Abstract
Background
From an early age, humans look longer at preferred stimuli and also typically look longer at facial expressions of emotion, particularly happy faces. Atypical gaze patterns towards social stimuli are common in autism spectrum conditions (ASC). However, it is unknown whether gaze fixation patterns have any genetic basis. In this study, we tested whether variations in the cannabinoid receptor 1 (CNR1) gene are associated with gaze duration towards happy faces. This gene was selected because CNR1 is a key component of the endocannabinoid system, which is involved in processing reward, and in our previous functional magnetic resonance imaging (fMRI) study, we found that variations in CNR1 modulate the striatal response to happy (but not disgust) faces. The striatum is involved in guiding gaze to rewarding aspects of a visual scene. We aimed to validate and extend this result in another sample using a different technique (gaze tracking).
Methods
A total of 30 volunteers (13 males and 17 females) from the general population observed dynamic emotional expressions on a screen while their eye movements were recorded. They were genotyped for the identical four single-nucleotide polymorphisms (SNPs) in the CNR1 gene tested in our earlier fMRI study.
Results
Two SNPs (rs806377 and rs806380) were associated with differential gaze duration for happy (but not disgust) faces. Importantly, the allelic groups associated with a greater striatal response to happy faces in the fMRI study were associated with longer gaze duration at happy faces.
Conclusions
These results suggest that CNR1 variations modulate the striatal function that underlies the perception of signals of social reward, such as happy faces. This suggests that CNR1 is a key element in the molecular architecture of perception of certain basic emotions. This may have implications for understanding neurodevelopmental conditions marked by atypical eye contact and facial emotion processing, such as ASC.
Similar content being viewed by others
Background
Vision is the primary sensory modality in primates, reflected by the visual cortex being the largest of all the sensory cortices. Our eyes perform quick orienting movements ('saccades') towards interesting features of stimuli in the external world [1]. In general, we tend to look longer at more rewarding stimuli [2]. This rationale lies behind the 'preferential looking' technique in infancy research, where gaze duration and direction are assumed to reflect visual preference [2–6]. Gaze not only informs us about normative variation in the visual processing of stimuli but also is relevant to the understanding of complex neurodevelopmental conditions such as autism spectrum conditions (ASC), which are characterised by atypical gaze fixation patterns [7, 8]. This has led to the suggestion that gaze fixation patterns could constitute potential endophenotypes for such conditions. Gaze patterns show high test-retest reliability as well as a moderate to high heritability when tested in twins [9–11], suggesting a significant genetic contribution. This raises the possibility that variation in candidate genes underlie normative variation in gaze patterns.
The measure of particular interest to us is the duration of gaze fixation, given the evidence that people with ASC show reduced gaze fixation towards social stimuli [8, 12–15]. Research in primates suggests that the striatal region plays a major role in directing gaze [16]. The striatum is thought to encode a 'value map' of the visual stimuli. Both ventral striatal neurons as well as a subpopulation of caudate neurons encode reward magnitude of the stimuli [17, 18]. This 'value map', in addition to further frontal cortical inputs, is then passed to the lateral intraparietal area (LIP), where a fine-tuned map of 'relative expected subjective value' is created. The LIP projects to the frontal eye fields, which send excitatory projections to the caudate nucleus. A subset of neurons from the caudate nucleus inhibit the substantia nigra and consequently disinhibit the superior colliculus, which in turn controls the gaze control nuclei in the brainstem, leading to a gaze shift [19].
One of the key molecular systems involved in the functioning of the striatal circuit is the endocannabinoid system. It is a neuropeptidergic circuit involved in reward processing and works in tandem with the mesolimbic dopaminergic system [20]. Expressed selectively in the brain, the cannabinoid receptor 1 (CNR1) is the best-studied molecule of this system. Immunolocalisation studies in rats and humans have indicated high CNR1 expression levels in the striatum, a region known for its central role in reward processing [20–24]. CNR1 is believed to modulate striatal dopamine release through a trans-synaptic mechanism involving both GABAergic and glutamatergic synapses and is expressed strongly in the caudate, putamen, globus pallidus internal and substantia nigra, as well as in the shell of the nucleus accumbens [25]. Phasic release of striatal dopamine is the primary mechanism encoding for reward [26].
Recent studies have suggested abnormalities in ASC in striatal volume [27, 28], connectivity [29] and activity in response to social stimuli [30]. In addition, a gene expression study of postmortem brain tissue of people with ASC found reduced expression of CNR1[31]. In view of the atypical gaze behaviour of people with ASC, together with the observed striatal atypicalities, it is reasonable to examine the phenotype of gaze patterns as a function of variation in genes expressed in the striatum.
As gaze fixation is linked to striatal activity [16, 17, 19], we might expect that molecular variation in the genes involved in striatal function would be associated with differences in gaze towards socially rewarding stimuli. Using functional magnetic resonance imaging (fMRI), we previously found genetic variation in CNR1 modulated activity in the striatal region while watching happy (but not disgust) faces [32]. This result has been independently replicated in larger samples [33]. In the current study, we aimed to conduct an identical experiment using gaze-tracking in a new sample of volunteers. Specifically, we tested whether CNR1 genetic variation influences gaze duration towards happy faces. To ensure that this was closely matched to the original fMRI experiment, we also analysed gaze fixation duration for disgust expressions as a function of CNR1 genetic variation. Disgust faces are potential signals of 'nonreward', in contrast to rewarding happy faces, and hence provide a high-level control condition (that is, matched for face-specific qualities, such as configural features, as well as more general visual qualities of the stimuli, such as colour, shape and luminosity) for our experiment. We predicted that variation (single-nucleotide polymorphisms (SNPs)) in the CNR1 gene would be significantly associated with individual variability in gaze duration towards happy but not disgust faces.
Methods
Participants
A total of 30 student volunteers (13 males and 17 females; 29 right-handed and 1 left-handed; mean age ± SD, 24.1 ± 3.41 years old) were recruited by advertisement from the local universities. Participants were included only if all four grandparents were of Caucasian European ancestry to avoid genetic heterogeneity between different populations. Participants were also excluded if they reported any history of psychiatric diagnoses or regular drug abuse. They were equated for educational background in that all had completed high school and were studying towards a college degree. All had normal (or corrected to normal) vision. The study was approved by the Psychology Research Ethics Committee of the University of Cambridge.
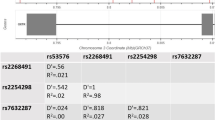
Buccal swabs were collected from all participants, and DNA was extracted. The four SNPs of choice were identical to those selected in our earlier fMRI study (rs1049353, rs806377, rs806380 and rs6454674), chosen to ensure a minor allele frequency > 0.2 in a Caucasian population and to cover as much of the gene as possible (see Figure 1) [32]. The DNA was genotyped by Geneservice, Inc. (Cambridge, UK) using standard TaqMan™ assays (Applied Biosystems, Inc., California, USA). Genotyping for these SNPs failed for two of these participants, resulting in a sample size of 28 participants for the final analysis.
Schematic structure of the cannabinoid receptor CNR1 gene with all four genotyped single-nucleotide polymorphisms indicated. Top: White boxes indicate untranslated regions, black boxes indicate translated regions and intervening straight line indicates an intronic region. Bottom: The linkage disequilibrium structure of the gene in the Caucasian (CEU) population is shown (using the publicly available HapMap version 3, release R2, database available at http://hapmap.ncbi.nlm.nih.gov/.
Procedure
The stimuli were taken from the Mindreading™ set developed by Baron-Cohen et al. [34], since dynamic facial expressions of emotion are assumed to be more ecologically valid than static photographs. The Mindreading set consists of video, audio and textual examples for 412 different emotions arranged into 24 emotion groups and organised according to six different developmental levels (based on emotions recognised in childhood through adulthood). These stimuli have been validated in typical populations and in people with ASC [35–38]. These stimuli were chosen over other existing available stimuli because Mindreading stimuli comprise dynamic emotional expressions whilst alternatives (such as the Ekman and Friesen set [39], the Karolinska Directed Emotional Faces set [40] and the NimStim set [41]) comprise static expressions. The Mindreading stimuli have excellent interrater reliability and external validity [36, 38] (stimuli are available at http://www.jkp.com/mindreading/).
Participants were seated comfortably at a fixed distance of 60 cm from the screen and were instructed to keep movement to a minimum. Participants watched 80 videoclips (three seconds each and sixteen clips for each of the five emotions) presented in a pseudorandom order using GazeTracker™ software (DynaVox Inc., Virginia, USA) with an interstimulus interval of six seconds. Participants were shown a fixation cross during the interstimulus interval. All stimuli were centred on a 19-inch monitor and occupied 70% of the screen area. To ensure that participants were attending to the stimulus, they were asked to say aloud what emotion they thought was being displayed (choosing one of five emotion words: 'happy', 'sad', 'angry', 'disgust' or 'fear'). Their responses were recorded by the experimenter.
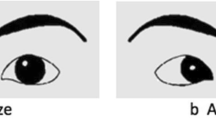
The Eye Response Interface Computer Aid camera (ERICA; http://www.eyeresponse.com/) was used to measure fixation time at each point at 60 Hz. ERICA uses reflected low-frequency infrared rays (λ = 880 nm) to map macrosaccades and fixation times at each point. The data were preprocessed using GazeTracker™ software. To ensure that the measured gaze duration was specific to the socially informative regions of the emotion expressions [35], 'look zones' were manually drawn around the eyes (the eyebrows and lower eyelids) and mouth region (the region from the bottom of the nose to the bottom of the lower lip) of all stimuli (see Figure 2). All look zones were 'dynamic'; that is, they tracked the eyes and the mouth regions while allowing for head movement of the actors.
Example gaze trail from a single participant and group mean gaze durations for happy faces classified by genotype. (a) Example gaze trail from a single participant on a still frame from a video stimulus showing a happy expression. The black circles represent fixation points and the amount of time (in seconds) spent in each. The dotted lines demarcate each look zone (eyes region and mouth region). (b) Gaze duration for happy faces grouped by genotype for rs806380 (top) and rs806377 (bottom), respectively. Unfilled circles indicate mean gaze duration, and error bars represent ± 1 SEM. From the Mindreading™ set developed by Baron-Cohen et al. [34]
The sum of duration of all fixations was recorded for each look zone. A fixation was defined as a continuous gaze for 100 ms within a 40-pixel diameter (corresponding to a 1.3° visual angle), which was in line with parameters used in similar studies [15]. Gaze duration for each expression was calculated by summing the mean fixation time for eyes and mouth regions. Different regions of the face (that is, eyes and mouth) are relevant for processing different basic emotions [12, 42–44]. Thus it is not ideal to compare the fixation time to the eyes region for happy and disgust faces, since disgust faces are associated with greater gaze duration to the mouth region. Hence, total fixation time across eyes and mouth regions was used as the dependent variable.
Results
Both happy and disgust expressions were recognised with > 80% accuracy. Genetic association was measured using the UNPHASED programme (http://www.mrc-bsu.cam.ac.uk/personal/frank/software/unphased), which computes the retrospective likelihood, that is, the probability of observing different genotypes given an observed distribution of a quantitative trait [45]. The two dependent variables (gaze duration for all happy and all disgust faces) and the genotypes for all four SNPs were included in a single analysis. This analysis revealed a significant association of the gaze duration for happy faces with rs806377 (χ2 = 8.88, df = 2, P = 0.011) and rs806380 (χ2 = 8.46, df = 2, P = 0.014). No significant associations (at P ≤ 0.05) were noted for gaze duration for disgust faces (nominal prs806377 = 0.104 and nominal prs806380 = 0.086). To test whether the observed lack of significant association with disgust faces was due to one video clip that was misclassified by a majority of the participants, the data were reanalysed after removing all fixation data associated with this one video clip. This revealed an identical pattern of results, with a nominal prs806377 = 0.111 and a nominal prs806380 = 0.105.
However, when multiple SNPs are in linkage disequilibrium (LD), hypothesis tests in single-locus analyses are not independent. To take this into account, Li and Ji [46] proposed a method for estimating the true number of independent tests (Meff), which takes into consideration the LD between SNPs. This method was implemented using the SNPSpD software programme [47], which revealed that Meff was 3 in the current sample. The Bonferroni correction for three independent tests gave a corrected P = 0.033 for rs806377 and a corrected P = of 0.042 for rs806380 for association with gaze duration for happy faces.
To further analyse genotypic differences for each SNP that were significantly associated with gaze duration for happy faces, post hoc t-tests were conducted. In rs806377, the CC genotype was associated with longer gaze duration than the CT genotype (t = 2.92, P < 0.025 with the Bonferroni correction). In rs806380, the GG genotype was associated with longer gaze duration than the AA genotype (t = 2.78, P < 0.05 with the Bonferroni correction) (see Figure 2).
The main effects of all possible haplotypes were tested with various possible window sizes (two, three and four marker combinations) using UNPHASED software. None of these haplotypic association tests were significant at P < 0.05. While the small sample size did not allow for a robust test of sex differences in this genetic association, we report the nominal P values for these tests for the sake of completeness. rs806377 was significantly associated with the gaze duration for happy faces in both females (P = 0.021) and males (P = 0.004). Additionally, in males, rs806380 (P = 0.019) and rs1049353 (P = 0.004) were found to be associated with gaze duration for happy faces.
Discussion
In this experiment, we predicted that CNR1 genetic variations would be associated with differences in gaze fixation duration towards happy faces. This prediction was confirmed: two SNPs in this gene (rs806377 and rs806380) were associated with differences in gaze duration for happy (but not disgust) faces. This finding fits well with the established role of the CNR1 gene in reward processes [20] and is consistent with the results of fMRI studies [32, 33] in showing that this gene is a component of the molecular architecture of social reward processing. Social reward processing has been suggested to be impaired in people with ASC [48–50], particularly as reflected in atypical gaze patterns towards social stimuli. Hence the current results could be relevant to understanding the genetic underpinning of the social behavioural symptoms in people with ASC.
A comparison of these results with those from our earlier fMRI study reveals that for the SNP rs806377, the allelic group (CC) associated with the highest striatal response is also associated with the longest gaze duration for happy faces. For rs806380, the allelic group associated with the highest striatal response (GG) is also associated with the longest gaze duration for happy faces. rs806377 is located in an untranslated region (UTR) of the gene (Figure 1), and rs806380 was found to be in significant LD with a 5'-UTR SNP (rs78074274) using CandiSNPer [51]. The observed effects can thus be mediated by either or both of these UTR SNPs by potentially altering gene transcription and/or mRNA stability. Since the fMRI and gaze duration data come from largely independent samples (only three of thirty participants were common to both studies), it is likely that the observed genetic differences reflect real effects.
We interpret the genetically linked biasing of visual perception in terms of individual differences in the reward circuitry. The two processes of increased visual preference (indicated by longer gaze duration) and increased striatal response for happy faces are linked in a positive feedback loop [5]. We tend to look longer at preferred stimuli, which in turn increases our preference/'reward value' for these stimuli. Consequently, we interpret the observed effect in biasing visual perception of social stimuli in terms of differences in the individual reward circuitry. Whether such intrinsic differences in reward circuitry change the formation and nature of 'saliency/value maps' formed during gaze fixation is a question for future research [19, 52].
A second broader question for future research is whether the observed CNR1 genotypic differences in fixation duration for happy faces is specific to social rewards or whether this holds true for all classes of rewards. Variation in CNR1 has been linked to polysubstance abuse and associated with increased activity in reward-processing areas of the brain in response to drug cues for both marijuana and alcohol addicts [53, 54]. Hence it is possible that the observed genotypic differences in the general population may extend to other classes of rewards. Crucially, however, a reduced experience of rewards in response to social stimuli such as happy faces (as has been suggested by Dawson et al. [48] to apply to ASC) has more far-reaching consequences, since if an infant is looking less at happy faces and is finding them less rewarding, this will make social interactions less reinforcing, which in turn can exacerbate the social difficulties observed in people with ASC.
It is possible that a number of genes, each of small to medium effect size, determine the striatal response to social stimuli such as happy faces. Other potential candidate genes might include those involved in the oxytocin-vasopressin system (OXTR, AVPR1A and AVPR1B) as well as those coding for key proteins involved in neutrotransmission (for example, MAOA and GABRB3) [55]. We speculate that these genes have an additive effect and might potentially underlie complex traits related to social functioning [56]. In a larger population-based genetic association study of empathy, we found a nominally significant association of CNR1 genetic variation with the Empathy Quotient [55, 57]. Additionally, reduced expression of CNR1 was found in postmortem brain tissue of individuals with ASC [31]. Together, these findings further support the implication that variation in the CNR1 gene modulates the response to social stimuli such as happy faces.
However, the current findings should be interpreted with caution, since, in the absence of any expression data, any functional role for the SNPs can only be speculative; that is, the observed SNP effects may be caused by being in LD with other functional polymorphisms and/or through mechanisms that affect mRNA stability or splicing as mentioned earlier. However, the observation of genetic differences in two separate (albeit small) samples using an identical paradigm with two different techniques points towards a putative role played by CNR1 in the response to happy faces.
Conclusions
In this study, we tested whether common variants in the CNR1 gene modulate gaze duration towards happy faces. We found that two SNPs in this gene were significantly associated with gaze duration for happy (but not disgust) faces. This result is consistent with that of our previous fMRI study [32]. Specifically, the allelic groups that were found to be associated with the strongest striatal response in our fMRI study were associated with the longest gaze duration for happy faces in the current sample. This finding suggests a role for CNR1 in social reward processing and could have significance for clinical conditions such ASC, which are marked by a deficit in social reward processing as well as atypical responses to facial expressions of emotion [35, 36, 49].
Abbreviations
- CNR1 :
-
cannabinoid receptor 1
- mRNA:
-
messenger RNA
- SNP:
-
single-nucleotide polymorphism.
References
Moschovakis A, Highstein SM: The anatomy and physiology of primate neurons that control rapid eye movements. Annu Rev Neurosci. 1994, 17: 465-488. 10.1146/annurev.ne.17.030194.002341.
Calvo M, Avero P: Time course of attentional bias to emotional scenes in anxiety: gaze direction and duration. Cogn Emot. 2005, 19: 433-451. 10.1080/02699930441000157.
Butterworth G, Jarrett N: What minds have in common is space: spatial mechanisms serving joint visual attention in infancy. Br J Dev Psychol. 1991, 9: 55-72.
Calvo M, Lang P: Gaze patterns when looking at emotional pictures: motivationally biased attention. Motiv Emot. 2004, 28: 221-243.
Shimojo S, Simion C, Shimojo E, Scheier C: Gaze bias both reflects and influences preference. Nat Neurosci. 2003, 6: 1317-1322. 10.1038/nn1150.
Connellan J, Baron-Cohen S, Wheelwright S, Batki A, Ahluwalia J: Sex differences in human neonatal social perception. Infant Behav Dev. 2001, 23: 113-118.
Fletcher-Watson S, Leekam SR, Benson V, Frank MC, Findlay JM: Eye-movements reveal attention to social information in autism spectrum disorder. Neuropsychologia. 2009, 47: 248-257. 10.1016/j.neuropsychologia.2008.07.016.
Klin A, Jones W, Schulz R, Volkmar F, Cohen DJ: Visual fixation patterns during viewing of naturalistic social situations as predictors of social competence in individuals with autism. Arch Gen Psychiatry. 2002, 9: 809-816.
Katsanis J, Taylor J, Iacono WG, Hammer MA: Heritability of different measures of smooth pursuit eye tracking dysfunction: a study of normal twins. Psychophysiology. 2000, 37: 724-730. 10.1111/1469-8986.3760724.
Iacono WG: Eye tracking in normal twins. Behav Genet. 1982, 12: 517-526. 10.1007/BF01073782.
Greenwood TA, Braff DL, Light GA, Cadenhead KS, Calkins ME, Dobie DJ, Freedman R, Green MF, Gur RE, Gur RC, Mintz J, Nuechterlein KH, Olincy A, Radant AD, Seidman LJ, Siever LJ, Silverman JM, Stone WS, Swerdlow NR, Tsuang DW, Tsuang MT, Turetsky BI, Schork NJ: Initial heritability analyses of endophenotypic measures for schizophrenia: the Consortium on the Genetics of Schizophrenia. Arch Gen Psychiatry. 2007, 64: 1242-1250. 10.1001/archpsyc.64.11.1242.
Baron-Cohen S, Wheelwright S, Jolliffe T: Is there a "language of the eyes"? Evidence from normal adults and adults with autism or Asperger syndrome. Vis Cogn. 1997, 4: 311-331. 10.1080/713756761.
Joliffe T, Mortimore C, Robertson M: Another advanced test of theory of mind: evidence from very high functioning adults with autism or Asperger syndrome. J Child Psychol Psychiatry. 1997, 38: 813-822. 10.1111/j.1469-7610.1997.tb01599.x.
Swettenham J, Baron-Cohen S, Charman T, Cox A, Baird G, Drew A, Rees L, Wheelwright S: The frequency and distribution of spontaneous attention shifts between social and non-social stimuli in autistic, typically developing, and non-autistic developmentally delayed infants. J Child Psychol Psychiatry. 1998, 9: 747-753.
Senju A, Southgate V, White S, Frith U: Mindblind eyes: an absence of spontaneous theory of mind in Asperger syndrome. Science. 2009, 325: 883-885. 10.1126/science.1176170.
Hikosaka O: Basal ganglia mechanisms of reward-oriented eye movement. Ann N Y Acad Sci. 2007, 1104: 229-249. 10.1196/annals.1390.012.
Lau B, Glimcher PW: Action and outcome encoding in the primate caudate nucleus. J Neurosci. 2007, 27: 14502-14514. 10.1523/JNEUROSCI.3060-07.2007.
Cai X, Kim S, Lee D: Heterogeneous coding of temporally discounted values in the dorsal and ventral striatum during intertemporal choice. Neuron. 2011, 69: 170-182. 10.1016/j.neuron.2010.11.041.
Trommershäuser J, Glimcher PW, Gegenfurtner KR: Visual processing, learning and feedback in the primate eye movement system. Trends Neurosci. 2009, 32: 583-590. 10.1016/j.tins.2009.07.004.
Gardner EL, Vorel SR: Cannabinoid transmission and reward-related events. Neurobiol Dis. 1998, 5: 502-533. 10.1006/nbdi.1998.0219.
Freund TF, Katona I, Piomelli D: Role of endogenous cannabinoids in synaptic signaling. Physiol Rev. 2003, 83: 1017-1066.
Fusco FR, Martorana A, Giampà C, De March Z, Farini D, D'Angelo V, Sancesario G, Bernardi G: Immunolocalization of CB1 receptor in rat striatal neurons: a confocal microscopy study. Synapse. 2004, 53: 159-167. 10.1002/syn.20047.
Hurley MJ, Mash DC, Jenner P: Expression of cannabinoid CB1 receptor mRNA in basal ganglia of normal and parkinsonian human brain. J Neural Transm. 2003, 110: 1279-1288. 10.1007/s00702-003-0033-7.
Haber SN, Knutson B: The reward circuit: linking primate anatomy and human imaging. Neuropsychopharmacology. 2010, 35: 4-26. 10.1038/npp.2009.129.
van der Stelt M, Di Marzo V: The endocannabinoid system in the basal ganglia and in the mesolimbic reward system: implications for neurological and psychiatric disorders. Eur J Pharmacol. 2003, 480: 133-150. 10.1016/j.ejphar.2003.08.101.
Schultz W: Getting formal with dopamine and reward. Neuron. 2002, 36: 241-263. 10.1016/S0896-6273(02)00967-4.
Langen M, Schnack HG, Nederveen H, Bos D, Lahuis BE, de Jonge MV, van Engeland H, Durston S: Changes in the developmental trajectories of striatum in autism. Biol Psychiatry. 2009, 66: 327-333. 10.1016/j.biopsych.2009.03.017.
Estes A, Shaw DW, Sparks BF, Friedman S, Giedd JN, Dawson G, Bryan M, Dager SR: Basal ganglia morphometry and repetitive behavior in young children with autism spectrum disorder. Autism Res.
Di Martino A, Kelly C, Grzadzinski R, Zuo XN, Mennes M, Mairena MA, Lord C, Castellanos FX, Milham MP: Aberrant striatal functional connectivity in children with autism. Biol Psychiatry. 2011, 69: 847-856. 10.1016/j.biopsych.2010.10.029.
Weng SJ, Carrasco M, Swartz JR, Wiggins JL, Kurapati N, Liberzon I, Risi S, Lord C, Monk CS: Neural activation to emotional faces in adolescents with autism spectrum disorders. J Child Psychol Psychiatry. 2011, 52: 296-305. 10.1111/j.1469-7610.2010.02317.x.
Purcell A, Jeon O, Zimmerman A, Blue M, Pevsner J: Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001, 57: 1618-1628.
Chakrabarti B, Kent L, Suckling J, Bullmore E, Baron-Cohen S: Variations in the human cannabinoid receptor (CNR1) gene modulate striatal responses to happy faces. Eur J Neurosci. 2006, 23: 1944-1948. 10.1111/j.1460-9568.2006.04697.x.
Domschke K, Dannlowski U, Ohrmann P, Lawford B, Bauer J, Kugel H, Heindel W, Young R, Morris P, Arolt V, Deckert J, Suslow T, Baune BT: Cannabinoid receptor 1 (CNR1) gene: impact on antidepressant treatment response and emotion processing in major depression. Eur Neuropsychopharmacol. 2008, 18: 751-759. 10.1016/j.euroneuro.2008.05.003.
Baron-Cohen S, Golan O, Wheelwright S, Hill JJ: Mindreading: The Interactive Guide to Emotions. 2004, London: Jessica Kingsley Ltd
El Kaliouby R, Robinson P, Keates S: Temporal context and the recognition of emotion from facial expression. Proceedings of HCI International Conference. 2003, Springer-Verlag
Golan O, Baron-Cohen S: Systemizing empathy: teaching adults with Asperger's syndrome/high functioning autism to recognize emotions using interactive multimedia. Dev Psychopathol. 2006, 18: 591-617.
Chakrabarti B, Bullmore E, Baron-Cohen S: Empathizing with basic emotions: common and discrete neural substrates. Soc Neurosci. 2006, 1: 364-384. 10.1080/17470910601041317.
Golan O, Baron-Cohen S, Hill J: The Cambridge Mindreading (CAM) Face-Voice Battery: testing complex emotion recognition in adults with and without Asperger syndrome. J Autism Dev Disord. 2006, 36: 169-183. 10.1007/s10803-005-0057-y.
Ekman P, Friesen W: Pictures of Facial Affect. 1976, Palo Alto: Consulting Psychologists Press
Lundqvist D, Flykt A, Öhman A: The Karolinska Directed Emotional Faces-KDEF, CD ROM from Department of Clinical Neuroscience, Psychology section, Karolinska Institutet. 1998, Stockholm: Karolinska Institutet
Tottenham N, Tanaka JW, Leon AC, McCarry T, Nurse M, Hare TA, Marcus DJ, Westerlund A, Casey BJ, Nelson C: The NimStim set of facial expressions: judgments from untrained research participants. Psychiatry Res. 2009, 168: 242-249. 10.1016/j.psychres.2008.05.006.
Chakrabarti B: Individual differences in human emotion perception: neuroimaging, genetic and behavioural investigations. PhD thesis. 2007, Department of Psychiatry, School of Clinical Medicine, University of Cambridge
Gamer M, Büchel C: Amygdala activation predicts gaze toward fearful eyes. J Neurosci. 2009, 29: 9123-9126. 10.1523/JNEUROSCI.1883-09.2009.
Kliemann D, Dziobek I, Hatri A, Steimke R, Heekeren HR: Atypical reflexive gaze patterns on emotional faces in autism spectrum disorders. J Neurosci. 2010, 30: 12281-12287. 10.1523/JNEUROSCI.0688-10.2010.
Dudbridge F: Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008, 66: 87-98. 10.1159/000119108.
Li J, Ji L: Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005, 95: 221-227. 10.1038/sj.hdy.6800717.
Nyholt DR: A simple correction for multiple testing for SNPs in linkage disequilibrium with each other. Am J Hum Genet. 2004, 74: 765-769. 10.1086/383251.
Dawson G, Carver L, Meltzoff AN, Panagiotides H, McPartland J, Webb SJ: Neural correlates of face and object recognition in young children with autism spectrum disorder, developmental delay, and typical development. Child Dev. 2002, 73: 700-717. 10.1111/1467-8624.00433.
Scott-Van Zeeland AA, Dapretto M, Ghahremani DG, Poldrack RA, Bookheimer SY: Reward processing in autism. Autism Res. 2010, 3: 53-67.
Kohls G, Peltzer J, Herpertz-Dahlmann B, Konrad K: Differential effects of social and non social reward on response inhibition in children and adolescents. Dev Sci. 2009, 12: 614-625. 10.1111/j.1467-7687.2009.00816.x.
Schmitt AO, Aßmus J, Bortfeldt RH, Brockmann GA: CandiSNPer: a web tool for the identification of candidate SNPs for causal variants. Bioinformatics. 2010, 26: 969-970. 10.1093/bioinformatics/btq068.
Henderson J: Human gaze control during real-world scene perception. Trends Cogn Sci. 2003, 7: 498-504. 10.1016/j.tics.2003.09.006.
Filbey FM, Schacht JP, Myers US, Chavez RS, Hutchison KE: Individual and additive effects of the CNR1 and FAAH genes on brain response to marijuana cues. Neuropsychopharmacology. 2009, 35: 967-975.
Hutchison KE, Haughey H, Niculescu M, Schacht J, Kaiser A, Stitzel J, Horton WJ, Filbey F: The incentive salience of alcohol: translating the effects of genetic variant in CNR1. Arch Gen Psychiatry. 2008, 65: 841-850. 10.1001/archpsyc.65.7.841.
Chakrabarti B, Dudbridge F, Kent L, Wheelwright S, Hill-Cawthorne G, Allison C, Banerjee-Basu S, Baron-Cohen S: Genes related to sex steroids, neural growth, and social-emotional behavior are associated with autistic traits, empathy, and Asperger syndrome. Autism Res. 2009, 2: 157-177. 10.1002/aur.80.
Ebstein RP, Israel S, Chew SH, Zhong S, Knafo A: Genetics of human social behavior. Neuron. 2010, 65: 831-844. 10.1016/j.neuron.2010.02.020.
Baron-Cohen S, Wheelwright S: The Empathy Quotient: an investigation of adults with Asperger syndrome or high functioning autism, and normal sex differences. J Autism Dev Disord. 2004, 34: 163-175.
Acknowledgements
We are grateful to Zanna Szlachta, Siamac Rezaiezadeh, Kathleen Maura Linnane, and Stefania Cannella for help with the experiment and to Carrie Allison and Sally Wheelwright for volunteer database management. During the period of this work, BC was funded by a scholarship from Trinity College Cambridge and held a Research Fellowship at Darwin College Cambridge. SBC was funded by Target Autism Genome, the Nancy Lurie Marks Family Foundation and the Medical Research Council, UK during the period of this work. This work was submitted in partial fulfilment of the doctoral degree by BC to the University of Cambridge and was conducted in association with the National Institute for Health Research Collaboration, Collaboration for Leadership in Applied Health Research and Care for Cambridgeshire and Peterborough, NHS Mental Health Foundation Trust. We are also grateful to Lindsey Kent, Frank Dudbridge, Ofer Golan, Tom Hutchinson, Mike Lombardo, Caroline Robertson, Bonnie Auyeung and Chris Ashwin for valuable discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
BC designed and ran the experiment, analysed the data and wrote the paper. SBC provided intellectual input at all stages and cowrote the paper. Both authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Chakrabarti, B., Baron-Cohen, S. Variation in the human cannabinoid receptor CNR1 gene modulates gaze duration for happy faces. Molecular Autism 2, 10 (2011). https://doi.org/10.1186/2040-2392-2-10
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2040-2392-2-10