Abstract
Background
Previous studies have suggested that DNA methylation contributes to coronary artery disease (CAD) risk variability. DNA hypermethylation at the ATP-binding cassette transporter A1 (ABCA1) gene, an important modulator of high-density lipoprotein cholesterol and reverse cholesterol transport, has been previously associated with plasma lipid levels, aging and CAD, but the association with CAD has yet to be replicated.
Results
ABCA1 DNA methylation levels were measured in leucocytes of 88 men using bis-pyrosequencing. We first showed that DNA methylation at the ABCA1 gene promoter locus is associated with aging and CAD occurrence in men (P < 0.05). The latter association is stronger among older men with CAD (≥61 years old; n = 19), who showed at least 4.7% higher ABCA1 DNA methylation levels as compared to younger men with CAD (<61 years old; n = 19) or men without CAD (n = 50; P < 0.001). Higher ABCA1 DNA methylation levels in older men were also associated with higher total cholesterol (r = 0.34, P = 0.03), low-density lipoprotein cholesterol (r = 0.32, P = 0.04) and triglyceride levels (r = 0.26, P = 0.09). Furthermore, we showed that acetylsalicylic acid therapy is associated with 3.6% lower ABCA1 DNA methylation levels (P = 0.006), independent of aging and CAD status of patients.
Conclusions
This study provides new evidence that the ABCA1 epigenetic profile is associated with CAD and aging, and highlights that epigenetic modifications might be a significant molecular mechanism involved in the pathophysiological processes associated with CAD. Acetylsalicylic acid treatment for CAD prevention might involve epigenetic mechanisms.
Similar content being viewed by others
Background
The ATP-binding cassette transporter A1 (ABCA1) catalyzes the transfer of lipids from various tissues and cells to apolipoprotein A1 containing lipoproteins[1]. This reaction is the rate-limiting step in the biogenesis of high-density lipoprotein particles and reverse cholesterol transport[1]. Mutations within the ABCA1 gene in humans are responsible for Tangier disease (OMIM: 2054000) and familial hypoalphalipoproteinemia (OMIM: 604091)[2–4]. These two genetic disorders are characterized by markedly reduced plasma high-density lipoprotein cholesterol (HDL-C) levels, the accumulation of cholesterol esters in peripheral tissues and an increased risk of coronary artery disease (CAD)[2–6].
Previous candidate gene and genome-wide studies have suggested that DNA methylation contributes to CAD risk variability[7–13]. Indeed, we have recently shown that a higher DNA methylation level at the ABCA1 gene promoter locus was associated with lower HDL-C levels and a previous history of CAD in familial hypercholesterolemia (FH)[7]. Moreover, our group and others have shown that higher ABCA1 DNA methylation levels were associated with a lower ABCA1 gene expression[14, 15]. All these previous results suggest that perturbations of the ABCA1 epigenetic profile might be a new molecular mechanism involved in CAD. However, these results have not yet been replicated.
DNA methylation is a non-traditional heritable factor occurring at cytosines located upstream of a guanine (CpG dinucleotides). It is involved in gene expression regulation[16]. This epigenetic modification is mitotically stable, and several environmental factors modulate its levels across the genome[16]. Interestingly, we recently observed that ABCA1 DNA methylation level variability in newborns is associated with maternal glycemic and HDL-C status, suggesting that the in utero environment might modulate the ABCA1 epigenetic profile[14]. Moreover, previous epigenetic studies performed by researchers from The Netherlands showed that aging and prenatal famine exposure are associated with DNA hypermethylation at the ABCA1 gene promoter locus[17, 18]. Overall, these results suggest that both the in utero and postnatal environments might modulate the ABCA1 epigenetic profile and trigger a long-term susceptibility to cardiovascular diseases (CVDs)[14, 17, 18].
Environmental cardiovascular risk factors, such as smoking, a high-fat diet and physical activity, have been previously associated with DNA methylation variability in humans[19–22]. More interestingly, statins and acetylsalicylic acid (ASA), two drugs frequently prescribed to patients with a high cardiovascular risk profile, have been shown to be associated with the induction or attenuation of epigenetic marks in vitro[23, 24]. However, no study has yet determined whether environmental cardiovascular risk factors or medication might be associated with the ABCA1 DNA methylation profile in humans.
The aims of this study were thus to replicate the association between ABCA1 DNA methylation and CAD in a non-FH population, as well as assess whether aging and environmental factors, especially tobacco smoking and medication, might be associated with ABCA1 DNA methylation in a sample of 88 French-Canadian men.
Results
Table 1 shows the characteristics of subjects according to their CAD status and median age (61 years old). We first assessed whether mean DNA methylation levels at 8 CpG dinucleotides located at the ABCA1 gene promoter locus might be associated with CAD occurrence and aging in men (Figure 1). We observed that men with a previous history of CAD (n = 38) showed higher mean ABCA1 DNA methylation levels than men without CAD (n = 50) (38.7 ± 1.2 versus 36.0 ± 1.0, P = 0.04; after consideration of age and current treatments) (Figure 1A). Moreover, older men (age ≥61 years old) had higher mean ABCA1 DNA methylation levels than younger men (age <61 years old) (38.0 ± 1.2 versus 35.2 ± 1.0, P = 0.02; after consideration of CAD occurrence and current treatments) (Figure 1B).
ABCA1 DNA methylation levels in leucocytes according to coronary artery disease occurrence and aging. Men with a previous history of coronary artery disease (CAD) (A) and older men (B) showed higher mean ATP-binding cassette transporter A1 (ABCA1) DNA methylation levels compared to men without history of CAD and younger men, respectively. aP-values were obtained after consideration of patients’ current treatments and age. bP-values were obtained after consideration of patients’ current treatments, CAD status and age.
Interestingly, we also observed a significant interaction between CAD status and age on mean ABCA1 DNA methylation levels (P = 0.01; Figure 2). Older men with CAD (age ≥61 years old; n = 19) showed significantly higher DNA methylation levels at the ABCA1 gene promoter locus compared to younger men without CAD (age <61 years old), older men without CAD (age ≥61 years old) and younger men with CAD (age <61 years old) (Table 1 and Figure 2), independently of current treatment. No significant mean ABCA1 DNA methylation level difference was observed between younger men with or without CAD (P = 0.67). In older men (age ≥61 years old), we also observed that a higher ABCA1 DNA methylation level was positively correlated with total cholesterol (r = 0.34; P = 0.03), low-density lipoprotein cholesterol (r = 0.32; P = 0.04), as well with fasting triglyceride levels (trend; r = 0.26; P = 0.09) after consideration of age, CAD status and medication. In younger men (age <61 years old), no significant association was observed between mean ABCA1 DNA methylation levels and plasma lipid profile (data not shown).
Interaction between coronary artery disease and aging on ABCA1 DNA methylation levels in leucocytes. Older men (>61 years old) with coronary artery disease (CAD) had the highest mean ATP-binding cassette transporter A1 (ABCA1) DNA methylation levels. CAD occurrence and aging interact to increase ABCA1 DNA methylation levels in leucocytes (P = 0.01). P-values were obtained after consideration of acetylsalicylic acid treatment.
Next, we further assessed whether smoking and medication were associated with the ABCA1 epigenetic profile in men. No significant association was observed between ABCA1 DNA methylation levels and tobacco smoking status (data not showed). However, we observed that subjects under ASA treatment (n = 42) had significantly lower mean DNA methylation levels at the ABCA1 gene promoter locus compared to subjects not taking ASA (n = 46), even when the statistical model was adjusted for age and CAD (Figure 3). ABCA1 DNA methylation levels measured at all eight CpGs were significantly associated with ASA treatment (all P < 0.008). No significant association was observed between ABCA1 DNA methylation levels and the use of other medications (data not shown).
Association between ABCA1 DNA methylation levels and acetylsalicylic acid therapy. Subjects on acetylsalicylic acid (ASA) therapy had lower mean ATP-binding cassette transporter A1 (ABCA1) DNA methylation levels (Δ = −3.6%) compared to subjects not on ASA therapy, independent of age and CAD. In subjects without CAD, we observed the same significant association between ASA and a lower mean ABCA1 DNA methylation level (Δ = −4.7%; P = 0.01). Although not significant, we observed that in subjects with CAD, those under an ASA treatment exhibited a similar decrease in mean ABCA1 DNA methylation levels (Δ = −3.9%; P = 0.14). aP-values were obtained after consideration of age. bP-value was obtained after consideration of age and CAD status.
Discussion
Previous work from our group and others has shown that ABCA1 DNA methylation is associated with aging, dyslipidemia and CAD[7, 18]. In the current study, we provide the first confirmatory evidence that a higher DNA methylation level at the ABCA1 gene is independently associated with CAD, an association that might be specific to older men. We also provide additional validation of the independent associations with age and a proatherogenic lipid profile. As previously described, the relationship between dyslipidemia, CAD and higher DNA methylation levels at the ABCA1 gene promoter locus (associated with a lower ABCA1 gene expression) suggests that the ABCA1-driven reverse cholesterol transport is compromised when ABCA1 DNA methylation increases[7, 14, 15]. This could lead to an increased susceptibility to CAD as seen in Tangier disease or familial hypoalphalipoproteinemia[2, 4]. However, the higher ABCA1 DNA methylation levels observed in older men with CAD could also be related to an increased severity of CAD or a longer disease history in these patients, but this relation will have to be confirmed. Nonetheless, the association between ABCA1 DNA methylation and CAD that we first observed in FH subjects has now been replicated in men with common hypercholesterolemia.
Recently, results reported by Tobi and colleagues and our group also suggested that adverse fetal environmental conditions might have persistent epigenetic consequences on the ABCA1 epigenetic profile and predispose newborns to develop obesity, diabetes and CVD[14, 17]. However, it was additionally suggested that the postnatal environment might also modulate the DNA methylome[19–22]. The increased ABCA1 DNA methylation that we observed with aging in men with CAD could thus illustrate the effects of an accumulation of exposures to pro-CAD environmental factors (such as a high-fat diet, physical inactivity or tobacco smoking) that induces ABCA1 DNA methylation variability over time and predisposes to CVD. In the current study, we did not observe a significant association between the tobacco smoking status of men and ABCA1 DNA methylation although smoking has already been associated with DNA methylation changes at other loci[19]. Unfortunately, it has not been possible to assess the association with other postnatal environmental exposures (diet or physical activity). Nevertheless, these results provide evidence in support of longitudinal and interventional studies.
Interestingly, we observed that subjects on ASA treatment had lower ABCA1 DNA methylation levels compared to those not under this non-steroidal anti-inflammatory drug (NSAID) treatment. It has been previously suggested that ASA treatment is associated with a decrease of the concentration of atherogenic lipids, an increase of HDL-C levels, as well as the variability of gene expression levels of ABCA1 and other key genes involved in reverse cholesterol transport in humans[25–28]. Previous epigenetic studies have shown that ASA and other NSAIDs might induce DNA methylation changes in humans[29–31]. Indeed, it has been suggested that ASA use might reduce the methylation rate associated with aging, especially at cancer-related genes[32]. More recently, Chang and colleagues suggested that ASA lowers the CAD risk by an epigenetic mechanism that protects against atherogenic electronegative low-density lipoprotein particles[23]. However, the mechanism by which ASA induces DNA methylation variability is still unknown, but likely involves its anti-inflammatory properties[30]. Indeed, inflammation has been known to promote de novo methylation, and ASA might therefore decrease DNA methylation levels at inflammatory-sensitive loci through its anti-inflammatory properties[30]. Based on these preliminary results and the current literature, it is tempting to speculate that ASA treatment lowers the CAD risk by reducing inflammation and ABCA1 gene DNA methylation levels, leading in turn to the stimulation of the reverse cholesterol transport. Although this hypothesis will have to be confirmed in larger cohorts before strong conclusions can be drawn, our study brings new and promising research hypotheses to the fields of lipid research and pharmacoepigenetics.
One of the strengths of the study is the use of a robust and reproducible technology for DNA methylation quantification (that is, pyrosequencing of bisulfite-treated DNA). In addition, our sample size, although small for association studies, was fairly large compared with that of other epigenetic studies. Importantly, it allowed us to take into account possible confounding factors[11–13]. A limitation of this study relates to the impossibility to infer the causal relationship observed between ABCA1 DNA methylation and CAD. Indeed, a patient’s methylome can either be modulated by CAD occurrence or be a molecular mechanism leading to CAD. Also, our study focused only on men in order to control for the possible confounding effects of sex hormones. It follows that our conclusions might not apply to women considering that the CAD risk profile might be associated with different epigenetically modulated loci in men and women. Finally, a candidate gene approach might seem less attractive compared to recent genome-wide methylation analyses[10, 33]. However, we would like to point out that the ABCA1 gene promoter locus analyzed in the current study is not assessed on the Infinium HumanMethylation27 and HumanMethylation450 BeadChips, the most widely used genome-wide methylation technologies. This clearly illustrates that candidate gene approaches are still essential, even in the current microarray era.
Conclusions
This study confirms that higher DNA methylation levels at the ABCA1 gene promoter locus are associated with aging and CAD in men. It supports the recently uncovered epigenetics-related cardioprotective effects of ASA treatment. It also provides the results of one of the few - though greatly needed - replication studies in epigenetic epidemiology. Overall, these findings could help our understanding of the molecular mechanisms involved in the pathophysiological processes leading to CAD in men and, in time, the development of new therapeutic strategies.
Methods
Sample and clinical data
This study included 88 men with (n = 38) and without CAD (n = 50) recruited from patients who underwent heart surgery at the Institut universitaire de cardiologie et de pneumologie de Québec (IUCPQ). All patients with and without CAD were caucasians (of French Canadian origin). Men with CAD were selected from patients undergoing coronary artery bypass grafting with a pre-operative angiogram showing the presence of at least one lesion of 50%. The average number of grafts per patient was 2.95 ± 1.55 (range 1 to 6). Only two patients had been diagnosed with acute coronary syndrome. Patients with a normal angiogram were selected as controls among individuals undergoing isolated valve surgery. All subjects were normoglycemic, and it was unlikely that they were FH based on their clinical history and the low prevalence of this inherited condition in the Quebec City area where they were recruited (prevalence of 1 FH subject on 208 inhabitants[34]. The subjects’ current treatments/medications (aldosterone receptor blocker, ASA, β-blocker, diuretic, calcium channel blocker, lipid-lowering drugs, angiotensin-converting-enzyme inhibitor and nitroglycerine), as well as their smoking status (current, former or never) were recorded. Patients signed an informed consent to include biological material and corresponding clinical data in our local biobank of the Centre de Recherche de l’IUCPQ (CRIUCPQ). This project received the approval of the IUCPQ ethics committee.
Fasting blood samples were obtained preoperatively. Plasma cholesterol, triglyceride and glucose concentrations were enzymatically measured using standard procedures.
Nucleic acid extraction and DNA methylation level measurement
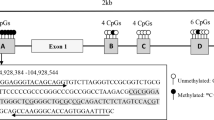
DNA was purified from buffy coat samples using the Qiagen QIAamp Blood Midi kit (Qiagen, CA, USA). ABCA1 gene promoter DNA methylation levels were measured using pyrosequencing of sodium bisulfite-treated DNA, as previously described[7]. Briefly, this gold standard technology for DNA methylation analysis is a simple, accurate quantitative sequencing assay that combines sodium bisulfite DNA conversion, PCR amplification and sequencing by synthesis (PyroMark Q24, Qiagen). PCR and sequencing primers were selected using the PyroMark Assay Design v2.0.1.15 (Qiagen), as previously described[7] (Additional file1: Figure S1). DNA methylation levels were measured at eight CpG dinucleotides upstream from the first exon of the ABCA1 gene (ABCA1-CpG1 to -CpG8; Additional file1: Figure S1). Considering that DNA methylation levels were found to be well correlated among these eight CpGs (r > 0.70; P < 0.001), a mean of ABCA1 DNA methylation levels of these eight CpG dinucleotides was thus computed for each subject and subsequently used in the statistical analyses.
Statistical analyses
The normal distribution of all variables was assessed using the Kolmogorov-Smirnov test. Only fasting triglyceride levels were not normally distributed and therefore log-10 transformed. Men were stratified into two groups according to the median age: younger men (age <61 years old) and older men (age ≥61 years old). Categorical variables were compared using a Pearson χ2-statistic, whereas mean differences between groups for continuous variables were compared with an analysis of covariance including the following potential confounding factors: age, medication and CAD status, and followed by Bonferroni’s post-hoc comparison test. Partial Pearson’s correlation was used to assess the association between ABCA1 DNA methylation levels and plasma lipid concentrations after consideration of the same confounding factors. Results were considered statistically significant when P-values were <0.05 (two sided). All statistical analyses were performed with the IBM SPSS Statistic 20 software (release 20.0.0, SPSS, International Business Machines (IBM) Corp., NY, USA).
Abbreviations
- ABCA1:
-
ATP-binding cassette transporter A1
- ASA:
-
acetylsalicylic acid
- CAD:
-
coronary artery disease
- CRIUCPQ:
-
Centre de Recherche de l’IUCPQ
- CVD:
-
cardiovascular disease
- FH:
-
familial hypercholesterolemia
- HDL-C:
-
high-density lipoprotein cholesterol
- IUCPQ:
-
Institut universitaire de cardiologie et de pneumologie de Québec
- LDL-C:
-
low-density lipoprotein cholesterol
- NSAID:
-
non-steroidal anti-inflammatory drug
- PCR:
-
polymerase chain reaction.
References
Joyce C, Freeman L, Brewer HB, Santamarina-Fojo S: Study of ABCA1 function in transgenic mice. Arterioscler Thromb Vasc Biol. 2003, 23: 965-971.
Serfaty-Lacrosniere C, Civeira F, Lanzberg A, Isaia P, Berg J, Janus ED, Smith MP, Pritchard PH, Frohlich J, Lees RS, Barnard GF, Ordovas JM, Schaefer EJ: Homozygous Tangier disease and cardiovascular disease. Atherosclerosis. 1994, 107: 85-98.
Wang J, Burnett JR, Near S, Young K, Zinman B, Hanley AJ, Connelly PW, Harris SB, Hegele RA: Common and rare ABCA1 variants affecting plasma HDL cholesterol. Arterioscler Thromb Vasc Biol. 2000, 20: 1983-1989.
Clee SM, Kastelein JJ, van Dam M, Marcil M, Roomp K, Zwarts KY, Collins JA, Roelants R, Tamasawa N, Stulc T, Suda T, Ceska R, Boucher B, Rondeau C, DeSouich C, Brooks-Wilson A, Molhuizen HO, Frohlich J, Genest J, Hayden MR: Age and residual cholesterol efflux affect HDL cholesterol levels and coronary artery disease in ABCA1 heterozygotes. J Clin Invest. 2000, 106: 1263-1270.
Frikke-Schmidt R, Nordestgaard BG, Jensen GB, Steffensen R, Tybjaerg-Hansen A: Genetic variation in ABCA1 predicts ischemic heart disease in the general population. Arterioscler Thromb Vasc Biol. 2008, 28: 180-186.
Francis GA, Knopp RH, Oram JF: Defective removal of cellular cholesterol and phospholipids by apolipoprotein A-I in Tangier disease. J Clin Invest. 1995, 96: 78-87.
Guay SP, Brisson D, Munger J, Lamarche B, Gaudet D, Bouchard L: ABCA1 gene promoter DNA methylation is associated with HDL particle profile and coronary artery disease in familial hypercholesterolemia. Epigenetics. 2012, 7: 464-472.
Guay SP, Brisson D, Lamarche B, Gaudet D, Bouchard L: Epipolymorphisms within lipoprotein genes contribute independently to plasma lipid levels in familial hypercholesterolemia. Epigenetics. 2014, 9: 718-729.
Breton CV, Park C, Siegmund K, Gauderman WJ, Whitfield-Maxwell L, Hodis HN, Avol E, Gilliland FD: NOS1 methylation and carotid artery intima-media thickness in children. Circ Cardiovasc Genet. 2014, 7: 116-122.
Sharma P, Garg G, Kumar A, Mohammad F, Kumar SR, Tanwar VS, Sati S, Sharma A, Karthikeyan G, Brahmachari V, Sengupta S: Genome wide DNA methylation profiling for epigenetic alteration in coronary artery disease patients. Gene. 2014, 541: 31-40.
Zhuang J, Peng W, Li H, Wang W, Wei Y, Li W, Xu Y: Methylation of p15INK4b and expression of ANRIL on chromosome 9p21 are associated with coronary artery disease. PLoS One. 2012, 7: e47193-
Jiang D, Zheng D, Wang L, Huang Y, Liu H, Xu L, Liao Q, Liu P, Shi X, Wang Z, Sun L, Zhou Q, Li N, Le Y, Ye M, Shao G, Duan S: Elevated PLA2G7 gene promoter methylation as a gender-specific marker of aging increases the risk of coronary heart disease in females. PLoS One. 2013, 8: e59752-
Xu L, Zheng D, Wang L, Jiang D, Liu H, Liao Q, Zhang L, Liu P, Shi X, Wang Z, Sun L, Zhou Q, Li N, Huang Y, Le Y, Ye M, Shao G, Duan S: GCK gene-body hypomethylation is associated with the risk of coronary heart disease. Biomed Res Int. 2014, 2014: 151723-
Houde AA, Guay SP, Desgagne V, Hivert MF, Baillargeon JP, St-Pierre J, Perron P, Gaudet D, Brisson D, Bouchard L: Adaptations of placental and cord blood ABCA1 DNA methylation profile to maternal metabolic status. Epigenetics. 2013, 8: 1289-1302.
Liang Y, Yang X, Ma L, Cai X, Wang L, Yang C, Li G, Zhang M, Sun W, Jiang Y: Homocysteine-mediated cholesterol efflux via ABCA1 and ACAT1 DNA methylation in THP-1 monocyte-derived foam cells. Acta Biochim Biophys Sin (Shanghai). 2013, 45: 220-228.
Bird A: DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16: 6-21.
Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, Slagboom PE, Heijmans BT: DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009, 18: 4046-4053.
Talens RP, Christensen K, Putter H, Willemsen G, Christiansen L, Kremer D, Suchiman HE, Slagboom PE, Boomsma DI, Heijmans BT: Epigenetic variation during the adult lifespan: cross-sectional and longitudinal data on monozygotic twin pairs. Aging Cell. 2012, 11: 694-703.
Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H: Tobacco-smoking-related differential DNA methylation: 27 K discovery and replication. Am J Hum Genet. 2011, 88: 450-457.
Barres R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, Caidahl K, Krook A, O'Gorman DJ, Zierath JR: Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012, 15: 405-411.
Ronn T, Volkov P, Davegardh C, Dayeh T, Hall E, Olsson AH, Nilsson E, Tornberg A, Dekker Nitert M, Eriksson KF, Jones HA, Groop L, Ling C: A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013, 9: e1003572-
Jacobsen SC, Gillberg L, Bork-Jensen J, Ribel-Madsen R, Lara E, Calvanese V, Ling C, Fernandez AF, Fraga MF, Poulsen P, Brons C, Vaag A: Young men with low birthweight exhibit decreased plasticity of genome-wide muscle DNA methylation by high-fat overfeeding. Diabetologia. 2014, 57: 1154-1158.
Chang PY, Chen YJ, Chang FH, Lu J, Huang WH, Yang TC, Lee YT, Chang SF, Lu SC, Chen CH: Aspirin protects human coronary artery endothelial cells against atherogenic electronegative LDL via an epigenetic mechanism: a novel cytoprotective role of aspirin in acute myocardial infarction. Cardiovasc Res. 2013, 99: 137-145.
Kodach LL, Jacobs RJ, Voorneveld PW, Wildenberg ME, Verspaget HW, van Wezel T, Morreau H, Hommes DW, Peppelenbosch MP, van den Brink GR, Hardwick JC: Statins augment the chemosensitivity of colorectal cancer cells inducing epigenetic reprogramming and reducing colorectal cancer cell ‘stemness’ via the bone morphogenetic protein pathway. Gut. 2011, 60: 1544-1553.
Vinals M, Bermudez I, Llaverias G, Alegret M, Sanchez RM, Vazquez-Carrera M, Laguna JC: Aspirin increases CD36, SR-BI, and ABCA1 expression in human THP-1 macrophages. Cardiovasc Res. 2005, 66: 141-149.
Ranga GS, Kalra OP, Tandon H, Gambhir JK, Mehrotra G: Effect of aspirin on lipoprotein (a) in patients with ischemic stroke. J Stroke Cerebrovasc Dis. 2007, 16: 220-224.
Sethi A, Parmar HS, Kumar A: The effect of aspirin on atherogenic diet-induced diabetes mellitus. Basic Clin Pharmacol Toxicol. 2011, 108: 371-377.
Charan J, Goyal JP, Saxena D: Effect of Pollypill on cardiovascular parameters: systematic review and meta-analysis. J Cardiovasc Dis Res. 2013, 4: 92-97.
Pereira MA, Tao L, Wang W, Li Y, Umar A, Steele VE, Lubet RA: Modulation by celecoxib and difluoromethylornithine of the methylation of DNA and the estrogen receptor-alpha gene in rat colon tumors. Carcinogenesis. 2004, 25: 1917-1923.
Tahara T, Shibata T, Nakamura M, Yamashita H, Yoshioka D, Okubo M, Maruyama N, Kamano T, Kamiya Y, Fujita H, Nagasaka M, Iwata M, Takahama K, Watanabe M, Hirata I, Arisawa T: Chronic aspirin use suppresses CDH1 methylation in human gastric mucosa. Dig Dis Sci. 2010, 55: 54-59.
Cheong HS, Park SM, Kim MO, Park JS, Lee JY, Byun JY, Park BL, Shin HD, Park CS: Genome-wide methylation profile of nasal polyps: relation to aspirin hypersensitivity in asthmatics. Allergy. 2011, 66: 637-644.
Noreen F, Röösli M, Gai P, Pietrzak J, Weis S, Urfer P, Regula J, Schär P, Truninger K: Modulation of age- and cancer-associated DNA methylation change in the healthy colon by aspirin and lifestyle. J Natl Cancer Inst. 2014, 106:
Guay SP, Voisin G, Brisson D, Munger J, Lamarche B, Gaudet D, Bouchard L: Epigenome-wide analysis in familial hypercholesterolemia identified new loci associated with high-density lipoprotein cholesterol concentration. Epigenomics. 2012, 4: 623-639.
Nordestgaard BG, Chapman MJ, Humphries SE, Ginsberg HN, Masana L, Descamps OS, Wiklund O, Hegele RA, Raal FJ, Defesche JC, Wiegman A, Santos RD, Watts GF, Parhofer KG, Hovingh GK, Kovanen PT, Boileau C, Averna M, Boren J, Bruckert E, Catapano AL, Kuivenhoven JA, Pajukanta P, Ray K, Stalenhoef AF, Stroes E, Taskinen MR, Tybjaerg-Hansen A, European Atherosclerosis Society Consensus Panel: Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur Heart J. 2013, 34: 3478-3490.
Acknowledgments
The authors are thankful to all participants and the staff of the ECOGENE-21 Laboratory and Clinical Research Center and CRIUCPQ. We particularly acknowledge the contribution of Céline Bélanger, Christine Racine and Nathalie Gaudreault for their dedicated work. S-PG was recipient of a doctoral research award from the Canadian Institutes for Health and Research (CIHR). CL was recipient of a financial support from the Faculté de médecine et des sciences de la santé (FMSS) de l’Université de Sherbrooke. A-AH was recipient of a Fonds de recherche du Québec - Santé (FRQS) doctoral training award and was supported by Diabète Québec. YB is the recipient of a Junior 2 Research Scholar award from the FRQS. PM is a senior research scholar from the FRQS. LB is a junior research scholar from the FRQS and a member of the FRQS-funded Centre de recherche clinique du CHUS.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
S-PG conceived the study design, participated in the data collection, performed the data analysis/interpretation and wrote the manuscript. CL performed data collection and revised the manuscript. A-AH contributed to data analysis/interpretation and revised the manuscript. YB and PM participated in the data collection, analysis and interpretation and revised the manuscript. LB conceived the study design, participated in the data analysis/interpretation process and revised the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
13148_2014_81_MOESM1_ESM.tiff
Additional file 1: Figure S1:ABCA1 gene promoter locus. The primary DNA sequence was numbered relative to the first ABCA1 codon in exon 2 (Ensembl release 61 [February 2011]). The first exon of ABCA1 is in red and bold type. The epigenotyped region is shown in green. Arrows indicate both PCR primer sequences. The analysed CpG dinucleotides have been numbered relative to the 5’ of the amplicon. (TIFF 159 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Guay, SP., Légaré, C., Houde, AA. et al. Acetylsalicylic acid, aging and coronary artery disease are associated with ABCA1 DNA methylation in men. Clin Epigenet 6, 14 (2014). https://doi.org/10.1186/1868-7083-6-14
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1868-7083-6-14