatic Potential. Int J Biol Sci. 2013, 9 (1): 67-77. 10.7150/ijbs.5577
Baldassarre M, Razinia Z, Burande CF, Lamsoul I, Lutz PG, Calderwood DA: Filamins regulate cell spreading and initiation of cell migration. PLoS One. 2009, 4 (11): e7830. 10.1371/journal.pone.0007830
Heuze ML, Lamsoul I, Baldassarre M, Lad Y, Leveque S, Razinia Z, Moog-Lutz C, Calderwood DA, Lutz PG: ASB2 targets filamins A and B to proteasomal degradation. Blood. 2008, 112 (13): 5130-5140. 10.1182/blood-2007-12-128744
Razinia Z, Baldassarre M, Bouaouina M, Lamsoul I, Lutz PG, Calderwood DA: The E3 ubiquitin ligase specificity subunit ASB2alpha targets filamins for proteasomal degradation by interacting with the filamin actin-binding domain. J Cell Sci. 2011, 124 (Pt 15): 2631-2641.
O'Connell MP, Fiori JL, Baugher KM, Indig FE, French AD, Camilli TC, Frank BP, Earley R, Hoek KS, Hasskamp JH, et al: Wnt5A activates the calpain-mediated cleavage of filamin A. J Invest Dermatol. 2009, 129 (7): 1782-1789. 10.1038/jid.2008.433
Guiet R, Verollet C, Lamsoul I, Cougoule C, Poincloux R, Labrousse A, Calderwood DA, Glogauer M, Lutz PG, Maridonneau-Parini I: Macrophage mesenchymal migration requires podosome stabilization by filamin A. J Biol Chem. 2012, 287 (16): 13051-13062. 10.1074/jbc.M111.307124
De Braekeleer M, Morel F, Le Bris MJ, Herry A, Douet-Guilbert N: The MLL gene and translocations involving chromosomal band 11q23 in acute leukemia. Anticancer Res. 2005, 25 (3B): 1931-1944.
De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Meyer C, Marschalek R, Ferec C, De Braekeleer M: FLNA, a new partner gene fused to MLL in a patient with acute myelomonoblastic leukaemia. Br J Haematol. 2009, 146 (6): 693-695. 10.1111/j.1365-2141.2009.07824.x
Fiori JL, Zhu TN, O'Connell MP, Hoek KS, Indig FE, Frank BP, Morris C, Kole S, Hasskamp J, Elias G, et al: Filamin A modulates kinase activation and intracellular trafficking of epidermal growth factor receptors in human melanoma cells. Endocrinology. 2009, 150 (6): 2551-2560. 10.1210/en.2008-1344
Byers HR, Etoh T, Doherty JR, Sober AJ, Mihm MC Jr: Cell migration and actin organization in cultured human primary, recurrent cutaneous and metastatic melanoma. Time-lapse and image analysis. Am J Pathol. 1991, 139 (2): 423-435.
Yue J, Lan S, Yuan C, Shen Z: Prognostic values of filamin-A status for topoisomerase II poison chemotherapy. Int J Biol Sci. 2012, 8 (4): 442-450. 10.7150/ijbs.4155
Yue J, Lu H, Liu J, Berwick M, Shen Z: Filamin-A as a marker and target for DNA damage based cancer therapy. DNA Repair (Amst). 2012, 11 (2): 192-200. 10.1016/j.dnarep.2011.10.019. 10.1016/j.dnarep.2011.10.019
Shen Z: Genomic instability and cancer: an introduction. J Mol Cell Biol. 2011, 3 (1): 1-3. 10.1093/jmcb/mjq057
Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, et al: A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005, 4 (12): 1920-1932. 10.1074/mcp.M500279-MCP200
Frierson HF Jr, El-Naggar AK, Welsh JB, Sapinoso LM, Su AI, Cheng J, Saku T, Moskaluk CA, Hampton GM: Large scale molecular analysis identifies genes with altered expression in salivary adenoid cystic carcinoma. Am J Pathol. 2002, 161 (4): 1315-1323. 10.1016/S0002-9440(10)64408-2
Guedj N, Zhan Q, Perigny M, Rautou PE, Degos F, Belghiti J, Farges O, Bedossa P, Paradis V: Comparative protein expression profiles of hilar and peripheral hepatic cholangiocarcinomas. J Hepatol. 2009, 51 (1): 93-101. 10.1016/j.jhep.2009.03.017
Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, et al: Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006, 9 (4): 287-300. 10.1016/j.ccr.2006.03.003
Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S: Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003, 63 (10): 2649-2657.
Smith SC, Oxford G, Baras AS, Owens C, Havaleshko D, Brautigan DL, Safo MK, Theodorescu D: Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin Cancer Res. 2007, 13 (13): 3803-3813. 10.1158/1078-0432.CCR-06-2419
Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C: Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006, 24 (5): 778-789. 10.1200/JCO.2005.03.2375
Notterman DA, Alon U, Sierk AJ, Levine AJ: Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001, 61 (7): 3124-3130.
Wang HY, Bakshi K, Frankfurt M, Stucky A, Goberdhan M, Shah SM, Burns LH: Reducing amyloid-related Alzheimer's disease pathogenesis by a small molecule targeting filamin a. J Neurosci. 2012, 32 (29): 9773-9784. 10.1523/JNEUROSCI.0354-12.2012
Wang HY, Burns LH: Naloxone's pentapeptide binding site on filamin A blocks Mu opioid receptor-Gs coupling and CREB activation of acute morphine. PLoS One. 2009, 4 (1): e4282. 10.1371/journal.pone.0004282
Jay D, Garcia EJ, Lara JE, Medina MA, de la Luz IM: Determination of a cAMP-dependent protein kinase phosphorylation site in the C-terminal region of human endothelial actin-binding protein. Arch Biochem Biophys. 2000, 377 (1): 80-84. 10.1006/abbi.2000.1762
Wang Y, Kreisberg JI, Bedolla RG, Mikhailova M, DeVere White RW, Ghosh PM: A 90 kDa fragment of filamin A promotes Casodex-induced growth inhibition in Casodex-resistant androgen receptor positive C4-2 prostate cancer cells. Oncogene. 2007, 26 (41): 6061-6070. 10.1038/sj.onc.1210435
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JY drafted the first version of the manuscript and figures; SH amended the first draft; ZS finalized the manuscript and figures. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Yue, J., Huhn, S. & Shen, Z. Complex roles of filamin-A mediated cytoskeleton network in cancer progression. Cell Biosci 3, 7 (2013). https://doi.org/10.1186/2045-3701-3-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2045-3701-3-7