Abstract
Background
From a clinical perspective, it is important to know which serogroups, virulence genes and antibiotic resistance patterns are present in Shiga toxin-producing Escherichia coli strains in pediatric patients suffering from diarrheic and non-diarrheic infections. This is the first study in Iran that has comprehensively investigated the Shiga toxin-producing Escherichia coli -related infection characteristics in diarrheic and non-diarrheic pediatric patients of 0–60 months of age.
Methods
Two-hundred and twenty four diarrheic and 84 non-diarrheic stool specimens were collected from the Baqiyatallah hospital of Tehran, Iran. The stool samples were cultured immediately and those that were E. coli-positive were analyzed for the presence of antibiotic resistance genes and bacterial virulence factors using PCR. Antimicrobial susceptibility testing was performed using disk diffusion method.
Results
One-hundred and fifty four out of 224 (68.75%) diarrheic stools and 31 out of 84 (36.90%) non-diarrheic stools harbored E. coli. In addition, children in 13–24 month-old age group had the highest incidence of infection with this bacterium (77.63%). A significant difference was found between the frequency of Attaching and Effacing Escherichia coli and Enterohaemorrhagic Escherichia coli (P =0.045). The genes encoding Shiga toxins and intimin were the most commonly detected virulence factors. Among all serogroups studied, O26 (27.04%) and O111 (18.85%) had the highest incidences in the diarrheic and non-diarrheic patients. The incidence of genes encoding resistance against sulfonamide (sul1), gentamicin (aac(3)-IV), trimethoprim (aadA1), cephalothin (blaSHV) and tetracycline (tetA) were 82.78%, 68.03%, 60.65%, 56.55% and 51.63%, respectively. High resistance levels against penicillin (100%), tetracycline (86.88%), gentamicin (62.29%) and streptomycin (54.91%) were observed. Marked seasonality in the serogroup distributions was evident, while STEC infections were more common in summer (P =0.041).
Conclusions
Our findings should raise awareness about antibiotic resistance in diarrheic pediatric patients in Iran. Clinicians should exercise caution when prescribing antibiotics, especially during the warmer months of the year.
Similar content being viewed by others
Background
Infection with Shiga toxin (Stx)-producing Escherichia coli (STEC) can result in a spectrum of outcomes, ranging from asymptomatic carriage to uncomplicated diarrhea, hemolytic uremic syndrome (HUS), bloody diarrhea, hemolytic anemia, thrombocytopenia, and acute renal failure [1–4]. High mortality and morbidity rates have been reported for HUS, which can occur from infection with STEC strains [1, 5]. The pathogenesis of STEC is related to several bacterial virulence factors [4, 6]. Some of the most important of these virulence factors include the intimin (eae) protein, two shiga toxins called stx1 and stx2, and the plasmid-encoded protein known as hemolysin (ehly) [4, 6].
Most outbreaks and sporadic cases of bloody and non-bloody diarrhea and HUS have been attributed to strains of the STEC serogroup O157 [7, 8]. However, non-O157 strains such as O26, O111, O145, O26, O91, O103, O113, O128, O121 and O45 have been shown to cause bloody and non-bloody diarrhea and HUS [7, 8]. If diarrheic patients do not receive effective treatment, they are susceptible to secondary infections and illnesses.
Diseases caused by E. coli often require antimicrobial therapy; however, antibiotic-resistant strains of this bacterium cause longer and more severe illnesses than their antibiotic-susceptible counterparts. Several studies have shown that antibiotic resistance in E. coli has increased over time [8–10]. In keeping with this, an epidemiological investigation in Iran revealed that STEC strains were the most commonly detected strains in pediatric patients with diarrhea and that there was a high incidence of resistance (85–100%) to commonly used antibiotics [11, 12]. Antibiotic resistance genes are known to cause antibiotic resistance in STEC strains isolated from diarrheic patients [13].
Data on the distribution of serogroups, virulence genes and the antimicrobial resistance properties of STEC strains isolated from pediatric patients are scarce in Iran [11]. Therefore, the aim of the present study was to characterize STEC strains isolated from Iranian diarrheic and non-diarrheic pediatric patients at the molecule level and investigate their susceptibility to 12 commonly used antibiotics, as well as investigating seasonal variation in the prevalence and serogroups distribution of E. coli.
Methods
Sample collection, preparation, and identification of E. coli serogroups
From March 2012 to March 2013, a period covering seasonal variation, 308 stool samples from diarrheic and non-diarrheic pediatric patients were collected from the Baqiyatallah hospital in Tehran, Iran. Stool samples were classified as either diarrheic or non-diarrheic. Individuals from the diarrheic group were placed into six groups based on their ages (1–12, 13–24, 25–36, 37–48 and 49–60 month-old children) (Table 1). Information on the clinical and epidemiological history of these patients was obtained through questionnaires. Patients presented at the hospital with symptoms such as nausea and fever, while others had dysentery-like or inconsequential symptoms. Stool specimens were collected using sterile rectal swabs, which were transferred to tubes containing Stuart medium (Merck, Germany). Samples were transferred to the Biotechnology and Microbiology Research Center of the Islamic Azad University of Shahrekord at 4°C. All samples were diluted in phosphate buffered saline (PBS, Merck, Germany). The samples were plated onto MacConkey’s agar (MC, Merck, Germany) and incubated overnight at 37°C. A lactose positive colony was selected from each sample, streaked onto Eosin Methylene Blue (EMB, Merck, Germany) plates and incubated overnight at 37°C. Green colonies with a metallic luster were considered as typical E. coli colonies. Such colonies were confirmed as E. coli using standard biochemical tests (e.g., Methyl red, Voges-Proskauer, Indole, and Citrate utilization tests). Colonies were confirmed as E. coli by PCR [14]. E. coli isolates were stored in Tryptic Soy Broth (TSB, Merck, Germany) containing 20% glycerol at -70°C for further characterization.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing of the isolates was performed using the Kirby–Bauer disc diffusion method and Mueller–Hinton agar (Merck, Germany) according to Clinical and Laboratory Standards Institute (CLSI) instructions [15]. Inoculated plates were incubated aerobically at 37°C for 18–24 h, after which antimicrobial susceptibility in the E. coli isolates were tested. Tetracycline (30 μg/disk), ampicillin (10 u/disk), penicillin (10 u/disk), sulfamethoxazole (25 μg/disk), streptomycin (10 μg/disk), sulfonamide (100 μg/disk), chloramphenicol (30 μg/disk), gentamicin (10 μg/disk), trimethoprim (5 μg/disk), ciprofloxacin (5 μg/disk), enrofloxacin (5 μg/disk), cephalothin (30 μg/disk), and nitrofurantoin (300 μg/disk) were tested. The results were interpreted in accordance with CLSI criteria [16]. E. coli ATCC 25922 was used as quality control for antimicrobial susceptibility determination.
DNA extraction
Bacterial strains were grown overnight in Trypticase Soy Agar (TSA, Merck, Germany) at 37°C. A single colony was suspended in 100 μL of sterile distilled water. After boiling the suspension for 13 min, the suspension was frozen and centrifuged at 14,000 rpm for 15 min to pellet the cell debris [17]. The supernatant was used as a template for PCR amplification.
PCR detection of serogroups, virulence factors and antibiotic resistance genes in STEC strains
To detect virulence factors, serogroups, and antibiotic resistance genes in the E. coli isolates, different PCR assays were used [16, 18–29]. All the PCR products were electrophoresed on 1.5% agarose gels that were strained with ethidium bromide and examined under ultraviolet illumination. E. coli O159:H20, O157:K88ac:H19, CAPM 6006, and CAPM 5933 strains were used as positive controls and distilled water was used as a negative control.
Statistical analyses
The data were analyzed using SPSS (Statistical Package for the Social Sciences) software and P values were calculated using Chi-square and Fisher's exact tests to identify statistically significant relationships between the following: patient ages, seasonal variations, patient symptoms and distribution of virulence genes, serogroups and antibiotic resistance properties of the STEC strains isolated from diarrheic and non-diarrheic pediatric patients. A P value < 0.05 was considered statistically significant.
Ethical considerations
The present study was authorized by the ethical committee of the Baqiyatallah hospital of Tehran, Iran and the Microbiology, Biotechnology and Infectious Diseases Center of the Islamic Azad University of Shahrekord Branch, Iran. All patients or their parents signed the written informed consent.
Results and discussion
All of the diarrheic and non-diarrheic stool samples were examined using culture and PCR techniques. From 308 diarrheic and non-diarrheic stool samples, 185 (60.06%) were positive for E. coli (Table 1). In addition, 154 out of 224 diarrheic stool samples (68.75%) and 31 out of 84 non-diarrheic stool samples (36.90%) were positive for E. coli. The age distribution of the pediatric patients with regard to infection with E. coli is shown in Table 1. We found that the 13–24 month-old patients had the highest incidence of E. coli (77.63%), while the 49–60 month-old children had the lowest incidence (57.57%).
The distribution of virulence genes in the E. coli subtypes isolated from diarrheic and non-diarrheic pediatric patients is shown in Table 2. We found that stx1 and eaeA were the most commonly detected virulence genes in stool samples from both groups of children. The Attaching and Effacing E. coli (AEEC) subtype was the most commonly detected. The EHEC subtype was only detected in the 13–24, 25–36, and 49–60 month-old patients.
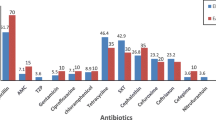
Table 3 shows the incidence of STEC serogroups isolated from diarrheic and non-diarrheic patients. O26 (27.04%) had the highest incidence, followed by O111 (18.85%). We also found that 13–24 month-old patients had the highest incidence of the STEC serogroups. The distribution of antimicrobial resistance genes within the STEC serogroup isolated from diarrheic and non-diarrheic pediatric patients is shown in Table 4. Genes that encode resistance to sulfonamide, gentamicin, trimethoprim, cephalothin and tetracycline antibiotics, i.e., sul1 (82.78%), aac(3)-IV (68.03%), aadA1 (60.65%), blaSHV (56.55%) and tetA (51.63%) were the most common antibiotic resistance genes in the diarrheic and non-diarrheic patients. Interestingly, we found that O26 had the highest frequency of antibiotic resistance genes. Antimicrobial resistance in the STEC serogroups isolated from the diarrheic and non-diarrheic patients is shown in Table 5. STEC strains exhibited the highest level of resistance to penicillin (100%), followed by tetracycline (86.88%), gentamicin (62.29%) and streptomycin (54.91%). Descriptions of the seasonal profiles and clinical signs in diarrheic children for each serogroup are shown in Table 6. Samples that were collected in the summer had the highest incidence of STEC serogroups, while those collected in autumn had the lowest incidence. Children with fever symptom had the highest incidence of STEC serogroups. Distribution of antimicrobial resistance pattern of STEC strains in various clinical samples are shown in Table 7. We found that the STEC strains of dysentery and fever samples had the highest levels of antibiotic resistance. The highest levels of antibiotic resistance of the STEC strains isolated from children with dysentery were found against streptomycin, sulfamethoxazole, gentamicin, cephalothin, nitrofurantoin and ampicillin.
Our work has identified marked seasonality in the incidence of STEC serogroup strains in diarrheic and non-diarrheic pediatric patients. There were significant differences (P =0.041) in the incidence of STEC serogroup strains between the hot and cold seasons of the year. One possible explanation for the high prevalence of STEC serogroups in summer in Iran is that climatic variables such as heat, rain and thunderstorms, together with variable barometric pressure may have affected the patients’ autonomic nervous systems. These variables could affect immunity, thus making people more susceptible to infections. Alternatively, the higher prevalence of STEC serogroups may be related to higher growth rates in the bacteria. Therefore, the highest levels of pediatric health care should be performed during the warmer months of year. Of the studies that have been conducted [30, 31] in this field, all have shown a seasonal distribution for E. coli with the highest numbers of cases occurring during the warmer months of the year [30, 31].
Data from 1996 to 2011 shows a seasonal increase in cases from April to November in Louisiana, USA [30]. Our results show that fever was most commonly associated with the highest incidence of STEC serogroup types (Table 7). This finding is in accordance with the results of Rivas et al. [32].
The most commonly infected group was 13–24 month-old age group (77.63%), but there were no significant differences in the incidence of STEC strains among the various age groups. In addition, 68.75% of diarrheic and 36.90% of non-diarrheic stool samples from the children were positive for E. coli strains; hence, the diarrheic patients showed the higher incidence of E. coli (P = 0.022). Lower incidence of STEC strains in Iranian diarrheic children were reported previously by Salmanzadeh-Ahrabi et al. [33] (15.5% incidence rate in pediatrics under 5 years old) and Alikhani et al. [34] (8.7% incidence rate in pediatrics under 10 years old).
Another important finding relates to the distributions of four bacterial virulence factors (stx1, stx2, ehly and eaeA) in the diarrheic and non-diarrheic patients. We found statistically significant (P =0.016) association between the incidence of stx1 and stx2 genes in all groups and between the EHEC and AEEC subtypes (P =0.045). In addition, there were significant differences amongst the incidence of virulence factors in diarrheic and non-diarrheic patients (P =0.047). Sang et al. [35] reported that 141 out of 380 diarrheic patients (37.1%) were positive for E. coli and of these, ETEC, STEC, EAEC, EIEC and EPEC strains comprised 29.8%, 24.1%, 14.2% 12.8% and 3.5%, respectively. The presence of multiple stx1, eaeA, and ehly genes was found in the EHEC strains isolated from 13–24 (100%), 25–36 (100%) and 49–60 month-old (100%) patients. Additionally, 13.33% of non- diarrheic, 11.11% of 49–60 month-old diarrheic, 8.33% of 37–48 month-old diarrheic, 11.76% of 25–36 month-old diarrheic, 10.25% of 13–24 month-old diarrheic and 8.33% of 1–12 month-old diarrheic patients harbored stx1, stx2 and eaeA genes. Similar findings have been reported in various parts of the world including Kenya, Korea, Australia and Brazil [35–38]. Of several studies which were conducted in this field in Iran. Shams et al. [39] showed that 2.8% of STEC isolates were positive for stx1 or stx2 genes, Aslani and Bouzari [40] showed that 96.5% of STEC isolates had stx1 gene, 44.8% had hly gene, 3.5% had stx2, and 24.1% had ast1 gene and Bonyadian et al. [41] reported that 27.6% of STEC isolates had stx1 gene, 20.7% had hly gene and 6.9% had stx2 which all was lower than our results. These studies indicate that the stx1, stx2, hly and eaeA genes are only present in some of the groups studied and that the eaeA and ehly genes are associated with diarrhea. Within the disease-associated strains, those containing stx1 and stx2 (especially stx2) appear to be more commonly responsible for serious complications bloody and non-bloody diarrhea and HUS than those containing other putative bacterial pathogenic genes [42, 43].
O26 was the most commonly detected serogroup of all of the diarrheic and non-diarrheic STEC strains. Statistical analyses showed a significant (P =0.028) association between the incidence of the O26 serogroup and other STEC serogroups in all of the study groups. High differences in the incidence of STEC serogroups between diarrheic and non-diarrheic patients was reported previously [44, 45]. Aslani and Alikhani [46] reported that O142, O111, O86 and O127 were the most commonly detected serogroups in Iranian diarrheic children under 5 years of age which was in contrast with our results. High presence of O55, O26, O111, O125, O128, O114, O86, O142, O119, O127, and O126 serogroups was reported in the STEC strains of Alikhani et al. investigation [47].
In two studies involving 11 sites in the USA in 1997 and four sites during 1998–1999, screening of 13,798 stool samples for STEC yielded a frequency of 0.9% (120 cases of STEC), and of these 120 cases, from the isolates available, 54% were O157 STEC, while 46% were non-O157 STEC [45]. Many studies have shown that O26 is the most clinically important genotype within the STEC serogroup isolated from diarrheic and even non-diarrheic patients [48, 49] because of its association with bloody diarrhea, non-bloody diarrhea and HUS.
Because inappropriate prescriptions of antibiotics causes antibiotic resistance, it was not surprising that our study found that resistance to penicillin was 100%, while resistance to tetracycline, gentamicin and streptomycin were 86.88%, 62.29%, 54.91%, respectively. In terms of bacterial antibiotic resistance genes, sul1 (82.78%), aac(3)-IV (68.03%), aadA1 (60.65%), blaSHV (56.55%) and tetA (51.63%) were the most commonly detected. There were statistically significant differences (P =0.029) amongst the incidences of sul1, cat1 and cmlA and among aac(3)-IV, cat1 and cmlA (P =0.041). Our results showed that the O26 serogroup had the highest incidence of antibiotic resistance genes and resistance to antibiotics. Our results showed that 1.63% of the STEC strains were resistance to chloramphenicol. Chloramphenicol is a banned antibiotic and the slight antibiotic resistance to this drug detected in our study indicates that irregular and unauthorized use of it may have occurred in Iran. Similar chloramphenicol resistance profiles to our own have been reported previously [50, 51]. Fazeli and Salehi [52] showed that 72.4%, 65.5% and 58.6% of STEC strains isolated from Iranian diarrheal patients were resistant to amoxicillin, trimethoprim-sulfamethoxazole and tetracycline, respectively.
There were statistically significant differences (P =0.040) amongst the incidences of antibiotic resistance in the STEC strains isolated from dysentery, fever, other clinical signs and non-diarrheic samples. The highest incidence of antimicrobial resistance in non-O157 STEC strains [9] was found for sulfisoxazole (36%), tetracycline (32%), streptomycin (29%), ampicillin (10%), trimethoprim (8%), co-trimoxazole (8%), chloramphenicol (7%), kanamycin (7%), piperacillin (6%) and neomycin (5%); these values are lower than the values reported in the present study. Jafari et al. [13] showed that diarrheagenic E. coli isolates of Iran had high resistance against amoxicillin and tetracycline (75.5%) which was similar to our results. Similar Iranian investigations have been done previously [52–54]. Käppeli et al. [8] reported that the STEC antibiotic resistance rates against ampicillin, amoxicillin/clavulanic acid, cephalothin, cefpodoxime, cefuroxime, gentamicin, and tetracycline were 3.1%, 12.4%, 1%, 1%, 2.1%, and 21.6%, respectively which was similar to our findings.
Our results showed that resistance to at least one of the 12 antimicrobial drugs tested was identified in all (100%) of the STEC strains, which is consistent with previous studies [8–10]. Our antimicrobial resistance data are in accordance with the results from a study in Spain, in which 238 (41%) of 581 non-O157 STEC strains were resistant to at least one out of the 26 antimicrobial drugs that were tested [9].
The above data highlight large differences in the prevalence of STEC strains in the different studies, as well as differences in virulence genes and antibiotic resistance properties in the clinical samples. This could be related to differences in the type of sample (stool, blood, urine, meat, milk, vegetable and other clinical samples) tested, number of samples, method of sampling, experimental methodology, geographical area, antibiotic prescription preference among clinicians, antibiotic availability, and climate differences in the areas where the samples were collected, which would have differed between each study.
Conclusions
In conclusion, we identified a large number of virulence factors, serogroups, and antibiotic resistance genes and resistance to more than one antibiotic in the STEC strains isolated from diarrheic and non-diarrheic patients. Our data indicate that O157 and especially non-O157 STEC strains are predominant in Iranian diarrheic children. Marked seasonal variation in the serogroup distribution was also found. Our data revealed that the O26 serogroup, the stx1, stx2, eaeA and ehly putative virulence genes, the sul1, aac(3)-IV, aadA1, blaSHV and tetA antibiotic resistance genes, and resistance to penicillin, tetracycline, gentamicin and streptomycin were the most commonly detected characteristics of the STEC strains isolated from Iranian diarrheic and non-diarrheic children. Hence, judicious use of antibiotics is required by clinicians.
Abbreviations
- E. coli:
-
Escherichia coli
- PCR:
-
Polymerase chain reaction
- EHEC:
-
Enterohaemorrhagic Escherichia coli
- AEEC:
-
Attaching and effacing Escherichia coli
- STEC:
-
Shiga toxin-producing Escherichia coli
- SPSS:
-
Statistical package for the social sciences
- CLSI:
-
Clinical and laboratory standards Institute.
References
Thorpe CM: Shiga toxin-producing Escherichia coli infection. Clin Infect Dis. 2004, 38 (9): 1298-1303. 10.1086/383473.
Karch H, Tarr PI, Bielaszewska M: Enterohaemorrhagic Escherichia coli in human medicine. Int J Med Microbiol. 2005, 295 (6–7): 405-418.
Wong CS, Jelacic S, Habeeb RL, Watkins SL, Tarr PI: The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N Engl J Med. 2000, 342 (26): 1930-1936. 10.1056/NEJM200006293422601.
Tarr PI, Gordon CA, Chandler WL: Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005, 365 (9464): 1073-1086.
Bielaszewska M, Friedrich AW, Aldick T, Schürk-Bulgrin R, Karch H: Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin Infect Dis. 2006, 43 (9): 1160-1167. 10.1086/508195.
Law D: Virulence factors of Escherichia coli O157 and other Shiga toxin-producing. E. coli. J Appl Microbiol. 2000, 88 (5): 729-745.
Schmidt H, Beutin L, Karch H: Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995, 63 (3): 1055-1061.
Käppeli U, Hächler H, Giezendanner N, Beutin L, Stephan R: Human infections with non-O157 Shiga toxin-producing Escherichia coli, Switzerland, 2000–2009. Emerg Infect Dis. 2011, 17 (2): 180-185. 10.3201/eid1702.100909.
Mora A, Blanco JE, Blanco M, Alonso MP, Dhabi G, Echeita A, González EA, Bernárdez MI, Blanco J: Antimicrobial resistance of Shiga toxin (verotoxin)-producing Escherichia coli O157:H7 and non-O157 strains isolated from humans, cattle, sheep and food in Spain. Res Microbiol. 2005, 156 (7): 793-806. 10.1016/j.resmic.2005.03.006.
Schroeder CM, Meng J, Zhao S, DebRoy C, Torcolini J, Zhao C, McDermott PF, Wagner DD, Walker RD, White DG: Antimicrobial resistance of Escherichia coli O26, O103, O111, O128, and O145 from animals and humans. Emerg Infect Dis. 2002, 8 (12): 1409-1414. 10.3201/eid0812.0200770.
Zali MR, Moez Ardalan K, Parcham Azad K, Nik-Kholgh B: Etiologies of acute diarrheal diseases in Iran. J Res Med Sci. 2003, 7 (4): 346-356.
Rezaie Homami M, Salmanzadeh Ahrabi S, Moez Ardalan K, Habibi E, Edalatkhah H, Jafari F, Moez Ardalan S, Zolfagharian K, Moghaddam Golmohammadi A, Azimi Rad M: Epidemiology of bacterial-induced acute diarrhea in Varamin. Pejouhandeh Q Res J. 2003, 8 (7): 467-474.
Jafari F, Hamidian M, Rezadehbashi M, Doyle M, Salmanzadeh-Ahrabi S, Derakhshan F, Reza Zali M: Prevalence and antimicrobial resistance of diarrheagenic Escherichia coli and Shigella species associated with acute diarrhea in Tehran, Iran. Can J Infect Dis Med Microbiol. 2009, 20 (3): e56-62.
Sabat G, Rose P, Hickey WJ, Harkin JM: Selective and sensitive method for PCR amplification of Escherichia coli 16S rRNA genes in soil. Appl Environ Microbiol. 2000, 66 (2): 844-849. 10.1128/AEM.66.2.844-849.2000.
Clinical and Laboratory Standards Institute (CLSI): Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved standard-Ninth Edition (M2-A9). Wayne, PA: Clinical and Laboratory Standards Institute, 2006.
Heuvelink AE, van de Kar NC, Meis JF, Monnens LA, Melchers WJ: Characterization of verocytotoxin-producing Escherichia coli O157 isolates from patients with haemolytic uraemic syndrome in Western Europe. Epidemiol Infect. 1995, 115 (1): 1-14. 10.1017/S0950268800058064.
Reischl U, Youssef MT, Kilwinski J, Lehn N, Zhang WL, Karch H, Strockbine NA: Real-time fluorescence PCR assays for detection and characterization of Shiga toxin, intimin, and enterohemolysin genes from Shiga toxin-producing Escherichia coli. J Clin Microbiol. 2002, 40 (7): 2555-2565. 10.1128/JCM.40.7.2555-2565.2002.
Brian MJ, Frosolono M, Murray BE, Miranda A, Lopez EL, Gomez HF, Cleary TG: Polymerase chain reaction for diagnosis of enterohemorrhagic Escherichia coli infection and hemolytic-uremic syndrome. J Clin Microbiol. 1992, 30 (7): 1801-1806.
Idress M, Mussarat U, Badshah Y, Qamar R, Bokhari H: Virulence factors profile of drug-resistant Escherichia coli isolates from urinary tract infections in Punjab, Pakistan. Eur J Clin Microbiol Infect Dis. 2010, 29 (12): 1533-1537. 10.1007/s10096-010-1036-6.
Possé B, De Zutter L, Heyndrickx M, Herman L: Metabolic and genetic profiling of clinical O157 and non-O157 Shiga-toxin-producing Escherichia coli. Res Microbiol. 2007, 158 (7): 591-599. 10.1016/j.resmic.2007.06.001.
DebRoy C, Fratamico PM, Roberts E, Davis MA, Liu Y: Development of PCR assays targeting genes in O-antigen gene clusters for detection and identification of Escherichia coli O45 and O55 serogroups. Appl Environ Microbiol. 2005, 71 (8): 4919-4924. 10.1128/AEM.71.8.4919-4924.2005.
Perelle S, Dilasser F, Grout J, Fach P: Identification of the O-antigen biosynthesis genes of Escherichia coli O91 and development of a O91 PCR serotyping test. J Appl Microbiol. 2002, 93 (5): 758-764. 10.1046/j.1365-2672.2002.01743.x.
DebRoy C, Roberts E, Kundrat J, Davis MA, Briggs CE, Fratamico PM: Detection of Escherichia coli serogroups O26 and O113 by PCR amplification of the wzx and wzy genes. Appl Environ Microbiol. 2004, 70 (3): 1830-1832. 10.1128/AEM.70.3.1830-1832.2004.
Fratamico PM, Briggs CE, Needle D, Chen CY, DebRoy C: Sequence of the Escherichia coli O121 O-antigen gene cluster and detection of enterohemorrhagic E. coli O121 by PCR amplification of the wzx and wzy genes. J Clin Microbiol. 2003, 41 (7): 3379-3383. 10.1128/JCM.41.7.3379-3383.2003.
Shao J, Li M, Jia Q, Lu Y, Wang PG: Sequence of Escherichia coli O128 antigen biosynthesis cluster and functional identification of an alpha-1,2-fucosyltransferase. FEBS Lett. 2003, 553 (1–2): 99-103.
Randall LP, Cooles SW, Osborn MK, Piddock LJ, Woodward MJ: Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J Antimicrob Chemother. 2004, 53 (2): 208-216. 10.1093/jac/dkh070.
Toro CS, Farfán M, Contreras I, Flores O, Navarro N, Mora GC, Prado V: Genetic analysis of antibiotic-resistance determinants in multidrug-resistant Shigella strains isolated from Chilean children. Epidemiol Infect. 2005, 133 (1): 81-86. 10.1017/S0950268804003048.
Mammeri H, Van De Loo M, Poirel L, Martinez-Martinez L, Nordmann P: Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob Agents Chemother. 2005, 49 (1): 71-76. 10.1128/AAC.49.1.71-76.2005.
Van TT, Chin J, Chapman T, Tran LT, Coloe PJ: Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int J Food Microbiol. 2008, 124 (3): 217-223. 10.1016/j.ijfoodmicro.2008.03.029.
Louisiana Office of Public Health: Infectious disease epidemiology section. E. coli-O157. 2012, H7: 3.
García-Aljaro C, Muniesa M, Jofre J, Blanch AR: Prevalence of the stx2 gene in coliform populations from aquatic environments. Appl Environ Microbiol. 2004, 70 (6): 3535-3540. 10.1128/AEM.70.6.3535-3540.2004.
Rivas M, Miliwebsky E, Balbi L, García B, Leardini N, Tous M, Chillemi G, Baschkier A, Strugo L: Intestinal bleeding and occlusion associated with Shiga toxin-producing Escherichia coli O127:H21. Med (B Aires). 2000, 60 (2): 249-252.
Salmanzadeh-Ahrabi S, Habibi E, Jaafari F, Zali MR: Molecular epidemiology of Escherichia coli diarrhoea in children in Tehran. Ann Trop Paediatr. 2005, 25 (1): 35-39. 10.1179/146532805X23335.
Alikhani MY, Mirsalehian A, Fatollahzadeh B, Pourshafie MR, Aslani MM: Prevalence of enteropathogenic and shiga toxin-producing Escherichia coli among children with and without diarrhoea in Iran. J Health Popul Nutr. 2007, 25 (1): 88-93.
Sang WK, Boga HI, Waiyaki PG, Schnabel D, Wamae NC, Kariuki SM: Prevalence and genetic characteristics of Shigatoxigenic Escherichia coli from patients with diarrhoea in Maasailand. Kenya. J Infect Dev Ctries. 2012, 6 (2): 102-108.
Kim YJ, Kim JH, Hur J, Lee JH: Isolation of Escherichia coli from piglets in South Korea with diarrhea and characteristics of the virulence genes. Can J Vet Res. 2010, 74 (1): 59-64.
Paton AW, Paton JC: Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998, 36 (2): 598-602.
Scaletsky IC, Aranda KR, Souza TB, Silva NP, Morais MB: Evidence of pathogenic subgroups among atypical enteropathogenic Escherichia coli strains. J Clin Microbiol. 2009, 47 (11): 3756-3759. 10.1128/JCM.01599-09.
Shams S, Haghi Ashtiani MT, Nasrollahi L, Shahsiah R, Monajemzadeh M, Tahbaz Lahafi B, Alaie Alamooti A: Frequency of Shiga toxin-producing genes of Escherichia coli isolated from diarrheic stools of Iranian children by PCR. Iranian J Pediatr. 2013, 23 (6): 637-642.
Aslani MM, Bouzari S: Characterization of virulence genes of non-O157 Shiga toxin-producing Escherichia coli isolates from two provinces of Iran. Jpn J Infect Dis. 2009, 62 (1): 16-19.
Bonyadian M, Momtaz H, Rahimi E, Habibian R, Yazdani A, Zamani M: Identification & characterization of Shiga toxin-producing Escherichia coli isolates from patients with diarrhoea in Iran. Indian J Med Res. 2010, 132 (3): 328-331.
Kleanthous H, Smith HR, Scotland SM, Gross RJ, Rowe B, Taylor CM, Milford DV: Haemolytic uraemic syndromes in the British isles, 1985–8: association with verocytotoxin producing Escherichia coli. Part 2: microbiological aspects. Arch Dis Child. 1990, 65 (7): 722-727. 10.1136/adc.65.7.722.
Ostroff SM, Tarr PI, Neill MA, Lewis JH, Hargrett-Bean N, Kobayashi JM: Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J Infect Dis. 1989, 160 (6): 994-998. 10.1093/infdis/160.6.994.
Johnson KE, Thorpe CM, Sears CL: The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin Infect Dis. 2006, 43 (12): 1587-1595. 10.1086/509573.
Andreoli SP, Trachtman H, Acheson DW, Siegler RL, Obrig TG: Hemolytic uremic syndrome: epidemiology, pathophysiology, and therapy. Pediatr Nephrol. 2002, 17 (4): 293-298. 10.1007/s00467-001-0783-0.
Aslani MM, Alikhani MY: Serotypes of Enteropathogenic Escherichia coli isolated from children under 5 years of age. Iranian J Publ Health. 2009, 38 (3): 70-77.
Alikhani MY, Mirsalehian A, Aslani MM: Detection of typical and atypical enteropathogenic Escherichia coli (EPEC) in Iranian children with and without diarrhoea. J Med Microbiol. 2006, 55 (Pt 9): 1159-1163.
Elliott EJ, Robins-Browne RM, O'Loughlin EV, Bennett-Wood V, Bourke J, Henning P, Hogg GG, Knight J, Powell H, Redmond D: Nationwide study of haemolytic uraemic syndrome: clinical, microbiological, and epidemiological features. Arch Dis Child. 2001, 85 (2): 125-131. 10.1136/adc.85.2.125.
Rivas M, Miliwebsky E, Chinen I, Roldán CD, Balbi L, García B, Fiorilli G, Sosa-Estani S, Kincaid J, Rangel J, Griffin PM: Characterization and epidemiologic subtyping of Shiga toxin-producing Escherichia coli strains isolated from hemolytic uremic syndrome and diarrhea cases in Argentina. Foodborne Pathog Dis. 2006, 3 (1): 88-96. 10.1089/fpd.2006.3.88.
Momtaz H, Karimian A, Madani M, Safarpoor Dehkordi F, Ranjbar R, Sarshar M, Souod N: Uropathogenic Escherichia coli in Iran: serogroup distributions, virulence factors and antimicrobial resistance properties. Ann Clin Microbiol Antimicrob. 2013, 12: 8-10.1186/1476-0711-12-8.
Momtaz H, Safarpoor Dehkordi F, Rahimi E, Ezadi H, Arab R: Incidence of Shiga toxin-producing Escherichia coli serogroups in ruminant's meat. Meat Sci. 2013, 95 (2): 381-388. 10.1016/j.meatsci.2013.04.051.
Fazeli H, Salehi R: Antibiotic resistance pattern in shiga toxin producing Escherichia coli isolated from diarrheal patients in Al-Zahra Hospital, Isfahan, Iran. Res Pharmacoceut Sci. 2007, 2 (1): 29-33.
Kalantar E, Alikhani MY, Naseri MH, Torabi V: Antibiotic resistance patterns of STEC and ETEC strains: a study on frozen foods of animal origin and children with acute diarrhea. J Microbiol Infect Dis. 2013, 3 (1): 31-35.
Alikhani MY, Hashemi SH, Aslani MM, Farajnia S: Prevalence and antibiotic resistance patterns of diarrheagenic Escherichia coli isolated from adolescents and adults in Hamedan, West Iran. Iran J Microbiol. 2013, 5 (1): 42-47.
Acknowledgements
This work was supported by the Islamic Azad University, Shahrekord Branch-Iran. The authors would like to thank M. Dodi at the Microbiology Research Center of the Islamic Azad University of Flavarjan, and Dr. E. Tajbakhsh and Mr. M. Momeni at the Biotechnology Research Center of the Islamic Azad University of Shahrekord for their important technical and clinical support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DNA extraction, PCR, manuscript preparation, statistical analysis and project support were all performed by HM and FSD. Sample collections and coordination was performed by MJH, MH and MS. All authors have read and approved the final manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Momtaz, H., Dehkordi, F.S., Hosseini, M.J. et al. Serogroups, virulence genes and antibiotic resistance in Shiga toxin-producing Escherichia coli isolated from diarrheic and non-diarrheic pediatric patients in Iran. Gut Pathog 5, 39 (2013). https://doi.org/10.1186/1757-4749-5-39
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1757-4749-5-39