Abstract
This paper presents a functional nanoparticle-enhanced enzyme-linked immunosorbent assay (FNP-ELISA) for detection of enterohemorrhagic Escherichia coli (EHEC) O157:H7. Immunomagnetic nanoparticles (IMMPs) conjugated with monoclonal anti-O157:H7 antibody were used to capture E. coli O157:H7. Beacon gold nanoparticles (B-GNPs) coated with polyclonal anti-O157:H7 and biotin single-stranded DNA (B-DNA) were then subjective to immunoreaction with E. coli O157:H7, which was followed by streptavidin-horseradish peroxidase (Strep-HRP) conjugated with B-GNPs based on a biotin-avidin system. The solutions containing E. coli O157:H7, IMMPs, B-GNPs, and Strep-HRP were collected for detecting color change. The signal was significantly amplified with detection limits of 68 CFU mL-1 in PBS and 6.8 × 102 to 6.8 × 103 CFU mL-1 in the food samples. The FNP-ELISA method developed in this study was two orders of magnitude more sensitive than immunomagnetic separation ELISA (IMS-ELISA) and four orders of magnitude more sensitive than C-ELISA. The entire detection process of E. coli O157:H7 lasted only 3 h, and thus FNP-ELISA is considered as a time-saving method.
Similar content being viewed by others
Introduction
The World Health Organization estimated that about 1.8 million people worldwide die every year from diarrheal diseases, which are often caused by consuming microbiologically contaminated food or by drinking water [1]. Among the pathogens causing diarrheal diseases, enterohemorrhagic Escherichia coli (EHEC) strains are prominently responsible for serious foodborne outbreaks [2, 3]. In particular, E. coli O157:H7, a predominant strain of EHEC that was first isolated and recognized as a new type of intestinal pathogenic bacterium in the United States in 1982 [4], has become a global public health problem. E. coli O157:H7 outbreaks have occurred in many developing and developed countries, causing huge health care costs and product recalls. The Center for Disease Control and Prevention of the United States estimated that 73,000 cases of illness and 61 deaths per year in the United States are caused by E. coli O157:H7 [5].
The development of a rapid and reliable detection of E. coli O157:H7 has become highly important for food safety and public health [6]. However, traditional methods for the detection of E. coli O157:H7 encompassing enrichment, plating, culturing, enumeration, biochemical testing, and microscopic examination can take up to 60 h, thereby being laborious and time-consuming [7]. Polymerase chain reactions (PCRs), including simple PCR [8], multiplex PCR [9, 10], and real-time PCR [11, 12], are commonly used for rapid detection of E. coli O157:H7, but require complex set-ups and well-trained personnel. In addition, some very sensitive and selective but expensive, complicated, and time-consuming methods have been applied in the detection of E. coli O157:H7, especially including immunomagnetic separation (IMS) analysis [13], flow cytometry [14], fluorescence in situ hybridization [15], DNA microarrays [16], and several label-free methods (such as surface plasmon resonance [17] and use of electrochemical impedance immunosensors [18, 19]).
Enzyme-linked immunosorbent assay (ELISA) was reported to quantitatively detect immunoglobulin G in 1971 [20]. Conventional ELISA (C-ELISA) has high reproducibility and possibility for the simultaneous quantification of a great number of assays, and is widely used to detect the presence of substances, including bacteria [21], viruses [22], proteins [23], and pesticides [24]. However, the detection limit of C-ELISA to E. coli O157:H7 is only 105 to 107 CFU mL-1[25], which is inadequate when the infectious dose is lower than 100 cells [26].
In recent years, the emergence of nanotechnology is opening new horizons for high detection limits in biological fields [27–30]. Nanoparticles of various shapes, sizes, and compositions have broad applications in microorganism detection [31, 32]. Much attention has been focused on amplifying the detection signal using nanoparticles [33, 34], which can enhance enzyme activity [35, 36]. Magnetic and gold particles have been used to improve the detection limit of ELISA [30, 37].
In this study, we developed a functional nanoparticle-enhanced ELISA (FNP-ELISA) using immunomagnetic nanoparticles (IMMPs) and beacon gold nanoparticles (B-GNPs) for detecting E. coli O157:H7. The detection limit of E. coli O157:H7 by the developed FNP-ELISA is much higher than that of C-ELISA or immunomagnetic separation ELISA (IMS-ELISA), and thus FNP-ELISA had the highest sensitivity compared to the other ELISA methods.
Materials and methods
Reagents and materials
Rabbit polyclonal anti-E. coli O157:H7 antibody and mouse monoclonal anti-O157:H7 antibody were prepared and purified in our laboratory. Single-stranded DNA 5′(biotin)-GCTAGTGAACACAGTT-GTGTAAAAAAAAAA (SH)-3′ was synthesized by Sangon Biotech Co., Ltd. (China). Streptavidin-horseradish peroxidase (Strep-HRP) and peroxidase-conjugated affinipure goat anti-rabbit IgG (IgG-HRP) were purchased from Beijing Biosynthesis Biological Technology Co., Ltd. (China). Bovine serum albumin (BSA), 3,3′,5,5′- tetramethylbenzidine (TMB-H2O2), and hydrogen tetrachloroaurate (III) trihydrate (HAuCl4 · 3H2O, 99.9%) were purchased from Sigma-Aldrich (USA). Dextran with a molecular weight of 40,000 (T-40) was obtained from Pharmacia (GE Healthcare, USA). Sorbitol-MacConkey agar (SMAC) and xylose-lysine-tergitol 4 (XLT4) agar were purchased from Difco (Becton Dickinson, USA). Ferric chloride hexahydrate (FeCl3 · 6H2O), ferrous chloride tetrahydrate (FeCl2 · 4H2O), and other chemicals were of analytically pure grade or better quality. The buffer solutions were prepared in our laboratory. All aqueous solutions were prepared using ultrapure water (18.0 MΩ/cm) as required.
Preparation of microbial samples
E. coli O157:H7 strain 35150 and E. coli K12 were obtained from the American Type Culture Collection (ATCC, USA). Salmonella senftenberg 50315, Shigella sonnei 51081, and E. coli O157:Hund strain 21531 (Hund indicated that H antigen was not determined) [38] were obtained from the Institute of Epidemiology and Microbiology, Academy of Preventive Medical Sciences of China. Pure cultures of bacteria were grown in nutrient broth at 37°C for 24 h before use. The concentrations of E. coli O157:H7, O157:Hund, and K12 were determined by the conventional surface plate count method using SMAC. S. senftenberg and S. sonnei were enumerated using XLT4 agar. The cultured bacteria were divided into two portions. The first portion was placed in a boiling water bath for 20 min to kill the bacterial cells, and diluted to the desired concentration with PBS (0.01 M, pH 7.4) for ELISA detection. The second portion was not heated because the number of living cells was counted.
Milk, vegetable, and ground beef were purchased from a local market in Tianjin (China), and each weighed 25 g (mL) for detection. The killed E. coli O157:H7 solution was transferred into a small vial equipped with an atomizer. The mists of E. coli O157:H7 inoculums were sprayed onto the three samples, and the samples were then stored at 4 ± 1°C for 1 h. Each sample was added to 0.25 mL of E. coli O157:H7 solution. The samples were placed into sterile filter stomacher bags, and macerated in 225 mL of PBS with a stomacher blender (Bilon-8 Bilang Co. Ltd., Beijing, China) at 200 rpm for 2 min. The homogenate was serially diluted in PBS for ELISA detection. The negative samples that were not added to E. coli O157:H7 solution were analyzed according to the Chinese National Standard Method GB/T 4789.36-2008 [39].
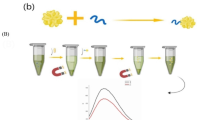
Preparation of IMMPs
Magnetic nanoparticles (MPs) were prepared from FeCl3, FeCl2, ammonia solution, and dextran (T-40), and oxidized with NaIO4 as described previously [40]. Mouse monoclonal anti-E. coli O157:H7 antibody (0.5 mg/mL) was added to the oxidized MP suspension at a ratio of 0.3:1, mixed thoroughly, and incubated in the dark at 4°C for approximately 24 h. IMMPs were washed three times with PBS by placing a magnetic plate against the side wall of the tubes for 5 min to concentrate the particles into the pellets on the side walls. The supernatant was discarded using a transferpettor. The pellets were resuspended in 1 mL of PBST (0.05% Tween-20 in 0.01 M PBS, pH 7.4). BSA was then added to a final concentration of 1% to block any unreacted or nonspecific site. The amount of IMMPs, incubation time, and separation time varied to determine their effects on the recovery of O157:H7 (details in Supplementary Materials).
Preparation of GNPs
GNPs were prepared according to the literature with slight modifications [41]. In brief, 2 mL of 1% HAuCl4 was mixed with 198 mL of fresh ultrapure water. The mixture was stirred vigorously with a magnetic agitator while being heated in a boiling water bath for 20 min, followed by the rapid addition of 5 mL of 1% sodium citrate solution. After the color finally turned to full red, the mixture was stirred again for 10 min before cooling to room temperature. The GNP solution was filtered through a 0.22 μm cellulose nitrate filter to remove any floating aggregates. The prepared GNPs were characterized using a transmission electron microscope (TEM; Tecnai G2 F20, FEI, Netherlands) and ultraviolet spectrophotometer (UV 2500, Shimadzu, Japan).
Preparation of various B-GNPs
B-GNPs were prepared following a previously reported procedure [42, 43] with slight modifications. Rabbit polyclonal anti-E. coli O157:H7 antibody (7 μg) was added to 1 mL of pH-adjusted GNP solution (pH 8.2) and incubated at room temperature for 30 min. The mixture was added with 30 μL of different concentrations of B-DNA, and incubated in the dark at 4°C for more than 16 h. Approximately 100 μL of 1% sodium chloride solution was then added to the mixture, and incubated at 4°C for 3 h. BSA was added to a final concentration of 1% to block any unreacted or nonspecific site. The prepared B-GNP solution was centrifuged at 20,000 g for 1 h at 4°C. The final deposition was suspended in 0.5 mL of storage buffer (PBS, 0.01 M, pH 7.4, 1% BSA, 0.02% NaN3) and stored at 4°C.
C-ELISA
Mouse monoclonal anti-E. coli O157:H7 antibody (100 μL of 5 mg L-1) was added to a 96-well plate and incubated at 37°C for 2 h. The plate was rinsed with PBST (0.05% Tween-20 in 0.01 M PBS, pH 7.4) three times to remove unbound antibodies, followed by the addition of 100 μL of PBS-BSA (1% BSA in 0.01 M PBS, pH 7.4) and incubation at 4°C for 12 h. Different concentrations of E. coli O157:H7 (100 μL) were added to each well and reacted at 37°C for 1 h. After rinsing three times, 100 μL of 5 mg L-1 rabbit polyclonal anti-E. coli O157:H7 antibody was added to the plate incubated at 37°C for 1 h. Subsequently, 100 μL of 0.02 mg L-1 IgG-HRP was added to the plate and incubated for 1 h at 37°C. The plate was rinsed three times to remove unbound IgG-HRP. Finally, 100 μL of TMB-H2O2 solution was added to each well and incubated at 37°C for 15 min. The reaction was terminated using 100 μL of 0.5 M sulfuric acid, and the absorbance at 450 nm was measured using a microplate reader.
IMS-ELISA
E. coli O157:H7 was separated using IMMPs according to the previous procedure in Supplementary Materials, and the particle-bacteria complex (100 μL) was finally resuspended. Rabbit polyclonal anti-O157:H7 antibody (100 μL of 5 mg L-1) was added to the complex, and incubated at room temperature for 30 min. The unbound antibody was removed by the magnetic plate method. The particle-bacteria-antibody complex was resuspended using 100 μL of 0.02 mg L-1 IgG-HRP, and incubated for 1 h at 37°C. The complex was resuspended and transferred to a 96-well plate after excess IgG-HRP was removed by the magnetic plate method. Finally, TMB-H2O2 and sulfuric acid were subsequently added, and the plate was read at 450 nm using a microplate reader.
FNP-ELISA
IMMPs (10 μL) were added to 1 mL of E. coli O157:H7 suspension at 106 CFU mL-1 in a 1.5 mL Eppendorf tube. The tube was carefully inverted several times and incubated at room temperature for 10 min. Approximately 100 μL of the complex of E. coli O157:H7 and IMMPs was obtained by the magnetic plate method. Various B-GNPs (100 μL) were added to 100 μL of the complex, and incubated at room temperature for 30 min. The unbound B-GNPs were removed by the magnetic plate method, and the complex was rinsed three times with PBST. Subsequently, 100 μL of Strep-HRP (0.01 mg L-1) solution was added to the Eppendorf tube and incubated at 37°C for 1 h. The unbound Strep-HRP was removed by the magnetic plate method. The final complex was washed three times with PBST by the magnetic plate method, and resuspended in 100 μL of PBS. The suspension was then transferred to a 96-well plate. Finally, TMB-H2O2 and sulfuric acid were subsequently added, and the plate was read at 450 nm using a microplate reader.
Experimental replicates and statistical methods
All the experiments were done with at least biological replicates, and the values were expressed as mean ± standard deviation. A conventionally used positive control to negative control (P/N) value ≥2.1 was considered positive in the three ELISA methods [44]. When needed, paired Student’s t- test was performed to determine statistically significant differences; P <0.01 was considered to indicate statistical significance.
Results and discussion
Properties of IMMS
We prepared MPs that were roughly spherical in shape with diameters ranging from 40–60 nm, and contained an electron-dense core of 5 nm (Additional file 1: Figure S1). We also prepared IMMPs, and the recovery of IMMPs increased with increasing amount of IMMPs and incubation time. Relatively high recovery was obtained with 10 μL of IMMPS (Additional file 1: Table S1) and 10 min of incubation (Additional file 1: Table S2). The optimal time for separation was 2 min (Additional file 1: Table S3). This finding was supported by a previous study, which showed that target cells can be separated from samples using MPs coupled with aptamer/nucleic acid/antibody [45, 46].
Properties of B-GNPs
We successfully prepared GNPs with an average diameter of approximately 18 nm as measured by TEM (Figure 1). The maximum peak of GNP solution was 518 nm as determined by UV scanning. We prepared a new type of GNP, namely, B-GNPs, which could target E. coli O157:H7 through the polyclonal antibody and amplify signals through the biotin-avidin Strep-HRP system.
Experimental design for FNP-ELISA
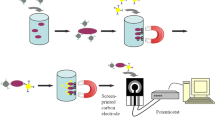
In this study, a novel ELISA method namely FNP-ELISA was established to detect E. coli O157:H7, in comparison with C-ELISA, and IMS-ELISA. A schematic diagram illustrating the detection of E. coli O157:H7 by FNP-ELISA is presented in Figure 2.
Flow chart of FNP-ELISA. Preparation of IMMPs and B-GNPs (part A); (part B) comprises five steps, namely, magnetic separation of target cells (step 1), conjugation of B-GNPs (step 2), removal of free B-GNPs (step 3), conjugation of Strep-HRP (step 4), and removal of free Strep-HRP (step 5); and FNP-ELISA detection (part C).
First, functional monoclonal anti-E. coli O157:H7-conjugated IMMPs and polyclonal anti-E. coli O157:H7 antibody and B-GNPs are prepared (A). Second, IMMPs are mixed with a sample to target E. coli O157:H7, and the O157:H7-IMMP complex is separated using the magnetic plate method (step 1, B). Third, B-GNPs are added to target E. coli O157:H7 in the E. coli O157:H7-IMMP complex (step 2, B), and the unbound B-GNPs are removed by the magnetic plate method (step 3, B). Fourth, Strep-HRP is added to react with polyclonal anti-E. coli O157:H7 (step 4, B), and unbound Strep-HRP is removed by the magnetic plate method. Finally, the remaining routine ELISA steps are completed (C).
Optimized amounts of B-DNA, Strep-HRP and B-GNPs for FNP-ELISA
The optimal amounts of B-DNA, Strep-HRP, and B-GNPs for FNP-ELISA were investigated (Figure 3). To obtain an optimized concentration of B-DNA, we added 100 μL of different B-GNPs conjugated with different concentrations of B-DNA to the complex of E. coli O157:H7 and IMMPs after separating E. coli O157:H7. The aforementioned procedure was then repeated.
The optimal volume of B-GNPs was determined by adding different volumes of the optimized B-GNPs to the complex of E. coli O157:H7 and IMMPs after separating E. coli O157:H7. The detection procedure was then operated as above.
The optimized volume of Strep-HRP was determined when the optimized volume of B-GNPs was applied in the procedure. After optimizing the parameters, various concentrations of E. coli O157:H7 were detected in PBS using FNP-ELISA. The negative samples included K12, S. senftenberg, and S. sonnei in FNP-ELISA. This method was then used to determine the concentration of E. coli O157:H7 in the milk, vegetable, and ground beef samples.
We found that the FNP-ELISA signal increased with increasing B-DNA concentration (Figure 3a), and the optimal concentration of B-DNA was 40 μmol L-1. This finding was also observed in a previous study, which reported that excess B-DNA molecules cause excessive HRP molecules to bind with GNPs, posing steric hindrance to hamper antigens from access to antibodies on GNPs [47].
The signal of FNP-ELISA was strongly dependent on the amount of B-GNPs. The ELISA signal increased as the amount of B-GNPs increased, and reached a plateau at 100 μL (Figure 3b). Thus, we used 100 μL of B-GNPs in subsequent experiments. The optimal amount of Strep-HRP was also obtained at 100 μL (Figure 3c).
Comparison of FNP-ELISA to C-ELISA and IMS-ELISA
Functional nanoparticles were used to improve the sensitivity of ELISA, and the results were compared simultaneously. The blank, positive, and negative controls were PBS, E. coli O157:H7 (106 CFU mL-1), and E. coli O157:Hund (106 CFU mL-1), respectively. These controls were included on each plate in the experiments.
The surface plate counts showed that the concentrations of the test solutions of E. coli O157:H7, E. coli O157: Hund, K12, S. senftenberg, and S. sonnei were 6.8 × 10°–6.8 × 108 CFU mL-1, 6.1 × 106 CFU mL-1, 7.1 × 10°–7.1 × 108 CFU mL-1, 8.1 × 10°–8.1 × 108 CFU mL-1, and 2.3 × 10°–2.3 × 108 CFU mL-1, respectively. The detection limits of C-ELISA, IMS-ELISA, and FNP-ELISA were 6.8 × 105 (n = 3, 1.8 ≤ RSD ≤ 4.7), 6.8 × 103 (n = 3, 0.6 ≤ RSD ≤ 5.2), and 68 CFU mL-1 (n = 3, 0.3 ≤ RSD ≤ 4.7) in PBS, respectively (Figure 4). The three ELISA methods had high reproducibility, and all RSD values were lower than 10.
The specificity of the three ELISA methods was dependent on the quality of the antibody. The specificity of the rabbit polyclonal anti-E. coli O157:H7 antibody and mouse monoclonal anti-E. coli O157:H7 antibody was tested using 61 bacteria, and false negative and false positive results were not observed (data not shown). E. coli O157:H7 is a common intestinal pathogen, so S. senftenberg and S. sonnei, which are also common intestinal pathogens, were selected as negative samples. E. coli K12, which represented E. coli, was another negative sample. The P/N values of the negative signals were all significantly lower than 2.1 (Figure 4), which shows that FNP-ELISA had high specificity.
E. coli O157:H7 was not detected in the raw vegetable, milk, and ground beef samples using GB/T method 4789.36-2008. The detection limits of FNP-ELISA were 6.8 × 102 CFU mL-1 in vegetable and milk, and 6.8 × 103 CFU mL-1 in ground beef. The reduction in sensitivities may be attributed to the loss of some functional nanoparticles and targeting bacteria in the food residues, particularly grease foods.
Concluding remarks
Multiple ultrasensitive methods for detecting E. coli O157:H7 have been reported [48, 49], but they usually require expensive equipment or skilled personnel and thus have difficulty in wide use. C-ELISA has been widely established to detect microorganisms, proteins, pesticides, and heavy metals because of its simplicity and low cost, but its application is limited because of its low detection limit. Data presented here indicated that FNP-ELISA had a high sensitivity in detecting E. coli O157:H7, with a detection limit of 68 CFU mL-1 in PBS and 6.8 × 102 to 103 CFU mL-1 in foods. The detection limit of FNP-ELISA was about two or four orders of magnitude lower than that of IMS-ELISA or C-ELISA, respectively. Moreover, the total analysis time of FNP-ELISA was only approximately 3 h. Therefore, FNP-ELISA may be used for detection of E. coli O157:H7 in foods, and also for other microorganisms if appropriate antibodies are available.
References
Velusamy V, Arshak K, Korostynska O, Oliwa K, Adley C: An overview of foodborne pathogen detection: in the perspective of biosensors. Biotechnol Adv. 2010, 28: 232-254. 10.1016/j.biotechadv.2009.12.004.
Abadias M, Usall J, Anguera M, Solsona C, Vinas I: Microbiological quality of fresh, minimally-processed fruit and vegetables, and sprouts from retail establishments. Int J Food Microbiol. 2008, 123: 121-129. 10.1016/j.ijfoodmicro.2007.12.013.
Wu CJ, Hsueh PR, Ko WC: A new health threat in Europe: Shiga toxin-producing Escherichia coli O104:H4 infections. J Microbiol Immunol Infect. 2011, 44: 390-393. 10.1016/j.jmii.2011.07.001.
Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, Hebert RJ, Olcott ES, Johnson LM, Hargrett NT, Blake PA, Cohen ML: Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983, 308: 681-685. 10.1056/NEJM198303243081203.
Park S, Kim H, Paek SH, Hong JW, Kim YK: Enzyme-linked immuno-strip biosensor to detect Escherichia coli O157:H7. Ultramicroscopy. 2008, 108: 1348-1351. 10.1016/j.ultramic.2008.04.063.
Dweik M, Stringer RC, Dastider SG, Wu Y, Almasri M, Barizuddin S: Specific and targeted detection of viable Escherichia coli O157:H7 using a sensitive and reusable impedance biosensor with dose and time response studies. Talanta. 2012, 94: 84-89.
Fedio WM, Jinneman KC, Yoshitomi KJ, Zapata R, Wendakoon CN, Browning P, Weagant SD: Detection of E. coli O157:H7 in raw ground beef by Pathatrix immunomagnetic-separation, real-time PCR and cultural methods. Int J Food Microbiol. 2011, 148: 87-92. 10.1016/j.ijfoodmicro.2011.05.005.
Johnston LM, Elhanafi D, Drake M, Jaykus LA: A simple method for the direct detection of Salmonella and Escherichia coli O157:H7 from raw alfalfa sprouts and spent irrigation water using PCR. J Food Prot. 2005, 68: 2256-2263.
Paton AW, Paton JC: Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J Clin Microbiol. 2002, 40: 271-274. 10.1128/JCM.40.1.271-274.2002.
Kumar A, Grover S, Kumar Batish V: Application of multiplex PCR assay based on uidR and fliCH7 genes for detection of Escherichia coli O157:H7 in milk. J Gen Appl Microbiol. 2013, 59: 11-19. 10.2323/jgam.59.011.
Wong LY, Cao Y, Balachandran P, Zoder P, Furtado MR, Petrauskene OV, Tebbs RS: Validation of the Applied Biosystems MicroSEQ real-time PCR system for detection of E. coli O157:H7 in food. J AOAC Int. 2012, 95: 1495-1504. 10.5740/jaoacint.11-289.
Wasilenko JL, Fratamico PM, Narang N, Tillman GE, Ladely S, Simmons M, Cray WC: Influence of primer sequences and DNA extraction method on detection of non-O157 Shiga toxin-producing Escherichia coli in ground beef by real-time PCR targeting the eae, stx, and serogroup-specific genes. J Food Prot. 2012, 75: 1939-1950. 10.4315/0362-028X.JFP-12-087.
Wang L, Li Y, Mustaphai A: Rapid and simultaneous quantitation of Escherichia coli 0157:H7, Salmonella, and Shigella in ground beef by multiplex real-time PCR and immunomagnetic separation. J Food Prot. 2007, 70: 1366-1372.
Wilkes JG, Tucker RK, Montgomery JA, Cooper WM, Sutherland JB, Buzatu DA: Reduction of food matrix interference by a combination of sample preparation and multi-dimensional gating techniques to facilitate rapid, high sensitivity analysis for Escherichia coli serotype O157 by flow cytometry. Food Microbiol. 2012, 30: 281-288. 10.1016/j.fm.2011.11.002.
Garcia-Armisen T, Servais P: Enumeration of viable E. coli in rivers and wastewaters by fluorescent in situ hybridization. J Microbiol Methods. 2004, 58: 269-279. 10.1016/j.mimet.2004.04.014.
Donhauser SC, Niessner R, Seidel M: Sensitive quantification of Escherichia coli O157:H7, Salmonella enterica, and Campylobacter jejuni by combining stopped polymerase chain reaction with chemiluminescence flow-through DNA microarray analysis. Anal Chem. 2011, 83: 3153-3160. 10.1021/ac2002214.
Wang Y, Ye Z, Si C, Ying Y: Monitoring of Escherichia coli O157:H7 in food samples using lectin based surface plasmon resonance biosensor. Food Chem. 2013, 136: 1303-1308. 10.1016/j.foodchem.2012.09.069.
Wang L, Liu Q, Hu Z, Zhang Y, Wu C, Yang M, Wang P: A novel electrochemical biosensor based on dynamic polymerase-extending hybridization for E. coli O157:H7 DNA detection. Talanta. 2009, 78: 647-652. 10.1016/j.talanta.2008.12.001.
Viswanathan S, Rani C, Ho JA: Electrochemical immunosensor for multiplexed detection of food-borne pathogens using nanocrystal bioconjugates and MWCNT screen-printed electrode. Talanta. 2012, 94: 315-319.
Engvall E, Perlmann P: Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry. 1971, 8: 871-874. 10.1016/0019-2791(71)90454-X.
Chunglok W, Wuragil DK, Oaew S, Somasundrum M, Surareungchai W: Immunoassay based on carbon nanotubes-enhanced ELISA for Salmonella enterica serovar Typhimurium. Biosens Bioelectron. 2011, 26: 3584-3589. 10.1016/j.bios.2011.02.005.
Zhu J, Yang Q, Cao L, Dou X, Zhao J, Zhu W, Ding F, Bu RE, Suo S, Ren Y, Li G, Ren X: Development of porcine rotavirus vp6 protein based ELISA for differentiation of this virus and other viruses. Virol J. 2013, 10: 91-10.1186/1743-422X-10-91.
Jalallou N, Bandehpour M, Khazan H, Haghighi A, Kazemi B: Evaluation of recombinant sag1 protein for detection of toxoplasma gondii specific immunoglobulin M by ELISA test. Iran J Parasitol. 2012, 7: 17-21.
Byer JD, Struger J, Sverko E, Klawunn P, Todd A: Spatial and seasonal variations in atrazine and metolachlor surface water concentrations in Ontario (Canada) using ELISA. Chemosphere. 2011, 82: 1155-1160. 10.1016/j.chemosphere.2010.12.054.
Strachan NJ, Ogden ID: A sensitive microsphere coagulation ELISA for Escherichia coli O157:H7 using Russell’s viper venom. FEMS Microbiol Lett. 2000, 186: 79-84. 10.1111/j.1574-6968.2000.tb09085.x.
Tuttle J, Gomez T, Doyle MP, Wells JG, Zhao T, Tauxe RV, Griffin PM: Lessons from a large outbreak of Escherichia coli O157:H7 infections: insights into the infectious dose and method of widespread contamination of hamburger patties. Epidemiol Infect. 1999, 122: 185-192. 10.1017/S0950268898001976.
Shen ZQ, Wang JF, Qiu ZG, Jin M, Wang XW, Chen ZL, Li JW, Cao FH: QCM immunosensor detection of Escherichia coli O157:H7 based on beacon immunomagnetic nanoparticles and catalytic growth of colloidal gold. Biosens Bioelectron. 2011, 26: 3376-3381. 10.1016/j.bios.2010.12.035.
Su XL, Li Y: A QCM immunosensor for Salmonella detection with simultaneous measurements of resonant frequency and motional resistance. Biosens Bioelectron. 2005, 21: 840-848. 10.1016/j.bios.2005.01.021.
Cheng HY, Lai LJ, Ko FH: Rapid and sensitive detection of rare cancer cells by the coupling of immunomagnetic nanoparticle separation with ELISA analysis. Int J Nanomedicine. 2012, 7: 2967-2973.
Zhou F, Wang M, Yuan L, Cheng Z, Wu Z, Chen H: Sensitive sandwich ELISA based on a gold nanoparticle layer for cancer detection. Analyst. 2012, 137: 1779-1784. 10.1039/c2an16257a.
Tamer U, Cetin D, Suludere Z, Boyaci IH, Temiz HT, Yegenoglu H, Daniel P, Dincer I, Elerman Y: Gold-coated iron composite nanospheres targeted the detection of Escherichia coli. Int J Mol Sci. 2013, 14: 6223-6240. 10.3390/ijms14036223.
Li F, Zhou R, Zhao K, Chen H, Hu Y: Magnetic beads-based electrochemical immunosensor for detection of pseudorabies virus antibody in swine serum. Talanta. 2011, 87: 302-306.
Wang J, Moore J, Laulhe S, Nantz M, Achilefu S, Kang KA: Fluorophore-gold nanoparticle complex for sensitive optical biosensing and imaging. Nanotechnology. 2012, 23: 095501-10.1088/0957-4484/23/9/095501.
Gan N, Jin H, Li T, Zheng L: Fe(3)O(4)/Au magnetic nanoparticle amplification strategies for ultrasensitive electrochemical immunoassay of alfa-fetoprotein. Int J Nanomedicine. 2011, 6: 3259-3269.
Huang SH, Liao MH, Chen DH: Direct binding and characterization of lipase onto magnetic nanoparticles. Biotechnol Prog. 2003, 19: 1095-1100. 10.1021/bp025587v.
Pandey P, Singh SP, Arya SK, Gupta V, Datta M, Singh S, Malhotra BD: Application of thiolated gold nanoparticles for the enhancement of glucose oxidase activity. Langmuir. 2007, 23: 3333-3337. 10.1021/la062901c.
Cudjoe KS, Thorsen LI, Sorensen T, Reseland J, Olsvik O, Granum PE: Detection of Clostridium perfringens type A enterotoxin in faecal and food samples using immunomagnetic separation (IMS)-ELISA. Int J Food Microbiol. 1991, 12: 313-321. 10.1016/0168-1605(91)90145-F.
Bai L, Liu X, Fu P, Guo Y: Serotyping and virulence genes of suspected Escherichia coli O157 strains in food from 2005 to 2007. Wei Sheng Yan Jiu. 2010, 39: 335-338.
Ministry of Health P.R. China: Microbiological examination of food hygiene-examination of Escherichia coli O157:H7/NM. Standardization Administration PR China. 2008, GB/T 4789.36-2008
Duan HL, Shen ZQ, Wang XW, Chao FH, Li JW: Preparation of immunomagnetic iron-dextran nanoparticles and application in rapid isolation of E. coli O157:H7 from foods. World J Gastroenterol. 2005, 11: 3660-3664.
Ambrosi A, Castaneda MT, Killard AJ, Smyth MR, Alegret S, Merkoci A: Double-codified gold nanolabels for enhanced immunoanalysis. Anal Chem. 2007, 79: 5232-5240. 10.1021/ac070357m.
Kong XL, Qiao FY, Qi H, Li FR: One-step preparation of antibody and oligonucleotide dual-labeled gold nanoparticle bio-probes and their properties. Biotechnol Lett. 2008, 30: 2071-2077. 10.1007/s10529-008-9802-6.
Nam JM, Thaxton CS, Mirkin CA: Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003, 301: 1884-1886. 10.1126/science.1088755.
Sherwani S, Farleigh L, Agarwal N, Loveless S, Robertson N, Hadaschik E, Schnitzler P, Bugert JJ: Seroprevalence of Molluscum contagiosum virus in German and UK populations. PLoS One. 2014, 9: e88734-10.1371/journal.pone.0088734.
Cutler JI, Zheng D, Xu X, Giljohann DA, Mirkin CA: Polyvalent oligonucleotide iron oxide nanoparticle “click” conjugates. Nano Lett. 2010, 10: 1477-1480. 10.1021/nl100477m.
Kumar A, Jena PK, Behera S, Lockey RF, Mohapatra S: Multifunctional magnetic nanoparticles for targeted delivery. Nanomedicine. 2010, 6: 64-69. 10.1016/j.nano.2009.04.002.
Liu M, Jia C, Huang Y, Lou X, Yao S, Jin Q, Zhao J, Xiang J: Highly sensitive protein detection using enzyme-labeled gold nanoparticle probes. Analyst. 2010, 135: 327-331. 10.1039/b916629g.
Almeida C, Sousa JM, Rocha R, Cerqueira L, Fanning S, Azevedo NF, Vieira MJ: Detection of Escherichia coli O157 by peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) and comparison to a standard culture method. Appl Environ Microbiol. 2013, 79: 6293-6300. 10.1128/AEM.01009-13.
Chan KY, Ye WW, Zhang Y, Xiao LD, Leung PH, Li Y, Yang M: Ultrasensitive detection of E. coli O157:H7 with biofunctional magnetic bead concentration via nanoporous membrane based electrochemical immunosensor. Biosens Bioelectron. 2013, 41: 532-537.
Acknowledgments
This study was supported by National Natural Science Foundation of China (81270041 and 21377061) and Natural Science and Technology Supporting Program of Tianjin (11ZCKFSF01100). The English writing was polished by EnPapers.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
DZ and JL conceived and designed the experiments. ZS, NH, MJ, ZQ, JW, BZ, XW and JW performed the experiments. ZS, DZ and JL analyzed the data. ZS, DZ and JL drafted the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
13099_2014_133_MOESM1_ESM.doc
Additional file 1: Table S1: Effect of amount of IMMPs on recovery. Table S2 Effect of incubation time on recovery. Table S3 Effect of separation on recovery. Figure S1 Transmission electron micrograph of IMMPs. The electron dense core was about 5 nm. (DOC 4 MB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Shen, Z., Hou, N., Jin, M. et al. A novel enzyme-linked immunosorbent assay for detection of Escherichia coli O157:H7 using immunomagnetic and beacon gold nanoparticles. Gut Pathog 6, 14 (2014). https://doi.org/10.1186/1757-4749-6-14
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1757-4749-6-14