Abstract
Breast cancer is the most prevalent malignancy in women in Western countries, currently accounting for one third of all female cancers. Familial aggregation is thought to account for 5–10 % of all BC cases, and germline mutations in BRCA1 and BRCA2 account for less of the half of these inherited cases. In Lebanon, breast cancer represents the principal death-causing malignancy among women, with 50 % of the cases diagnosed before the age of 50 years.
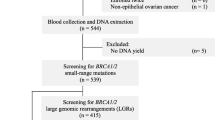
In order to study BRCA1/2 mutation spectra in the Lebanese population, 72 unrelated patients with a reported family history of breast and/or ovarian cancers or with an early onset breast cancer were tested. Fluorescent direct sequencing of the entire coding region and intronic sequences flanking each exon was performed.
A total of 38 BRCA1 and 40 BRCA2 sequence variants were found. Seventeen of them were novel. Seven confirmed deleterious mutations were identified in 9 subjects providing a frequency of mutations of 12.5 %. Fifteen variants were considered of unknown clinical significance according to BIC and UMD-BRCA1/BRCA2 databases.
In conclusion, this study represents the first evaluation of the deleterious and unclassified genetic variants in the BRCA1/2 genes found in a Lebanese population with a relatively high risk of breast cancer.
Similar content being viewed by others
Introduction
In the western countries the lifetime risk of developing breast cancer is around 12 %. About 5 %–10 % of breast cancer (BC) and ovarian cancer (OC) are hereditary and 30 %–50 % of these are due to mutations with an autosomal dominant inheritance [1]. Deleterious mutations in the BRCA1 and BRCA2 genes are the principal known cause of hereditary BC. They are localized on chromosomes 17q21 and 13q12 respectively [2, 3]. They are found in 1/400-1/800 people in the general population [4–6].
For women with a personal or family history of breast and/or ovarian cancer, BRCA1/2 full-sequencing and analysis for large genomic rearrangements (LGR) are routinely used to quantify the genetic component of cancer risk. The spectrum of mutations found in BRCA1/2 depends on the studied population, ranging from few founder mutations in some, to a wide spectrum of mutations in others [7].
In Lebanon, breast cancer remains the most frequent type of cancer among women since 1960. In 2005, its age adjusted incidence rate was estimated at 76 new cases per 100.000 by the Public Health Ministry of Lebanon and it is still increasing (http://www.public-health.gov.lb). In France, during the same period, it was of 101 new cases per 100.000 according to the National Institute of Cancer (http://www.e-cancer.fr/). On the other hand, the median age at diagnosis for BC in Lebanon is 52.5 years, approximately 9 years younger than the European/North American median [8].
Given that a woman with breast cancer is more likely to carry a susceptibility gene mutation the younger she is at the time of diagnosis [5, 9, 10], one possibility is that the allelic frequency of high penetrance genes in the Lebanese population may be higher than that in Caucasian populations.
In order to determine the spectra and frequency of BRCA1/2 mutations within the Lebanese population, we studied a cohort of 72 Lebanese unrelated patients with breast cancer.
Subjects and Methods
Participants
From 2001 to 2011, 72 unrelated patients were selected to undergo sequence DNA testing for mutations in BRCA1/2. They were referred from a wide variety of settings from all over the country, ranging from private physicians’ offices to major academic medical centers, because of elevated risk of a BRCA1/2 mutation: a personal history of invasive breast cancer and at least one of the following criteria: A) breast cancer at any age at onset in ≥ 2 first- and/or second-degree relatives, B) breast cancer <50 years in a first- or second-degree relative, C) ovarian cancer in ≥ 2 first- and/or second-degree relatives, D) breast and ovarian cancer in ≥ 2 first- and/or second-degree relatives, E) both breast and ovarian cancer in a single first- or second-degree relative, F) male breast cancer.
Two cases in men with a personal history of sporadic breast cancer were also included.
BRCA1 and BRCA2 analysis
After an informed consent was signed, DNA was extracted from 10 ml of peripheral blood using standard methods [11]. All exons and > 50 bp of each flanking intron were amplified in a final volume of 50 μl containing 100 ng DNA, 0.25 mM dNTPs, 100 ng of each primer and 0.02 unit of Taq polymerase (Invitrogen Life Technologies, Carlsbad, CA, USA). PCR was performed in an ABI9700 thermocycler (Applied Biosystems, Foster City, CA) with initial denaturation at 94 °C for 5 min, followed by 35 cycles of (95 °C for 30s, specific annealing temperature for 30s, 72 °C for 30s), except for BRCA1 exons 14 and 15 where 5 cycles were added to the initial 35 cycles. Primers sequences are available on request as well as annealing temperatures of each exon.
PCR products were purified using the illustraTM GFX PCR DNA and Gel Band Purification Kit (GE Healthcare, Buckinghamshire, UK), and resuspended in 40 μl of deionized water. Both strands of the resultant products were sequenced using the BigDye® Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) under standard conditions. The labeled products were subjected to electrophoresis on an ABI3130 Genetic Analyzer sequencing system (Applied Biosystems, Foster City, CA, USA). Electropherograms were analyzed using Sequence Analysis Software version 5.2 (Applied Biosystems, Foster City, CA, USA) and compared to reference sequences (BRCA1: ADNg L78833; ADNc NM_007294.3 and BRCA2: ADNg Z74739; ADNc NM_000059.3) using ChromasPro version 1.5 (Technelysium, Queensland, Australia). All deleterious mutations were confirmed first on an independent second amplification and second on a second blood specimen.
Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequences.
The effect of missense mutations was predicted using the BIC database (http://research.nhgri.nih.gov) and the UMD-BRCA1/BRCA2 databases and the prediction programs: SIFT (Sorting Intolerant From Tolerant) (http://sift.jcvi.org/www/SIFT_Blink_submit.html), PolyPhen (http://genetics.bwh.harvard.edu/pph/index.html), and Align-GVGD from the International Agency for research on Cancer (http://agvgd.iarc.fr) [12, 13]. Prediction of splicing effect was performed with MaxEntScan using a 15 % variation threshold as proposed by Houdayer et al.[14, 15].
Identified variations were classified into three categories: deleterious, unclassified variant, and neutral.
Results
The mean age at diagnosis of BC for the seventy-two patients was 41 years old. Thirty-seven patients provided us with their histopathological results. Thirty-one patients had a positive estrogen-receptor (ER) and a positive progesterone-receptor (PR) disease, four patients had a positive ER and a negative PR disease and one patient had a negative ER and a positive PR disease. Only one patient had a triple negative disease and was not found to have any deleterious mutation.
Within this cohort, we identified a total of 38 BRCA1 and 40 BRCA2 sequence variants, of which 17 (21.8 %) were novel (not reported in the databases BIC and UMD-BRCA1/BRCA2): 11 (14.1 %) in BRCA1 and 6 (7.7 %) in BRCA2 (Tables 1 and 2). Five disease-associated BRCA1 mutations and two disease-associated BRCA2 mutations were found in this cohort (Figure 1).
Six out of the 38 variants found in BRCA1 (15.8 %) were found to be of uncertain clinical significance according to BIC and UMD. Nineteen variants were known polymorphisms, 14 were intronic variants, 1 was a nonsense mutation (p.W1815X), and 23 missense variations (Table 1), out of which two were found of interest: p.C44F and p.P142A [16, 17]. The first one was associated with a strong family history of breast and ovarian cancer in two unrelated families and the second one was found in a large family with many affected women of different age of onset. One patient presented the association of the three missense variations: p.Y179C, p.F486L and p.N550H, and one patient the variations p.N550H and p.F486L. In 41 patients (57 %) a haplotype presenting the combination of the following variations: p.S694S, p.L771L, p.P871L, p.E1038G, p.K1183R, p.S1436S, p.S1613G and IVS18 + 66 G > A, was found. In 9 of those, an additional variation, p.Q356R, was found. The BRCA1 p.Y1552H variation was the only mutation associated to the frequent haplotype found in our population.
Nine out of the 40 variants found in BRCA2 (22.5 %) were of uncertain clinical significance according to BIC and UMD databases. Twenty-three were known polymorphisms out of which one nonsense variation (p.K3326X). Eighteen variants were missense variations, 19 intronic variants, and 2 frameshift mutations (Table 2). For the latter, the first one, c.5576_5579delTTAA in exon 11, is a deletion of 4 bases pairs leading to a frameshift and a stop codon three amino acids further down (p.I1859KfsX3). This mutation was identified in a patient diagnosed at the age of 51 years and was considered as a truncating mutation. The family history of this individual included a breast cancer diagnosed in the mother at 63 years. The second mutation is at position c.9485-1 G > A leading to exon 26 skipping. It was identified in a patient diagnosed at the age of 33 years. The family history of this individual included a breast cancer diagnosis of the father and the paternal aunt.
No variant was predicted to affect splicing except the BRCA2 c.9485-1 G > A mutation which disrupts an acceptor splice site consensus sequence.
Discussion
In the present study, we have identified BRCA1 and BRCA2 germline deleterious mutations in 9 carriers among a cohort of 72 unrelated Lebanese patients with BC, providing a frequency of mutations which represents a prevalence of 12.5 %.
The low prevalence of mutations found in our population in comparison to other ones [5, 18–20], can be explained either by the genetic testing criteria, or by the possibility that some mutations were missed since we could not look for deletions or duplications of entire exons. Consanguinity might be also an equivocal risk-modifying factor [21, 22]. Indeed, the Lebanese population has a relatively high rate of consanguinity (around 15–20 %), and association studies on consanguinity and breast cancer, and the frequency of BRCA 1 and BRCA2 in highly consanguineous populations pointed that consanguinity might lead to decreased incidence of breast cancer [23]. Nevertheless, other studies obtained contradictory results and the question remains open [23].
Our findings could not contribute in understanding the cause of the high incidence of BC in Lebanon and the lower age of onset observed compared to western countries. The low age of onset was also reported in other Asian countries, and Arab countries in the Middle East [24–26]. It may be explained by the differences in exposure to female hormones, diet, physical activity, or other factors. For instance, the “Westernization” of these populations is due to delayed childbearing, lower parity, reduced breast-feeding, decreased exercise and dietary changes [24]. It can lead to a ‘cohort effect’ where the younger cohort have increased breast cancer risk compared to their mothers and grandmothers, thus giving rise to an apparent lower mean age of onset [25].
In BRCA1, a haplotype presenting the combination of the variations: p.S694S, p.L771L, p.P871L, p.E1038G, p.K1183R, p.S1436S, p.S1613G and IVS18 + 66 G > A was very often found in the Lebanese population. In fact, Dunning et al. reported a close to complete allelic association between the alleles at the 871, 1038 and 1613 residues [27]. Freedman et al. also reported this haplotype, except for the p.S694S variation, with a frequency of 32 % [28]. The latter study found no significant differences of frequencies between patients and control groups [28].
A major problem, the pathogenic status of several sequence variants that are novel or considered of unknown clinical significance, remains unresolved. For instance, a patient had the association of three missense variations: p.Y179C, p.F486L and p.N550H. Taken separately these variations were considered few years ago non or mildly pathogenic, while this haplotype has recently been considered to alter the protein function of BRCA1 [29]. In another patient, two variations were found: p.F486L and p.N550H. We considered the result as pathogenic. Analyzing the cosegregation of the variations with the cancer phenotype within the family can help resolving this problem, but the high cost and the reluctance of the families to undergo the screening prevent proceeding with this study.
To date, no other mutational analysis on breast cancer has been conducted in Lebanon. This report helps determining the spectrum of BRCA1/2 point mutations in our country and must be the beginning of other studies with a larger cohort in order to determine the prevalence of the mutations in the general population and to check for the existence of founder mutations given that each of the following mutations p.C44F and p.W1815X were found twice in this cohort. It will be also interesting to evaluate the differences of BRCA1/2 mutations frequencies among women of diverse ethnicities in Lebanon (Muslims and Christians) or more importantly, whether ethnicity should be taken into account in BRCA1/2 risk assessment. The answers are important considering the eligibility for genetic testing, and the adoption of targeted prevention strategies.
References
Ferla R, Calo V, Cascio S, Rinaldi G, Badalamenti G, Carreca I, Surmacz E, Colucci G, Bazan V, Russo A: Founder mutations in BRCA1 and BRCA2 genes. Ann Oncol 2007,18(6):93–98.
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al.: A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994, 266: 66–71. 10.1126/science.7545954
Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G: Identification of the breast cancer susceptibility gene BRCA2. Nature 1995, 378: 789–792. 10.1038/378789a0
Claus EB, Schildkraut JM, Thompson WD, Risch NJ: The genetic attributable risk of breast and ovarian cancer. Cancer 1996, 77: 2318–2324. 10.1002/(SICI)1097-0142(19960601)77:11<2318::AID-CNCR21>3.0.CO;2-Z
Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE: Breast Cancer Linkage Consortium: Risks of cancer in BRCA1-mutation carriers. Lancet 1994, 343: 692–695. 10.1016/S0140-6736(94)91578-4
Whittemore AS, Gong G, Itnyre J: Prevalence and contribution of BRCA1 mutations in breast cancer and ovarian cancer: results from three U.S. population-based case–control studies of ovarian cancer. Am J Hum Genet 1997, 60: 496–504.
Armaou S, Pertesi M, Fostira F, Thodi G, Athanasopoulos PS, Kamakari S, Athanasiou A, Gogas H, Yannoukakos D, Fountzilas G, Konstantopoulou I: Contribution of BRCA1 germ-line mutations to breast cancer in Greece: a hospital-based study of 987 unselected breast cancer cases. Br J Cancer 2009, 101: 32–37. 10.1038/sj.bjc.6605115
Lakkis NA, Adib SM, Osman MH, Musharafieh UM, Hamadeh GN: Breast cancer in Lebanon: incidence and comparison to regional and Western countries. Cancer Epidemiol. 2010 Jun,34(3):221–225. 10.1016/j.canep.2010.02.013
Langston AA, Malone KE, Thompson JD, Daling JR, Ostrander EA: BRCA1 mutations in a population-based sample of young women with breast cancer. N Engl J Med 1996, 334: 137–142. 10.1056/NEJM199601183340301
FitzGerald MG, MacDonald DJ, Krainer M, Hoover I, O'Neil E, Unsal H, Silva-Arrieto S, Finkelstein DM, Beer-Romero P, Englert C, Sgroi DC, Smith BL, Younger JW, Garber JE, Duda RB, Mayzel KA, Isselbacher KJ, Friend SH, Haber DA: Germ-line BRCA1 mutation in Jewish and non-Jewish women with early-onset breast cancer. N Engl J Med 1996, 334: 143–149. 10.1056/NEJM199601183340302
Miller SA, Dynes DD, Polesky HF: A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988, 16: 1215. 10.1093/nar/16.3.1215
Caputo S, Benboudjema L, Sinilnikova O, Rouleau E, Béroud C, Lidereau R: French BRCA GGC Consortium: Description and analysis of genetic variants in French hereditary breast and ovarian cancer families recorded in the UMD-BRCA1/BRCA2 databases. Nucleic Acids Res 2012, 40: D992-D1002. 10.1093/nar/gkr1160
Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, Hogervorst FB, Hoogerbrugge N, Spurdle AB, Tavtigian SV: IARC Unclassified Genetic Variants Working Group: Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat 2008, 29: 1282–1291. 10.1002/humu.20880
Yeo G, Burge CB: Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 2004, 11: 377–394. 10.1089/1066527041410418
Houdayer C: In silico prediction of splice-affecting nucleotide variants. Methods Mol Biol 2011, 760: 269–281. 10.1007/978-1-61779-176-5_17
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al.: A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994 Oct 7,266(5182):66–71. 10.1126/science.7545954
Meza JE, Brzovic PS, King MC, Klevit RE: Mapping the functional domains of BRCA1. Interaction of the ring finger domains of BRCA1 and BARD1. J Biol Chem 1999 Feb 26,274(9):5659–5665. 10.1074/jbc.274.9.5659
Hall MJ, Reid JE, Burbidge LA, Pruss D, Deffenbaugh AM, Frye C, Wenstrup RJ, Ward BE, Scholl TA, Noll WW: BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer 2009, 115: 2222–2233. 10.1002/cncr.24200
Uhrhammer N, Abdelouahab A, Lafarge L, Feillel V: Ben Dib A, Bignon YJ: BRCA1 mutations in Algerian breast cancer patients: high frequency in young, sporadic cases. Int J Med Sci 2008, 5: 197–202.
Malone KE, Daling JR, Neal C, Suter NM, O'Brien C, Cushing-Haugen K, Jonasdottir TJ, Thompson JD, Ostrander EA: Frequency of BRCA1/BRCA2 mutations in a population-based sample of young breast carcinoma cases. Cancer 2000, 88: 1393–1402. 10.1002/(SICI)1097-0142(20000315)88:6<1393::AID-CNCR17>3.0.CO;2-P
Liede A, Malik IA, Aziz Z, Rios Pd Pde L, Kwan E, Narod SA: Contribution of BRCA1 and BRCA2 Mutations to Breast and Ovarian Cancer in Pakistan. Am J Hum Genet 2002, 71: 595–606. 10.1086/342506
Bener A, Ayoubi HR, Ali AI, Al-Kubaisi A, Al-Sulaiti H: Does consanguinity lead to decreased incidence of breast cancer? Cancer Epidemiol 2010, 34: 413–418. 10.1016/j.canep.2010.04.004
Hamamy H, Antonarakis SE, Cavalli-Sforza LL, Temtamy S, Romeo G, Kate LP, Bennett RL, Shaw A, Megarbane A, van Duijn C, Bathija H, Fokstuen S, Engel E, Zlotogora J, Dermitzakis E, Bottani A, Dahoun S, Morris MA, Arsenault S, Aglan MS, Ajaz M, Alkalamchi A, Alnaqeb D, Alwasiyah MK, Anwer N, Awwad R, Bonnefin M, Corry P, Gwanmesia L, Karbani GA, Mostafavi M, Pippucci T, Ranza-Boscardin E, Reversade B, Sharif SM, Teeuw ME, Bittles AH: Consanguineous marriages, pearls and perils: Geneva International Consanguinity Workshop Report. Genet Med 2011, 13: 841–847. 10.1097/GIM.0b013e318217477f
Porter P: “Westernizing” Women’s Risk? Breast cancer in lower-income countries. N Engl J Med 2008, 358: 213–216. 10.1056/NEJMp0708307
Toh GT, Kang P, Lee SS, Lee DS, Lee SY, Selamat S, Mohd Taib NA, Yoon SY, Yip CH, Teo SH: BRCA1 and BRCA2 Germline Mutations in Malaysian Women with Early-Onset Breast Cancer without a Family History. PLoS One 2008, 3: e2024. 10.1371/journal.pone.0002024
Kadouri L, Bercovich D, Elimelech A, Lerer I, Sagi M, Glusman G, Shochat C, Korem S, Hamburger T, Nissan A, Abu-Halaf N, Badrriyah M, Abeliovich D, Peretz T: A novel BRCA-1 mutation in Arab kindred from east Jerusalem with breast and ovarian cancer. BMC Cancer 2007, 7: 14. 10.1186/1471-2407-7-14
Dunning AM, Chiano M, Smith NR, Dearden J, Gore M, Oakes S, Wilson C, Stratton M, Peto J, Easton D, Clayton D, Ponder BA: Common BRCA1 variants and susceptibility to breast and ovarian cancer in the general population. Hum Mol Genet 1997, 6: 285–289. 10.1093/hmg/6.2.285
Freedman ML, Penney KL, Stram DO, Riley S, McKean-Cowdin R, Le Marchand L, Altshuler D, Haiman CA: A haplotype-based case–control study of BRCA1 and sporadic breast cancer risk. Cancer Res 2005, 65: 7516–7522. 10.1158/0008-5472.CAN-05-0132
Augello C, Bruno L, Bazan V, Calò V, Agnese V, Corsale S, Cascio S, Gargano G, Terrasi M, Barbera F, Fricano S, Adamo B, Valerio MR, Colucci G, Sumarcz E, Russo A: Gruppo Oncologico dell'Italia Meridionale: Y179C, F486L and N550H are BRCA1 variants that may be associated with breast cancer in a Sicilian family: results of a 5-year GOIM (Gruppo Oncologico dell'Italia Meridionale) prospective study. Ann Oncol 2006,17(7):30–33.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interest
The authors declare that they have no competing interests.
Author’s contributions
NJ, JN, EC, NS, NU, DSL, YJB, AM carried out the molecular genetic studies, participated in the sequence alignment ; DSL, YJB, AM drafted the manuscript. YJB and AM participated in the design of the study All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Jalkh, N., Nassar-Slaba, J., Chouery, E. et al. Prevalance of BRCA1 and BRCA2 mutations in familial breast cancer patients in Lebanon. Hered Cancer Clin Pract 10, 7 (2012). https://doi.org/10.1186/1897-4287-10-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1897-4287-10-7