Abstract
Background
Ochrobactrum intermedium is an emerging opportunistic pathogen of humans that is closely related to members of the genus Brucella. Earlier, we reported the case of an Indian subject with non-ulcer dyspeptic symptoms whose urease positive gastric biopsy revealed the presence of Helicobacter pylori along with non-Helicobacter like bacteria, eventually cultured and identified as O. intermedium strain M86.
Results
Here, we describe the unclosed draft genome of the strain M86 with a length of 5,188,688 bp and mean G+C content of 57.9%. We have also identified many putative gene clusters that might be responsible for its persistence in the gastric mucosa.Comparative analysis of genomic features of Ochrobactrum intermedium strain M86 and Ochrobactrum intermedium LMG 3301T was also done.
Conclusions
This paper attempts to gain whole-genome based insights into the putative gene determinants of O. intermedium for survival in the highly acidic stomach lumen environment .Identification of genes putatively involved in the various metabolic pathways may lead to a better understanding of the survival of O. intermdedium in acidic condition.
Similar content being viewed by others
Background
The Alphaproteobacteria represent a biologically diverse group of bacteria with members like Brucella, Bartonella, Agrobacterium and Ochrobactrum that are capable of interacting with eukaryotic cells. Ochrobactrum intermedium is a Gram-negative, capsulating, aerobic bacilli belonging to the Alphaproteobacteria. It is the closest genetic relative of genus Brucella as evidenced by protein profiling, western blot, immunoelectrophoresis, amplified fragment length polymorphism, 16S rRNA gene and RecA gene sequence based studies [1]. Previous reports have suggested O. intermedium as an emerging pathogen in liver abscess post-liver transplantation and in the bladder cancer patient causing presumptive bacteremia [2, 3]. But, clinical manifestations and diseases caused by O. intermedium are poorly characterised.
Several bacteria other than Helicobacter pylori have been detected earlier in gastric biopsies. The presence of O. intermedium along with H. pylori was reported earlier in a subject from North-India diagnosed with non-ulcer dyspepsia [4]. A unique observation was the presence of severe fibrosis in the lamina propria of the gastric mucosa revealed during histological examination of the gastric antral biopsy. Whether this fibrosis was caused either partially or totally by O. intermedium was not clear. Other species of Ochrobactrum have also been associated along with H. pylori, e.g., O. anthropi with mild gastritis in squirrel monkeys [5]. Similarly, Gastrospirillum hominis[6], enterococcci [7] and staphylococci have been associated with gastric disorders [8]. In some cases, they have been isolated from antral biopsies from patients with or without H. pylori colonization [9].
Importantly, both H. pylori and O. intermedium produce urease, and thus the detection of H. pylori by urease test in the presence of Ochrobactrum may be confounded. The role of Ochrobactrum in gastric pathology remains uncertain and requires detailed pathologic, microbiological and genetic investigations in order to evaluate the link between H. pylori and O. intermedium in the gastric niche. This paper attempts to gain whole-genome based insights into the putative gene determinants of O. intermedium for survival in the highly acidic stomach lumen environment.
Methods
Genome sequencing
Genomic DNA was isolated by PureLink®Genomic DNA Kit. The draft genome sequence of strain M86 was determined by Ion Torrent Personal Genome Machine (PGM™) sequencer using a 316 chip with 200-bp single-end shotgun sequencing. A total of 2,602,696 reads were obtained. PGM sequencing resulted in about 67X genome coverage with 148 contigs.
Assembly and annotation
The de novo approach was applied to finalize the unclosed draft genome using MIRA 3.4.0 version using default parameters [10]. Prediction and annotation of genes were done using RAST [11] server with SEED database and ISGA pipeline [12]. The data were further validated using gene prediction tools such as Glimmer [13]. Functional annotation was also performed by PGAAP using public database of National Centre for Biotechnology Information (NCBI). Prophages and putative phage like elements in the genome were identified using prophage-predicting PHAST [14] Web server. Regions identified algorithmically as “intact” by PHAST, as well as regions sharing a high degree of sequence similarity and conserved synteny with predicted “intact” prophages, were identified as prophages.
Submission of genome sequence
The Ochrobactrum intermedium strain M86 whole genome shotgun (WGS) project was submitted to the GenBank and has the project accession AOGE00000000 and consists of sequences AOGE01000001-AOGE01000148
Quality assurance
The genomic DNA was isolated from pure bacterial isolate and was further confirmed with 16S rRNA gene sequencing. Bioinformatic assessment of potential contamination of the genomic library by allochthonous microorganisms was done using PGAAP and RAST annotation systems.
Initial findings
Genome characteristics
Genome of O. intermedium strain M86 was sequenced on the IonTorrent Personal Genome Machine (PGM™) using 316 chip that resulted in 2,602,696 total reads with a mean read length of 155 bp. de-novo assembly using the MIRA assembler v3.4.0 [10] with default parameters yielded ~67X coverage. A total of 148 contigs with >500 bp length were obtained. The unclosed draft genome sequence of strain M86 is of 5,188,688 bps and 5043 predicted coding DNA sequences (CDSs) and 66 RNA genes with mean G + C content of 57.9%. RAST server based annotation of the whole genome, showed the presence of 437 subsystems (related functional roles) [11]. Figure 1 describes the subsystem distribution of strain M86.
Putative gene clusters responsible for survival of Ochrobactrum intermedium strain M86 in the acidic environment of stomach lumen
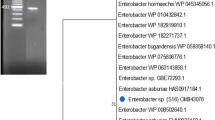
H. pylori have several genes for biosynthesis of cytosolic urease for its survival in the acidic environment of stomach lumen [15]. Genome of strain M86 contains urease gene cluster (see Figure 2): out of which, UreA, beta subunit, UreB gamma subunit, UreC, alpha subunit, are part of core Urease enzyme, While four accessory proteins: UreD, UreE, UreF and UreG play important role in Ni2+ uptake and insertion into active site of apo-enzyme. Genetic relatedness of urease gene cluster with phylogeneticaly closely related bacteria is shown in Figure 2. A complete operon encoding the VirB gene involved in conjugative transfer is present in strain M86. Genes encoding osmotic stress, oxidative stress HPIIb, cold shock GrpE, heat shock DnaK, periplasmic stress DegQ and protection from reactive oxygen species, sod are found. Genes predicated to encode flagellar biosynthesis protein FlhA and FlhB has been identified in genome of strain M86 which are likely elementary to adaptation of new lifestyle. Enterobactin synthesis clusters of entA, entB1, entB2, entC, entD, entE, entF, entG, entH genes were also observed in the genome sequence of strain M86 suggesting its ability of iron acquisition by siderophore production. Presence of membrane transport machinery with dominance of Dipeptide-binding ABC transporter, periplasmic substrate-binding component was detected in the genome of strain M86.It has been found that all the clinical isolates and the type strains of Ochrobactrum were highly resistant to all forms of ß-lactams except imipenem [16]. This resistance profile is consistent with the expression of the AmpC beta-lactamase characterized in O. anthropi[17, 18]. The genome of strain M86 shows the presence of AmpC beta-lactamase gene which supports its resistance to ß-lactams antibiotic observed by Dharne et al. [4] .Table 1 shows the comparison of genomic features of Ochrobactrum intermedium strain M86 and Ochrobactrum intermedium LMG 3301T.
Prediction of phage islands
The presence of prophage sequences may also allow some bacteria to acquire antibiotic resistance, to exist in new environmental niches, to improve adhesion or to become pathogenic [14]. In addition, phages play a crucial role in genome plasticity and chromosome remodelling. The intact phage of 32 kb in length together with phage-like proteins and hypothetical proteins were identified in the genome of strain M86 by prophage-predicting PHAST [14] Web server.
Future directions
The genomic properties of O. intermedium are poorly characterized and, as a consequence, their role in human health and disease remains unclear. Elucidation of the physiological properties and identification of genes putatively involved in the various metabolic pathways may lead to a better understanding of the survival of O. intermdedium in acidic condition. Further studies involving large scale genome sequencing and comparison of the O. intermedium strains isolated from several other non-ulcer dyspeptic individuals will help us apprehend the genomic features of its survival in the acidic condition of the stomach. Comparative genomic analyses of O. intemedium strain M86 and other environmental isolates of genus Ochrobactrum will permit us to understand its mechanisms for adaptation to new environments.
References
Velasco J, Romero C, Lopez-Goni I, Leiva J, Diaz R, Moriyon I: Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp. Int J Syst Bacteriol. 1998, 48: 759-768. 10.1099/00207713-48-3-759.
Moller LV, Arends JP, Harmsen HJ, Talens A, Terpstra PM, Slooff J: Ochrobactrum intermedium infection after liver transplantation. J Clin Microbiol. 1999, 37: 241-244.
Apisarnthanarak A, Kiratisin P, Mundy LM: Evaluation of Ochrobactrum intermedium bacteremia in a patient with bladder cancer. Diagn Microbiol Infect Dis. 2005, 53: 153-155. 10.1016/j.diagmicrobio.2005.05.014.
Dharne MS, Misra SP, Misra V, Dwivedi M, Patole MS, Shouche YS: Isolation of urease-positive Ochrobactrum intermedium in the stomach of a non-ulcer dyspeptic patient from north India. J Microbiol Immunol Infect. 2008, 41: 183-186.
Khanolkar-Gaitonde SS, Reubish GK, Lee CK, Stadtlander CT: Isolation of bacteria other than Helicobacter pylori from stomachs of squirrel monkeys (Saimiri spp.) with gastritis. Dig Dis Sci. 2000, 45: 272-80. 10.1023/A:1005496022881.
Heilmann KL, Borchard F: Gastritis due to spiral shaped bacteria other than Helicobacter pylori: clinical, histological, and ultrastructural findings. Gut. 1991, 32: 137-140. 10.1136/gut.32.2.137.
Hala MT, El-Zimaty , Ramchatesingh J, Clarrige JE, Abudayyeh S, Osato MS, Graham D: Enterococcal gastritis. Human Pathol. 2003, 34: 944-945. 10.1016/S0046-8177(03)00287-9.
Brandi G, Biavati B, Calabrese C, Granata M, Nannetti A, Mattarelli P, Di Febo G, Saccoccio G, Biasco G: Urease-positive bacteria other than Helicobacter pylori in human gastric juice and mucosa. Am J Gastroenterol. 2006, 101: 1756-61. 10.1111/j.1572-0241.2006.00698.x.
Sung L, Changsung K, Young C: Successful cultivation of a potentially pathogenic coccoid organism with tropism for gastric mucin. Infect Immun. 1997, 65: 49-52.
Chevreux B, Wetter T, Suhai S: Genome sequence assembly using trace signals and additional sequence information. Comput Sci Biol. 1999, 99: 45-56.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O: The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008, 8: 9-75.
Hemmerich C, Buechlein A, Podicheti R, Revanna KV, Dong Q: An Ergatis-based prokaryotic genome annotation web server. Bioinformatics. 2010, 26: 1122-1124. 10.1093/bioinformatics/btq090.
Delcher AL, Harmon D, Kasif S, White O, Salzberg SL: Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999, 27: 4636-4641. 10.1093/nar/27.23.4636.
Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS: PHAST: a fast phage search tool. Nucleic Acids Res. 2011, 39: 347-352. 10.1093/nar/gkq749.
Kusters JG, van Vliet AH, Kuipers EJ: Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006, 19: 449-490. 10.1128/CMR.00054-05.
Teyssier C, Marchandin H, Jean-Pierre H, Darbas H, Siméon De Buochberg M, Diego I, Gouby A, Jumas-Bilak E: Molecular and phenotypic features for identification of the opportunistic pathogens Ochrobactrum spp. J Med Microbiol. 2005, 54: 945-953. 10.1099/jmm.0.46116-0.
Higgins CS, Avison MB, Jamieson L, Simm AM, Bennett PM, Walsh TR: Characterization, cloning and sequence analysis of the inducible Ochrobactrum anthropi AmpC ß-lactamase. J Antimicrob Chemother. 2001, 47: 745-754. 10.1093/jac/47.6.745.
Nadjar D, Labia R, Cerceau C, Bizet C, Philippon A, Arlet G: Molecular characterization of chromosomal class C ß-lactamase and its regulatory gene in Ochrobactrum anthropi. Antimicrob Agents Chemother. 2001, 45: 2324-2330. 10.1128/AAC.45.8.2324-2330.2001.
Acknowledgements
Girish Kulkarni acknowledges the financial support received by the Council of Scientific and Industrial Research. This work was supported by funding from the Department of Biotechnology, Government of India, and Microbial Culture Collection project. We thank Dr. Kamlesh Jangid, Dr. Om Prakash for extensive review and useful comments on the manuscript. We also thank two anonymous reviewers for useful comments on earlier versions of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GK and DD were involved in the genome assembly, annotation and manuscript preparation. SC and SS generated ion PGM data. MD and MP were involved in isolation of M86 strain. SM and VM provided the human stomach biopsy. GK and YS were involved in overall experimental design. All authors have read the manuscript and approved.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kulkarni, G., Dhotre, D., Dharne, M. et al. Draft genome of Ochrobactrum intermedium strain M86 isolated from non-ulcer dyspeptic individual from India. Gut Pathog 5, 7 (2013). https://doi.org/10.1186/1757-4749-5-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1757-4749-5-7