Abstract
Background
Cancer-specific hypermethylation of (promoter) CpG islands is common during the tumorigenesis of colon cancer. Although associations between certain genetic aberrations, such as BRAF mutation and microsatellite instability, and the CpG island methylator phenotype (CIMP), have been found, the mechanisms by which these associations are established are still unclear. We studied genome-wide DNA methylation differences between colorectal tumors carrying a BRAF mutation and BRAF wildtype tumors.
Results
Using differential methylation hybridization on oligonucleotide microarrays representing 32,171 CpG-rich regions, we identified 1,770 regions with differential methylation between colorectal tumor and paired normal colon. Next, we compared the tumor/normal methylation ratios between different groups of patients. Related to CIMP, we identified 749 differentially methylated regions, of which 86% had a higher tumor/normal methylation ratio in the CIMP-positive group. We identified 758 regions with a BRAF mutation-specific methylation change, of which 96% had a higher tumor/normal methylation ratio in the BRAF mutant group. Among the genes affected by BRAF mutation-specific methylation changes, we found enrichment of several cancer-related pathways, including the PI3 kinase and Wnt signaling pathways. To focus on genes that are silenced in a tumor-specific rather than a lineage-specific manner, we used information on the epigenetic silencing mark H3K27me3 in embryonic stem (ES) cells. Among the genes showing BRAF mutation-specific promoter methylation but no H3K27me3 mark in ES cells were forkhead box (FOX) transcription factors associated with the PI3 kinase pathway, as well as MLH1 and SMO. Repression of FOXD3 gene expression in tumors could be related to its promoter hypermethylation.
Conclusions
We identified new BRAF mutation-specific methylation changes in colorectal cancer. Epigenetic downregulation of these targets may contribute to mutationally active BRAF-driven tumorigenesis, explaining its association with aberrant DNA methylation.
Similar content being viewed by others
Background
The CpG island methylator phenotype (CIMP) was introduced in 1999 by Toyota et al. to describe a subset of colorectal tumors with high levels of cancer-specific methylation [1]. Subsequent studies regarding CIMP in colon cancer described a strong association between this epigenetic phenotype, BRAF mutations, and microsatellite instability (MSI) [2–8]. As sporadic MSI colon cancer is caused by promoter methylation of a mismatch repair gene (MLH1, MSH2, or MSH6) the association between MSI and the high levels of DNA methylation in CIMP is considered a causative one [9, 10]. However, the association between activating BRAF mutations and CIMP remains unclear.
The field of epigenetic research has progressed from a candidate-gene to a genome-wide approach, which not only provides a plethora of new candidate targets of cancer-specific DNA methylation but also a better understanding of transcription regulation by DNA methylation [11]. Using such genome-wide DNA methylation approaches could help to identify new targets of BRAF mutation-specific promoter methylation. Hinoue et al. [2] examined the CIMP- and BRAF mutation-specific methylation status of 1,505 CpG sites, located at 807 genes, in 235 primary colorectal tumors and discovered specific methylation of genes mediating various signaling pathways involved in colon cancer tumorigenesis. In this study, we screened 32,171 CpG sites located at 10,537 genes in a selected cohort of 19 patients with right-sided colon cancer to obtain additional insight into the association between BRAF mutations and DNA methylation in colon cancer tumorigenesis. Recent studies have described a gradual increase in CIMP and BRAF mutation prevalence from the rectum to the ascending colon [12, 13]. To avoid tumor location as a factor that could possibly influence the levels of methylation, we specifically studied tumors originating from the ascending colon and cecum. The frequency of BRAF mutations in the CIMP-positive patients was comparable to those previously described in larger cohort studies [2, 8, 14, 15].
Recent publications have reported a possible pre-marking of cancer-specific hypermethylated genes by the inactivation mark histone H3 lysine 27 trimethylation (H3K27me3) and binding of the polycomb group member SUZ12 in both ES cells and differentiated normal colon mucosal tissue [16–18]. These studies led to the suggestion that colon cancer cells utilize a pre-existing repression program to target loci for cancer-specific promoter methylation [16, 18–20]. However, the presence of such repressive histone modifications at promoters during differentiation from ES to normal colon epithelium suggests that the associated genes are at a transcriptional silent state prior to tumor formation, reducing the relevance of the DNA methylation of pre-marked genes to tumorigenesis. In an attempt to identify biologically relevant BRAF mutation-specific promoter methylation, we excluded loci with H3K27me3 pre-marking in ES cells from the functional pathway analyses. By both extending the number of screened loci and filtering out pre-marked genes, we identified new targets of BRAF mutation-specific methylation that could either create a favorable setting for the acquisition of BRAF mutations or function as an addition to up-regulation of the RAS-RAF-MEK pathway.
Results
Colon cancer-specific CpG island methylation
We identified 1,770 CpG-rich regions with significant methylation differences between tumor and paired normal colon. Of these, 1,234 differentially methylated regions were associated with 816 genes, of which 531 were localized to gene promoters (Additional file 1). As expected, CpG islands were mostly found hypermethylated in tumors (78.8%) [11].
We compared our results with those of Irizarry et al. [11], who described 2,707 cancer-specific differentially methylated regions based on the comparison of 13 colorectal cancer tumor-normal pairs. Of the described differentially methylated regions, 1,203 overlapped with our CpG island array regions, of which 282 (23%) were also differentially methylated between tumor and normal in our analysis. This overlap is reasonable, considering the different, modest-sized patient groups, and different experimental approaches.
CIMP-specific methylation
Next, we compared the tumor/normal methylation ratios between different groups of patients. Between CIMP-positive (n = 11) and CIMP-negative (n = 8) patients, 749 CpG-rich regions showed methylation changes, of which 85.6% had a higher tumor/normal methylation ratio in the CIMP-positive group. Of these differentially methylated regions, 589 were associated with 508 genes, of which 244 were localized to gene promoters (Additional file 2). In 8 out of 11 CIMP-positive tumors, promoter methylation of MLH1, the cause of microsatellite instability in sporadic colon cancer, was observed, which was consistent with methylation-specific PCR (Additional file 3). We conclude that the hypermethylation in specific genomic regions used to define CIMP [6] is associated with methylation changes throughout the genome.
BRAF mutation-specific methylation
Activating BRAF mutations have been associated with high levels of CpG island methylation and MSI in colon cancer [2–8]. To investigate this association, we compared the tumor/normal methylation ratio profiles of BRAF wildtypes (n = 11) with those containing the BRAFV600E mutation (n = 8). We identified 758 regions with a BRAF mutation-specific methylation change, of which 96.3% had a higher tumor/normal methylation ratio in the BRAF mutant group. Out of these 758 regions, 579 were associated with 479 genes, of which 229 were localized to gene promoters (Additional file 4).
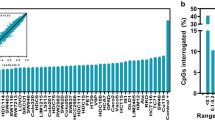
Since BRAF mutations and CIMP co-occurred in eight samples, as expected from other studies [8, 14, 15], there was a high level of overlap between CIMP- and BRAF mutation-specific methylation changes (Figure 1A). Comparable levels of overlap were found, focusing on promoter regions only (data not shown).
Regions with methylation changes are enriched for inactive chromatin marks
Regions binding the polycomb repressor complex 2 (PRC2) component SUZ12 in ES cells were found to be enriched among the loci differentially methylated between colon cancer and normal colon (Table 1). The histone mark H3K27me3 is mediated by the PRC2 complex [21], and the two marks have been reported to be highly correlated [17]. Enrichment of ES cell H3K27me3 binding regions among the regions with colon cancer-associated methylation changes was, therefore, expected and was indeed observed. Similarly, regions with CIMP- and BRAF mutation-associated differential methylation changes were also highly enriched for regions binding SUZ12 and H3K27me3 in ES cells (Table 1). Additionally, sites binding CTCF and the active chromatin mark H3K4me3 were underrepresented among the differentially methylated regions. Interestingly, although all colon cancer-, CIMP-, and BRAF mutation-specific differentially methylated regions are underrepresented for H3K4me3, this depletion is most evident for BRAF mutation-specific regions.
After exclusion of regions with H3K27me3 pre-marking in ES cells, the overlap between CIMP- and BRAF mutation-specific methylation changes for all loci (not shown) and promoters (Figure 1B) remained highly significant. Despite this high level of overlap, approximately 50% of BRAF mutation-specific methylation changes showed no overlap with CIMP. In our functional analysis, we focused on all promoter regions with BRAF mutation-specific methylation changes, regardless of overlap with CIMP.
BRAF mutation-associated methylation pathway analysis
To identify biological pathways affected by BRAF mutation-associated gene methylation, we used 186 promoter regions that did not bind H3K27me3 in ES cells representing 125 genes after exclusion of duplicates and annotation by Panther 6.0. We found five significantly enriched pathways (P < 0.01) containing 13 unique genes (Table 2).
With seven genes, the Wnt pathway contained the most BRAF mutation-specific methylation changes (Table 2). However, the tumor/normal log2 ratios (Figure 2) of AXIN1, CREBBP, GSK3A, and NKD2 in the BRAF wildtype samples were low (−0.26 median, 0.12 standard deviation) compared with those in the BRAF mutated samples (−0.02 median, 0.12 standard deviation). While this could indicate tumor hypomethylation in BRAF wildtype samples compared with normal and BRAF mutated samples, the high level of chromosomal instability among BRAF wildtype samples suggests that copy-number loss is the most plausible explanation. To filter for this phenomenon, we excluded regions with a log2 ratio below one standard deviation of the median log2 ratio of all BRAF mutation-specific regions in the BRAF wildtype group. A significant increase in the BRAF mutant log2 ratios, compared with those of the BRAF wildtypes, indicates BRAF mutation-specific hypermethylation in these colon cancer samples (Figure 2). After filtering out copy-number alterations, nine of the pathway-associated genes remained (SMO, FOXB1, FOXB2, FOXD3, CCND1, GNG4, LEF1, MTERF, TAF7) and the PI3 kinase pathway was the only statistically significant enriched (P = 0.005) pathway. Interestingly, besides promoter methylation of PI3 kinase pathway-associated forkhead box (FOX) genes, we identified promoter methylation of three other FOX transcription factors: FOXA1, FOXC1, and FOXF1. However, these promoters were bound by H3K27me3 and were excluded from our pathway analysis.
Scatter plots for nine unique genes with BRAF mutation-specific promoter methylation causing pathway enrichment. The y axis represents the tumor/normal log2 ratio for the median probe per region. Sample IDs are given below the x axis with BRAF wildtypes on the left of the black line and BRAF mutants on the right. Median log2 ratios and standard deviations (dotted lines) for the BRAF wildtype and BRAF mutant groups are given in dark green.
Validation of promoter methylation of FOX genes
FOX gene promoter hypermethylation in patient samples was validated by bisulfite sequencing analysis (BSA). For FOXB2 and FOXF1, which were found to be hypermethylated in BRAF mutated tumors compared with BRAF wildtypes, DNA methylation was validated using BSA. Bisulfite sequencing analysis was also attempted for FOXD3 but was unsuccessful, possibly as a result of the high guanine-cytosine (GC) content [22]. For both promoters, hypermethylation in BRAF mutant tumors was confirmed (Figure 3A and B). Methylation levels in normal tissue were below 8% and subtracted from the methylation levels in the corresponding tumors. The average methylation per sample for BRAF mutated tumors was significantly higher than that in BRAF wildtype tumors (Figure 3C, FOXB2 P = 0.0075, FOXF1 P = 0.001).
Tumor hypermethylation of FOXB2 and FOXF1.(A, B) Bisulfite sequence results for 12 BRAF wildtype tumors and 8 BRAF mutant tumors, separated by the horizontal red line. Schematic display of the BSA region with CpG locations therein shown on top (blue). Sample IDs are given on the 5' end of the BSA region. Each circle depicts a CpG. Color code: gray = ND, white = 0% to 20%, yellow = 20% to 40%, orange = 40% to 60%, red > 60% methylation. (C) Boxplot showing average methylation percentage over all detected CpGs in BRAF wildtype and BRAFV600E tumors for FOXB2 (left) and FOXF1 (right). The box indicates the 25 and 75 percentiles, with the median shown in bold. Upper and lower outliers are shown as red and purple asterisks, respectively. (D) Schematic display of FOXB2 (top) and FOXF1 (bottom) loci depicting the location of the BSA regions in relation to MseI fragments detected on the Agilent array (significant fragments marked red, not significant as black). CpG islands are shown in green, and the BSA region is shown in blue.
DNA methylation and gene expression
For a subset of nine tumor-normal pairs, the expression of FOXB2, FOXD3, and FOXF1 was determined using real-time reverse-transcription PCR (RT-qPCR). Only two tumors (both BRAF wildtype) had detectable levels of FOXB2 expression, and an additional three samples showed no detectable FOXB2 expression in either tumor or normal tissue. FOXD3 expression was detected in all but one of the normal tissue samples, and in three BRAF wildtype tumor samples. A decrease in FOXF1 expression was observed in all tumors except for sample 57. Next, we compared the tumor/normal expression ratios with the methylation measured by BSA (FOXB2 and FOXF1) or array (FOXD3) (Figure 4). FOXB2 showed loss of expression in tumors, independent of methylation status. Expression of FOXF1 was repressed in all tumors. In BRAF mutated tumors there appeared to be a negative correlation between DNA methylation and expression. The tumors with FOXD3 hypermethylation showed no detectable FOXD3 expression, suggesting methylation-related silencing.
Relation between methylation and expression. Tumor/normal expression ratios (y axis) were plotted against the average DNA methylation measured by bisulfite sequence analysis for FOXB2 (A) and FOXF1 (B), and the methylation log2 ratio measured by CpG array for FOXD3 (C). BRAF mutant samples are shown in red, wildtype samples in black.
Discussion
In this study, we extended the number of screened CpG loci compared with previous studies performed in context of BRAF mutations to identify new BRAF mutation-specific methylation changes in colorectal cancer. We validated hypermethylation of forkhead box transcription factors FOXB2 and FOXF1 in BRAFV600E tumors. Additionally, repression of FOXD3 gene expression in tumors could be related to promoter hypermethylation.
The association between DNA methylation and activating BRAF mutations in colon cancer has been identified in several studies [2–8]. Here, we attempted to identify additional targets of BRAF mutation-specific DNA methylation that could provide a favorable context, either to obtain a BRAF mutation or to attain the full potential of RAS-RAF-MEK-induced proliferation provided by this activating mutation. Identified targets of promoter methylation showing pre-marking by H3K27me3 in ES cells were excluded, to filter out methylation changes with minimal expected effects on transcription and thereby tumorigenesis [17]. We showed high levels of overlap between CIMP- and BRAF mutation-specific methylation changes, which remained after filtering out pre-marked loci. Although Rada-Iglesias et al. [17] showed a higher pre-marking of colon cancer-specific DNA methylation by H3K27me3 binding in normal colon epithelium compared with ES cells, we were restricted to using ES cell data, owing to the incompatibility between data formats in our analyses. Interestingly, the promoter region of MLH1, found methylated in both a CIMP- and BRAF mutation-specific manner, was not filtered out. Therefore, MLH1 promoter methylation, the cause of sporadic MSI colon cancer, is not established through utilization of a pre-existing repressive program in ES cells.
The study by Hinoue et al. [2] described BRAF mutation-specific DNA methylation of 60 genes in a comparison of 1,505 CpG sites between 33 BRAF mutated tumors and 202 BRAF wildtype tumors. The identification of promoter methylation of the mediator of BRAFV600E-induced senescence, IGFBP7, led them to suggest that this epigenetic silencing provides a favorable context for the acquisition of BRAF mutations [2, 23]. Despite differences in experimental techniques and coverage, 10 genes overlapped with our set of BRAF mutation-specific methylated regions, including the RAS-RAF hyperactivation-associated BMP3, receptor kinases EPHA3 and FLT3 as well as the hedgehog signaling protein SMO. However, no overlap was found for the mediator of RAS-RAF oncogene-induced senescence, IGFBP7, despite coverage of the IGFBP7 promoter CpG island with two fragments in our assay. Lack of overlap between these studies may be a consequence of different experimental methods as well as of different patient cohorts. Additionally, BMP3 and EPHA3 were pre-marked by H3K27me3 in our analysis suggesting minimal impact on gene expression and tumorigenesis.
We initially identified enrichment of five cancer-associated pathways by BRAF mutation-specific promoter methylation of nine unique genes. Our analysis took into account copy-number changes and filtered for this, as this could improve the reproducibility of differential methylation hybridization (DMH) based assays [24, 25]. After exclusion of these loci, the PI3 kinase pathway was the only pathway enriched in our analysis. Among the four genes enriched in this pathway were the FOX transcription factors FOXD3, FOXB1, and FOXB2. A recent study described FOXD3 as a TP53 and CDKN1A/p21cip1-dependent negative cell cycle regulator, which is suppressed by activated BRAF in melanoma cells [26]. Downregulation of FOXD3 levels by promoter methylation in colon cancer might provide a favorable setting for either acquisition of a BRAF mutation or proliferation by RAS-RAF-MEK over-activation, similar to IGFBP7[2].
Interestingly, the FOXO transcription factors have also been described as mediators of CDKN2A/p21cip1-dependent BRAF-induced senescence, indicating that multiple FOX genes are involved in this process [27]. We identified additional FOX genes with BRAF mutation-specific promoter methylation that were excluded from the pathway analysis as they were pre-marked by H3K27me3 in ES cells: FOXA1, FOXC1, and FOXF1. However, the promoters of these genes were also pre-marked with H3K4me3 indicating possible tissue-specific expression. All three are targets of inactivation in breast cancer and both FOXC1 and FOXF1 are subjected to promoter methylation [28–30]. Most intriguing is the description of FOXF1 as an inducer of G1-S and S-G2 cell cycle arrest, indicating a possible role in oncogene-induced senescence in breast cancer [29]. Our finding that FOXF1 was downregulated in the BRAF mutated cohort suggests that this gene might also play a role in oncogene-induced senescence in colon cancer. However, additional research is required to determine the role of these FOX genes in colon cancer-associated oncogene-induced senescence and the impact of their promoter methylation on this mechanism. Finally, research into the sequence of such events is required to provide a better insight in the association between activating BRAF mutations and DNA methylation in colon cancer.
Conclusions
In this study, we identified BRAF mutation-specific hypermethylation of CpG regions by DMH on high-density oligonucleotide microarrays. We found enrichment of several cancer-related pathways, including the PI3 kinase and Wnt signaling pathways. We validated differential methylation of forkhead box (FOX) transcription factors and found methylation-dependent silencing of FOXD3. By both extending the number of screened loci and filtering out genes pre-marked by H3K27me3, we identified new targets of BRAF mutation-specific methylation that could provide a favorable context to either obtain a BRAF mutation or to attain the full potential of this activating mutation.
Availability of supporting data
The data set supporting the results of this article is available in the Gene Expression Omnibus (GEO) repository under accession number GSE39334.
Materials and methods
Patient material
From anonymized tumor and normal fresh-frozen colon mucosa samples obtained from patients who underwent surgery between 2002 and 2005 at the Leiden University Medical Center (Leiden, The Netherlands) or at the Rijnland Hospital (Leiderdorp, The Netherlands), a cohort containing a high number of CIMP-positive patients was selected. Age, sex, histology, microsatellite instability, and BRAFV600E status for the 19 patients used for the array profiling are listed in Additional file 3. Prior to DNA isolation, frozen sections were micro-dissected to minimize the presence of normal epithelium and stromal cells. To correct for age-dependent methylation, we used normal mucosa, distant from the tumor, from the same individuals. The patients’ DNA was isolated by phenol and chloroform extraction and ethanol precipitation from 10 to 20 sections of 30 μm. This process yielded 10 to 50 μg of DNA. This study was approved by the Medical Ethics committee of the LUMC (protocol P01-019). Cases were analyzed following the medical ethical guidelines described in the Code Proper Secondary Use of Human Tissue established by the Dutch Federation of Medical Sciences.
BRAF mutation analysis
BRAFV600E mutations were detected using flanking primers that have been previously described [31]. The products of the PCR were purified with the QIAquick PCR Purification kit (#28106, Qiagen). Sequencing was performed at the Leiden Genome Technology Center (LGTC, Leiden, The Netherlands) using an ABI 3730 xl (Applied Biosystems). Mutational analysis was performed using Mutation Surveyor (SoftGenetics LLC). Results are summarized in Additional file 3.
Array hybridization
Differential methylation hybridization was performed according to Yan et al. [25] DNA (500 ng) was digested with MseI, ligated to linkers, and sequentially digested with two methylation-sensitive restriction enzymes (HpaII #R0171 and BstUI #R0518, New England Biolabs). Digested linker-ligated DNA was used as a template for PCR amplification (20 cycles) and coupled to fluorescent dyes. Cy5- or Cy3-labeled amplicons, representing methylated DNA fragments derived from tumor and normal samples, were co-hybridized to the Agilent 244 k human CpG island microarrays (#G4492A, Agilent Technologies) in a dye-swap setup. Detection was done on a G2565BA scanner (Agilent Technologies) and feature extraction using Feature Extraction Software version 9.5.3.1 (Agilent Technologies).
Array data analysis
Non-background corrected data were preprocessed by within-array LOESS normalization followed by between-array aquantile normalization using limma v3.2.1 [32] in R2.10.0 [33]. Data were corrected for gene-specific dye bias using R package dyebias v1.4.0 [34]. Raw data and preprocessed log2 ratios (tumor versus normal) per probe are available via the Gene Expression Omnibus (GEO) under accession number GSE39334. Probes mapping to the same MseI fragment were expected to show similar hybridization patterns and not to be independent. Therefore, we mapped probes to the human genome (UCSC assembly March 2006) cut in silico with MseI. Fragments of 150 to 3,000 bp mapping at least one complete probe and containing at least one BstUI or HpaII restriction site (n = 32,171) were selected. In total, 195,625 of the 244,000 array probes (80.2%) mapped to such informative fragments, mostly with 1 or 2 probes per fragment, up to 33. For statistical analysis and visualization, the median log ratio per fragment was used to represent the fragment. Methylation differences between tumor and normal samples and tumor subgroups were analyzed using a linear model in limma v3.2.1 [32]. The obtained P values were corrected for multiple testing [35] and fragments with a false discovery rate ≤0.01 were selected as significantly differentially methylated regions.
MLH1 and CIMP marker methylation
DNA samples (500 ng) were bisulfite converted using the EZ DNA methylation Gold kit (#D5006 Zymo Research). For validation of methylation changes, we performed a methylation-specific PCR on the MLH1 promoter using primers previously described (Additional file 3) [36]. Methylation of previously described CIMP markers: MINT1, MINT2, MINT12, MINT31, PRDM2/RIZ1, and TIMP3 were determined by methylation-specific PCR, while MINT27 and LRP2/megalin methylation were determined by Combined Bisulfite Restriction Analysis [6, 37]. Using the criteria suggested by Shen et al. [6, 37] including these methylation markers and mutation status of BRAF, KRAS, and TP53, tumors were determined to be CIMP-positive when two or more CIMP1 markers (BRAF mutation, methylated MLH1, TIMP3, MINT1, PRDM2/RIZ1), or three or more CIMP2 markers (KRAS mutation, methylated MINT27, MINT2, MINT31, LRP2/megalin) were present. Tumors were called CIMP-negative when two or more CIMP-negative markers (TP53 mutation, unmethylated MINT27, MINT2, MINT31, MINT1) were present. This CIMP marker set was previously validated with the CIMP loci proposed by Weisenberger et al. [8, 38]. Amplifications were carried out in a DNA Engine Dyad Peltier Thermal Cycler (Bio-Rad) using AmpliTaq Gold PCR buffer and enzyme (#4317742 Invitrogen). Amplified bands were visualized on a 2% agarose gel.
Bisulfite sequencing analysis
Bisulfite conversion was performed on 200 ng of DNA using the EZ DNA methylation Gold kit and eluted in 15 μl Milli-Q purified water. The PCR amplification was performed using primers designed using MethPrimer (Additional file 5) [39]. The PCR reaction mixture contained 1× iQ SYBR green supermix (#170-8884, Bio-Rad), 1 μl of the bisulfite-converted DNA and 5 nmol of forward and reverse primer. The PCR products were purified using the MinElute® 96 UF PCR Purification kit (#28051, Qiagen) and sent for sequencing at Macrogen (Macrogen, Europe). Sequence alignment and quantification of methylation was performed using ESME (Epigenomics Inc.) [40]. Statistical significance of hypermethylation in BRAFV600E tumors was determined using a one-sided Mann–Whitney test.
Gene expression analysis
Gene expression of FOXB2, FOXD3, and FOXF1 was determined in nine pairs of tumor and normal tissue of which RNA was available. cDNA was synthesized using 1 to 2 μg RNA, 50 ng oligo-dT, 1.6 μg random primer, 1 mM dNTPs, 5U AMV-RT transcriptase, and 10 U RNasin (#M5108 and #N2615, Promega). Gene expression was determined using TaqMan Gene Expression Assays (Applied Biosystems). Gene expression was normalized with CPSF6 and HNRNP[41], which were amplified using 0.8 pmol forward and reverse primer in a 1× iQ SYBR green supermix (#170-8884, Bio-Rad). For FOXB1, FOXB2, and FOXD3, 2 μl of 125× diluted cDNA was amplified in a mix containing 1× iQ supermix (#170-8862, Bio-Rad) and 1× TaqMan assay (#Hs00247213_s1, #Hs02386300_s1, Hs00255287_s1, Hs00230962_m1, Applied Biosystems). All qPCRs were performed in duplicate.
Exploratory data analysis
Differentially methylated regions were compared with publicly available data containing chromosomal regions identified in chromatin immunoprecipitation using antibodies against H3K27me3, H3K4me3, CTCF, and SUZ12 in ES cells followed by high-throughput sequencing [42–44]. By using the sqldf R package (version 0.3-5), we determined overlap of at least 20 bp between CpG regions represented on the array and these regions. Enrichment of chromatin domains among the differentially methylated regions was calculated by χ-squared test. Functional annotation clustering was performed in Panther 6.0 [45]. Filtering of the differential methylation datasets by H3K27me3 in ES cells using the dataset from Zhao et al. [44] was performed in R using the sqldf package.
Abbreviations
- BSA:
-
Bisulfite sequencing analysis
- CIMP:
-
CpG island methylator phenotype
- DMH:
-
Differential methylation hybridization
- ES cells:
-
Embryonic stem cells
- FOX:
-
Forkhead box
- H3K27me3:
-
Histone H3 lysine 27 trimethylation inactivation mark
- MSI:
-
Microsatellite instability
- PCR:
-
Polymerase chain reaction
- RT-qPCR:
-
Real-time reverse-transcription PCR.
References
Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP: CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999, 96: 8681-8686.
Hinoue T, Weisenberger DJ, Pan F, Campan M, Kim M, Young J, Whitehall VL, Leggett BA, Laird PW: Analysis of the association between CIMP and BRAFV600E in colorectal cancer by DNA methylation profiling. PLoS One. 2009, 4: e8357-
Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, Tanaka N, Higuchi T, Young J, Jass JR, Leggett BA: BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004, 53: 1137-1144.
Minoo P, Moyer MP, Jass JR: Role of BRAF-V600E in the serrated pathway of colorectal tumourigenesis. J Pathol. 2007, 212: 124-133.
Nagasaka T, Koi M, Kloor M, Gebert J, Vilkin A, Nishida N, Shin SK, Sasamoto H, Tanaka N, Matsubara N, Boland CR, Goel A: Mutations in both KRAS and BRAF may contribute to the methylator phenotype in colon cancer. Gastroenterology. 1960, 2008 (134): 1950-1960.
Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, Hernandez NS, Chen X, Ahmed S, Konishi K, Hamilton SR, Issa JP: Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci USA. 2007, 20: 18654-18659.
Velho S, Moutinho C, Cirnes L, Albuquerque C, Hamelin R, Schmitt F, Carneiro F, Oliveira C, Seruca R: BRAF, KRAS and PIK3CA mutations in colorectal serrated polyps and cancer: primary or secondary genetic events in colorectal carcinogenesis?. BMC Cancer. 2008, 8: 255-
Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW: CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006, 38: 787-793.
Boland CR, Goel A: Microsatellite instability in colorectal cancer. Gastroenterology. 2010, 138: 2073-2087.
Niv Y: Microsatellite instability and MLH1 promoter hypermethylation in colorectal cancer. World J Gastroenterol. 2007, 13: 1767-1769.
Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP: The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009, 41: 178-186.
Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower C, Chan AT, Giovannucci E, Fuchs C, Ogino S: Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012, 61: 847-854.
Yamauchi M, Lochhead P, Morikawa T, Huttenhower C, Chan AT, Giovannucci E, Fuchs C, Ogino S: Colorectal cancer: a tale of two sides or a continuum?. Gut. 2012, 61: 794-797.
Parsons MT, Buchanan DD, Thompson B, Young JP, Spurdle AB: Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet. 2012, 49: 151-157.
Samowitz WS, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, Wolff RK, Slattery ML: Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005, 129: 837-845.
Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, Berman DM, Jenuwein T, Pruitt K, Sharkis SJ, Watkins DN, Herman JG, Baylin SB: A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007, 39: 237-242.
Rada-Iglesias A, Enroth S, Andersson R, Wanders A, Pahlman L, Komorowski J, Wadelius C: Histone H3 lysine 27 trimethylation in adult differentiated colon associated to cancer DNA hypermethylation. Epigenetics. 2009, 4: 107-113.
Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, Bergman Y, Simon I, Cedar H: Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007, 39: 232-236.
Kondo Y: Epigenetic cross-talk between DNA methylation and histone modifications in human cancers. Yonsei Med J. 2009, 50: 455-463.
Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW: Epigenetic stem cell signature in cancer. Nat Genet. 2007, 39: 157-158.
Simon JA, Kingston RE: Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009, 10: 697-708.
Nelms BL, Labosky PA: A predicted hairpin cluster correlates with barriers to PCR, sequencing and possibly BAC recombineering. Sci Rep. 2011, 1: 106-
Lin J, Lai M, Huang Q, Ma Y, Cui J, Ruan W: Methylation patterns of IGFBP7 in colon cancer cell lines are associated with levels of gene expression. J Pathol. 2007, 212: 83-90.
Carmona FJ, Esteller M: Epigenomics of human colon cancer. Mutat Res. 2010, 693: 53-60.
Yan PS, Chen CM, Shi H, Rahmatpanah F, Wei SH, Caldwell CW, Huang TH: Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001, 61: 8375-8380.
Abel EV, Aplin AE: FOXD3 is a mutant B-RAF-regulated inhibitor of G(1)-S progression in melanoma cells. Cancer Res. 2010, 70: 2891-2900.
de Keizer PL, Packer LM, Szypowska AA, Riedl-Polderman PE, van den Broek NJ, de Bruin A, Dansen TB, Marais R, Brenkman AB, Burgering BM: Activation of forkhead box O transcription factors by oncogenic BRAF promotes p21cip1-dependent senescence. Cancer Res. 2010, 70: 8526-8536.
Muggerud AA, Ronneberg JA, Warnberg F, Botling J, Busato F, Jovanovic J, Solvang H, Bukholm I, Borresen-Dale AL, Kristensen VN, Sorlie T, Tost J: Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer. Breast Cancer Res. 2010, 12: R3-
Lo PK, Lee JS, Liang X, Han L, Mori T, Fackler MJ, Sadik H, Argani P, Pandita TK, Sukumar S: Epigenetic inactivation of the potential tumor suppressor gene FOXF1 in breast cancer. Cancer Res. 2010, 70: 6047-6058.
Nakshatri H, Badve S: FOXA1 in breast cancer. Expert Rev Mol Med. 2009, 11: e8-
Xu X, Quiros RM, Gattuso P, Ain KB, Prinz RA: High prevalence of BRAF gene mutation in papillary thyroid carcinomas and thyroid tumor cell lines. Cancer Res. 2003, 63: 4561-4567.
Wettenhall JM, Smyth GK: limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004, 20: 3705-3706.
Core Team RD: R: A Language and Environment for Statistical Computing. 2008, Vienna
Margaritis T, Lijnzaad P, van Leenen D, Bouwmeester D, Kemmeren P, van Hooff SR, Holstege FC: Adaptable gene-specific dye bias correction for two-channel DNA microarrays. Mol Syst Biol. 2009, 5: 266-
Benjamini Y, Hochberg Y: Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995, 57: 289-300.
Deng G, Chen A, Hong J, Chae HS, Kim YS: Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res. 1999, 59: 2029-2033.
Xiong Z, Laird PW: COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997, 25: 2532-2534.
van Roon EH, van Puijenbroek M, Middeldorp A, van Eijk R, de Meijer EJ, Erasmus D, Wouters KA, van Engeland M, Oosting J, Hes FJ, Tops CM, van Wezel T, Boer JM, Morreau H: Early onset MSI-H colon cancer with MLH1 promoter methylation, is there a genetic predisposition?. BMC Cancer. 2010, 10: 180-
Li LC, Dahiya R: MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002, 18: 1427-1431.
Lewin J, Schmitt AO, Adorjan P, Hildmann T, Piepenbrock C: Quantitative DNA methylation analysis based on four-dye trace data from direct sequencing of PCR amplificates. Bioinformatics. 2004, 20: 3005-3012.
van Wezel T, Lombaerts M, van Roon EH, Philippo K, Baelde HJ, Szuhai K, Cornelisse CJ, Cleton-Jansen AM: Expression analysis of candidate breast tumour suppressor genes on chromosome 16q. Breast Cancer Res. 2005, 7: R998-R1004.
Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, Micklem G, Piano F, Snyder M, Stein L, White KP, Waterston RH: Unlocking the secrets of the genome. Nature. 2009, 459: 927-930.
Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA: Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006, 125: 301-313.
Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, Bourque G, Oh S, Ruan Y, Ng HH, Wei CL: Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007, 1: 286-298.
Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A: PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003, 13: 2129-2141.
Acknowledgements
The authors would like to thank Michiel van Galen (Leiden Genome Technology Center Leiden, The Netherlands) for support with mapping fragments, and Philip Lijnzaad (Department of Physiological Chemistry, University Medical Center Utrecht, Utrecht, The Netherlands) for support with the dyebias R package.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
EHJvR carried out the methylation profiling study and data analysis, and drafted part of the manuscript. AB carried out the validation experiments and drafted part of the manuscript. AAD and RFE participated in data analysis. TvW participated in coordination, and interpretation of the data. HM conceived of the study and participated in its design. JMB participated in study design, and carried out array data analysis and critical revision of the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
13148_2012_36_MOESM4_ESM.pdf
Additional file 4:Regions with BRAF mutation-specific methylation changes (UCSC assembly: March 2006, NCBI36/hg18).(PDF 112 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
van Roon, E.H., Boot, A., Dihal, A.A. et al. BRAF mutation-specific promoter methylation of FOX genes in colorectal cancer. Clin Epigenet 5, 2 (2013). https://doi.org/10.1186/1868-7083-5-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1868-7083-5-2