Abstract
Background
It is important to understand the mechanisms by which the cells integrate signals from different receptors. Several lines of evidence implicate epidermal growth factor (EGF) receptor (EGFR) in the pathophysiology of hepatocarcinomas. Data also suggest a role of prostaglandins in some of these tumours, through their receptors of the G protein-coupled receptor (GPCR) family. In this study we have investigated mechanisms of interaction between signalling from prostaglandin receptors and EGFR in hepatocarcinoma cells.
Methods
The rat hepatocarcinoma cell line MH1C1 and normal rat hepatocytes in primary culture were stimulated with EGF or prostaglandin E2 (PGE2) and in some experiments also PGF2α. DNA synthesis was determined by incorporation of radiolabelled thymidine into DNA, phosphorylation of proteins in signalling pathways was assessed by Western blotting, mRNA expression of prostaglandin receptors was determined using qRT-PCR, accumulation of inositol phosphates was measured by incorporation of radiolabelled inositol, and cAMP was determined by radioimmunoassay.
Results
In the MH1C1 hepatocarcinoma cells, stimulation with PGE2 or PGF2α caused phosphorylation of the EGFR, Akt, and ERK, which could be blocked by the EGFR tyrosine kinase inhibitor gefitinib. This did not occur in primary hepatocytes. qRT-PCR revealed expression of EP1, EP4, and FP receptor mRNA in MH1C1 cells. PGE2 stimulated accumulation of inositol phosphates but not cAMP in these cells, suggesting signalling via PLCβ. While pretreatment with EP1 and EP4 receptor antagonists did not inhibit the effect of PGE2, pretreatment with an FP receptor antagonist blocked the phosphorylation of EGFR, Akt and ERK. Further studies suggested that the PGE2-induced signal was mediated via Ca2+ release and not PKC activation, and that it proceeded through Src and shedding of membrane-bound EGFR ligand precursors by proteinases of the ADAM family.
Conclusion
The results indicate that in MH1C1 cells, unlike normal hepatocytes, PGE2 activates the MEK/ERK and PI3K/Akt pathways by transactivation of the EGFR, thus diversifying the GPCR-mediated signal. The data also suggest that the underlying mechanisms in these cells involve FP receptors, PLCβ, Ca2+, Src, and proteinase-mediated release of membrane-associated EGFR ligand(s).
Similar content being viewed by others
Background
Malignant cells are exposed to a variety of active agents, including hormones, peptide growth factors, cytokines, and many other locally acting substances such as prostaglandins, which together control or modulate the cellular functions. It is of interest to understand the mechanisms by which the cells integrate signals from different bioactive molecules via their receptors. A notable example is the interaction between pathways from G protein-coupled receptors (GPCRs) and receptor tyrosine kinases (RTKs). Studies in many cells have shown that signals from GPCRs may involve interaction with the epidermal growth factor receptor (EGFR), an ErbB family RTK [1–5]. EGFR, which serves important functions in normal cells [6, 7], is involved in several malignancies [8, 9], and is a target of novel antitumour therapies [10, 11]. In studies including tumour cells from colon and pancreatic cancer, we have found that different mechanisms may be involved in the interaction of pathways from GPCRs and EGFR [12].
EGFR conveys strong mitogenic stimulation in normal hepatocytes [13–16], and several lines of evidence suggest a role of EGFR in hepatocarcinogenesis [17–20]. For example, overexpression of the EGFR agonist transforming growth factor alpha (TGFα) in mice causes hepatic hyperplasia and tumour formation [21, 22], and EGFR is upregulated in a majority of human hepatocarcinomas [23]. Inhibition of the EGFR by antibodies or tyrosine kinase blockers can attenuate the growth of hepatocarcinoma cells in vitro [24, 25], and are currently being tested in clinical trials in hepatocarcinomas [26].
Prostaglandins, acting through different receptors of the GPCR family, regulate many cellular functions [27]. In epithelial cells, prostaglandins often enhance proliferation and survival, and several lines of evidence implicate them in oncogenesis [28]. In many tumours, cyclooxygenases (COX-1 and COX-2), which catalyze the rate-limiting step in prostaglandin synthesis, are overexpressed, and the levels of prostaglandins, notably prostaglandin E2 (PGE2), are elevated [28–31]. In hepatocytes, PGE2 and other prostaglandins enhance DNA synthesis [15, 32–34], and COX-2 is overexpressed in many hepatocarcinomas [35, 36].
In the study presented here we examined the Morris hepatocarcinoma cell line MH1C1, which was chosen due to its responsiveness to both EGF and the prostaglandins PGE2 and PGF2α, and investigated the interaction between the pathways mediated by prostaglandin receptors and EGFR. We previously observed that while there was no evidence of transactivation of EGFR induced by prostaglandins or other GPCR agonists in hepatocytes, PGE2 induced phosphorylation of the EGFR in the MH1C1 cells [37, 38]. We have now investigated further the signalling mechanisms involved in this effect.
Methods
Chemicals
Dulbecco’s Modified Eagle's Medium, Dulbecco’s phosphate-buffered saline, William’s Medium E, glutamine, and Pen-Strep (10.000 U/ml) were from Lonza(Verviers, Belgium). HEPES was from Gibco (Grand Island, NY). Dexamethasone, insulin, bovine serum albumin, collagen (type I, rat tail), prostaglandin F2α (Tris salt) and epidermal growth factor (EGF) were obtained from Sigma-Aldrich (St.Louis, MO). GF109203X ([2-[1-(3-dimetylaminopropyl)-1 H-indol-3-yl]-male-imide]) and GM6001/Galardin (N-[(2R)-2 (hydroxamidocarbonylmethyl)-4-methylpentanoyl]-L-tryptophan methylamide) were from Calbiochem (San Diego, CA). Gefitinib was a gift from AstraZeneca (Cheshire, UK). [6-3 H]thymidine (20–30 Ci/mmol) and myo-[2-3 H]inositol (15.0 Ci/mmol) were from PerkinElmer (Boston, MA). AL8810 (9α,15R-dihydroxy-11β-fluoro-15-(2,3-dihydro-1 H-inden-2-yl)-16,17,18,19,20-pentanor-prosta-5Z,13E-dien-1-oic acid),L161982 (N-[[4'-[[3-butyl-1,5-dihydro-5-oxo-1-[2-(trifluoromethyl)phenyl]-4 H-1,2,4-triazol-4-yl]methyl][1,1'-biphenyl]-2-yl]sulfonyl]-3-methyl-2-thiophenecarboxamide), (+)fluprostenol, and prostaglandin E2 (PGE2) were from Cayman Chemical (Ann Arbor, MI). SC51322 (8-chloro-2-[3-[(2-furanylmethyl)thio]-1-oxopropyl]hydrazide, dibenz[b,f][1,4]oxazepine-10(11 H)-carboxylic acid) was obtained from BIOMOL Research Laboratories (Plymouth Meeting, PA). The Src inhibitor CGP77675 was a gift from Novartis Pharma AG (Basel, Switzerland). All other chemicals were of analytical quality.
Antibodies against phosphorylated AktSer473, total Akt, dually phosphorylated ERKThr202/Tyr204, GAPDH and phospho-ShcTyr239/240 were obtained from Cell Signaling Technology (Boston, MA). Antibody against phospho-EGF receptorTyr1173 was obtained from Invitrogen. Anti-ERK antibody was from Upstate/Millipore (Billerica, MA). Secondary antibodies were purchased from Bio-Rad Laboratories (Hercules, CA) and Licor Biosciences (Lincoln, NE).
Cells and culturing
The rat hepatocarcinoma cell line MH1C1, derived from a Morris hepatoma [39], was obtained from ATCC (Manassas, VA). The cells were seeded onto Costar plastic flasks and cultured in Dulbecco’s Modified Eagle’s medium. The Medium was supplemented with horse serum (10%), glutamine (2 mM), and 100 U/ml Pen-Strep. The cultures were kept in a humidified 5% CO2 incubator at 37°C. Cells were seeded onto culture wells at a density of 40 000–50 000 cells per cm2. After 24 hours, the medium was changed and the cells were cultured under serum-free conditions 24 h prior to stimulation.
Hepatocytes were isolated from male Wistar rats as previously described [40]. The hepatocytes were seeded onto Costar plastic culture wells at a density of 15 000–20 000 per cm2. The culture medium was a serum-free 1:1 combination of William’s Medium E and Dulbecco’s Modified Eagle’s Medium. The medium was supplemented with 100 U/ml Pen-Strep, collagen (3 μg/ml), insulin (100 nM) and dexamethasone (25 nM).
Immunoblotting
Aliquots containing ~30000 MH1C1 cells or hepatocytes (total cell lysate prepared in Laemmli or RIPA buffer) were electrophoresed on 6–12% (w/v) polyacrylamide gels (acrylamide: N’N’-bis-methylene acrylamide 30:1). This was followed by protein electrotransfer to nitrocellulose membranes and immunoblotting with antibodies against proteins as described in the figures. Usually the same membrane was stripped and reincubated with different antibodies, and then one single loading control was used as the final incubation. Immunoreactive bands were visualized with enhanced chemiluminescence using LumiGLO (KPL Protein research Products, Gaithersburg, MD) or by infrared imaging using Odyssey Infrared Imaging System, supplied by Licor Biosciences (Lincoln, NE).
RNA isolation and cDNA synthesis
RNA from MH1C1 cells was isolated with Qiagen RNeasy kit according to the manufacturer’s instructions, and was treated with DNAse. The integrity of RNA was evaluated by ethidium bromide agarose gel electrophoresis, and the quantity and purity was measured spectrophotometrically (OD 260/280). cDNA was synthesized from 1.0 μg RNA with Superscript® III reverse transcriptase (Invitrogen) according to manufacturer’s protocol. Reactions without reverse transcriptase were run in parallel to control for contamination with chromosomal DNA. Standard curves with RNA ranging from 0.25 to 2.0 μg of total RNA were made to control for the reverse transcription and PCR quantification.
Quantitative real-time PCR (qRT-PCR)
The cDNA was analyzed in triplicate by real time quantitative PCR on an ABI Prism 7900 HT Sequence detector (Applied Biosystems) with the following cycling parameters: 50°C for 2 min, 95°C for 10 min and 40 cycles of 15 s at 95°C and 60 s at 60°C, followed by melting point analysis when using SYBR green. Raw data were collected and analyzed in the Sequence Detector Software (SDS ver. 2.2, Applied Biosystems), and cycle of threshold value (Ct) was calculated from each amplification plot. Standard curves (Ct value versus log initial RNA concentration) were used to calculate the relative input amount of RNA for each sample based on the Ct value [41]. Satisfactory and comparable amplification efficiency was verified by the slopes of standard curves. Primers were designed using Primer Express® software v2.1 (ABI Prism, Applied Biosystems), and were validated by the production of single products of expected size on agarose gels, as well as uniformity of melting temperature, which was routinely performed. Prostaglandin receptor cDNA was detected with SYBR Green methodology and the following primers: EP1: forward 5’-CCT GCT GGT ATT GGT GGT GTT-3’ and reverse 5’-GGG GTA GGA GGC GAA GAA GTT-3’; EP2: forward 5’-GCT CCC TGC CTT TCA CAA TCT-3’ and reverse 5’-GGA CTG GTG GTC TAA GGA TGA CA-3’; EP3: forward 5’-GGT CGC CGC TAT TGA TAA TGA T-3’ and reverse 5’-CAG GCG AAC GGC GAT TAG-3‘; EP4: forward 5’-CTC GTG GTG CGA GTG TTC AT-3’ and reverse 5’-TGT AGA TCC AAG GGT CCA GGA T-3’; FP: forward 5’-GTC ATT CAG CTC CTG GCC ATA-3’ and reverse 5’-AGC GTC GTC TCA CAG GTC ACT-3’. GAPDH cDNA was quantified using the dual hybridization probe Double Dye oligonucleotide 5’ labelled with the fluorescent dye Yakima yellow and quenched with Dark Quencher, 5’-CTC ATG ACC ACA GTC CAT GCC ATC ACT-3’ and the following primers: forward 5’-CCA AGG TCA TCC ATG ACA ACT T-3’ and reverse 5’-AGG GGC CAT CCA CAG TCT T-3’. Results were normalized to GADPH.
Accumulation of inositol phosphates and cAMP
3 H]inositol, 5 μCi/well was added simultaneously with the serum-free medium. 30 minutes before agonist stimulation for 30 minutes in serum-starved cells, medium was removed and replaced with Krebs-Ringer-Hepes buffer pH 7.4, containing 10 mM glucose and 15 mM LiCl. MH1C1 cells were stimulated with PGE2, fluprostenol or isoproterenol as indicated, and the reaction was stopped by removing buffer and adding 1 ml ice-cold 0.4 M perchloric acid. Samples were harvested and neutralized with 1.5 M KOH, 60 mM EDTA and 60 mM Hepes, in the presence of Universal indicator. The neutralized supernatants were applied on columns containing 1 ml Dowex AG 1-X8 resin. The columns were washed with 20 ml distilled water and 10 ml 5 mM sodium tetraborate/60 mM ammonium formate, and inositol phosphates were eluted with 10 ml 1 M ammonium formate/0.1 M formic acid. cAMP was determined by radioimmunoassay as previously described [42].
Measurement of DNA synthesis
MH1C1 cells were seeded onto culture wells, and after 24 hours, the medium was changed and the cells were cultured under serum-free conditions. 24 h after change to serum-free medium, cells were treated with various concentrations of gefitinib and harvested at 48 hours, after three hours of pulsing with 3 H]thymidine. DNA synthesis was measured as the amount of radioactivity incorporated into DNA as previously described [34].
Results
In preliminary experiments we investigated the effect of PGE2 in the rat hepatocarcinoma cell lines MH1C1, McA7777, and M4IIE, and the human hepatocarcinoma cell line HepG2. Although some of these cell lines had strong responses to EGF (data not shown), the MH1C1 were the only cells showing consistent responses to both EGF and prostaglandins, and we therefore used these cells in further experiments.
Transactivation of EGFR induced by PGE2 and PGF2α in MH1C1 cells
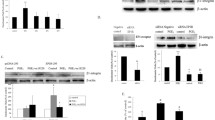
We previously observed that in the MH1C1 cells, unlike normal hepatocytes, PGE2 induced phosphorylation of the EGFR and activated ERK by a mechanism that was sensitive to EGFR inhibition [37]. Further investigation (Figure 1), showed that in addition to inducing phosphorylation of EGFR and ERK, PGE2 treatment also led to phosphorylation of Akt. All these effects were inhibited by gefitinib (1 μM) (Figure 1A), providing further support for a transactivation of EGFR in the MH1C1 cells. In contrast, the effects of PGE2 on ERK and Akt in hepatocytes were not dependent on the EGFR, since they were not inhibited by gefitinib (Figure 1B). We also observed that in the MH1C1 cells, the phosphorylation of the EGFR was somewhat slower after stimulation with PGE2 than with EGF (data not shown), suggesting an indirect mechanism consistent with PGE2-induced transactivation. As shown in Figure 1C, PGF2α also induced a gefitinib-sensitive phosphorylation of EGFR, Akt and ERK in these cells.
Effects of the EGFR inhibitor gefitinib on phosphorylation of signalling proteins and DNA synthesis. A) MH1C1 cells were treated with gefitinib (1 μM) for 30 min before stimulation with EGF (10 nM) or PGE2 (100 μM) for 5 min. B) Hepatocytes were treated with gefitinib (1 μM) for 30 min before stimulation with EGF (10 nM) or PGE2 (100 μM) for 5 min. C) Gefitinib (1 μM) was added 30 min prior to stimulation with either PGE2 (100 μM) or PGF2α (100 μM) for 5 min. Cells were harvested and subjected to SDS-PAGE followed by immunoblotting with antibodies and detection with enhanced chemiluminescence as described in Materials and Methods. All blots are representative of at least 3 independent experiments. D) Effect of gefitinib on DNA synthesis in MH1C1 cells. Increasing concentrations of gefitinib were added to serum-starved MH1C1 cells. [3 H]thymidine was added, and DNA synthesis was assessed as described under Materials and Methods. The results are presented as percent of control ± S.E.M of four independent experiments.
Figure 1D shows that the EGFR tyrosine kinase blocker gefitinib dose-dependently inhibited DNA synthesis in MH1C1, indicating that EGFR is involved in the growth in these cells. Most likely there is an autocrine release of EGFR agonist(s) in these long-term experiments (48 h culturing). This has not been explored further in the present study, as the experiments below focus on early receptor-mediated mechanisms.
Prostaglandin receptors and involvement of PLCβ
We next investigated which prostaglandin receptors are expressed in the MH1C1 cells. qRT-PCR analysis revealed mRNA expression of EP1, EP4, and FP subtypes of prostaglandin receptors, whereas only traces of EP3 receptor mRNA were present and no EP2 expression was detected (Figure 2A). The hepatocytes expressed EP2, EP3, EP4, and FP (Figure 2B).
Prostaglandin receptors and cAMP and PLCβ responses. A) and B) Expression of prostaglandin receptor mRNA in MH1C1 cells (data from three experiments, measured in triplicate) and hepatocytes (data from one experiment measured in triplicate). Quantitative RT-PCR of EP1, EP2, EP3, EP4 and FP normalized to GADPH. RNA was isolated as described in Materials and Methods. * not detected # low levels-not quantifiable. C) Left: Accumulation of cAMP in MH1C1 cells after stimulation with either PGE2 (100 μM) or isoproterenol (10 μM) in the presence of 0.5 mM IBMX. cAMP was measured after 3 minutes. Right: Accumulation of inositol phosphates in MH1C1 cells after stimulation with PGE2 (100 μM) for 30 minutes in the presence of 15 mM LiCl. The data shown are mean ± S.E.M of three independent experiments.
The available evidence indicates that the EP4 receptors are coupled to Gs proteins and adenylyl cyclase activity and thereby cAMP elevation, and that FP receptors couple to Gq proteins which mediate activation of phospholipase C-β (PLCβ) leading to formation of inositol trisphosphate (InsP3) and diacylglycerol (DAG) [27, 43]. The G proteins and signalling mechanisms stimulated by the EP1 receptors are not fully clarified [43, 44]. PGE2 has high affinity for EP1 and EP4 receptors, and while the FP receptor has the highest affinity for PGF2α, PGE2 also binds to this receptor [27]. In the MH1C1 cells no cAMP response to PGE2 could be detected, although the cells had a functional adenylyl cyclase, as shown by their marked cAMP elevation in response to the β-adrenergic agonist isoproterenol (Figure 2C left). In contrast, PGE2 stimulated accumulation of inositol phosphates (Figure 2C right). Thus, it is likely that PGE2 induces signalling through PLCβ activation in these cells.
To investigate which receptors are involved in the EGFR transactivation by PGE2, we studied the effect of pretreating the cells with selective inhibitors of different prostaglandin receptors. The results suggested that EP4 did not mediate this transactivation since the EP4 receptor antagonist L161982 did not inhibit the effect of PGE2 on the phosphorylation of EGFR, Akt, or ERK (Figure 3A), consistent with the lack of PGE2-induced cAMP response in these cells (Figure 2C). We then examined the roles of EP1 and FP receptors. Pretreatment of the cells with 10 μM of the EP1 receptor antagonist SC51322 did not affect PGE2-induced phosphorylation of EGFR, Akt, or ERK (Figure 3B). In contrast, the FP receptor antagonist AL8810 at 10 μM significantly inhibited the effect of PGE2 on the phosphorylation of ERK, while 100 μM inhibited phosphorylation of EGFR and Akt and blocked the effects on ERK almost completely (Figure 3C). These concentrations of AL8810 were not toxic to the cells. Although AL8810 is a less potent antagonist than L161982 or SC51322 [27, 45, 46], it was the only antagonist that had effect at 10 μM. It was previously shown that at 10 μM, AL8810 did not inhibit functional responses through other prostaglandin receptors, suggesting that it is a selective antagonist at the FP receptor [45]. Further support for a functional role of FP receptors in these cells was obtained in the results given in Figure 3D, demonstrating that AL8810 inhibited the inositol phosphate accumulation induced by the FP receptor agonist fluprostenol. Taken together, these results suggest that the PGE2-induced transactivation of EGFR in MH1C1 hepatoma cells is mediated primarily by FP receptors and signalling via Gq and PLCβ.
Effect of different prostaglandin receptor inhibitors in MH 1 C 1 cells. A) The EP4 inhibitor L-161982 (10 μM) was added 30 min prior to stimulation with PGE2 (100 μM) for 5 min. B) The EP1 inhibitor SC51322 (5 or 10 μM) was added 30 minutes prior to stimulation with PGE2 (100 μM) for 5 min. C) The FP inhibitor AL8810 (10 or 100 μM) was added 30 minutes prior to stimulation with PGE2 (100 μM) for 5 min. All blots are representative of three independent experiments. D) Effect of AL8810 (100 μM) on accumulation of inositol phosphates after stimulation with increasing concentrations of fluprostenol for 30 minutes in the presence of 15 mM LiCl. The data shown are mean ± S.E.M of four independent experiments.
Evidence of a role for Ca2+, but not PKC, in the PGE2-induced transactivation of EGFR
We next tried to determine which pathways downstream of PLCβ are mediating the PGE2-induced transactivation of EGFR. InsP3 and DAG stimulate cytosolic Ca2+ release and protein kinase C (PKC) activity, respectively. Pretreatment of the cells with the PKC inhibitor GF109203X did not prevent the effects of PGE2 on the phosphorylation of the EGFR, ERK, or Akt in the MH1C1 cells (Figure 4A). Furthermore, the data in Figure 4B, comparing PGE2 and the direct PKC activator tetradecanoylphorbol acetate (TPA), showed that TPA did not mimic the effect of PGE2 on Akt, and its stimulation of ERK, unlike the effect of PGE2, was blocked by GF109203X. Interestingly, pretreatment of the cells with GF109203X consistently increased basal and PGE2-induced Akt phosphorylation in the cells. This might result from a reduced feedback inhibition by PKC [47]. In contrast to TPA, thapsigargin, which increases the intracellular Ca2+ level by inhibiting the ‘sarco/endoplasmic reticulum Ca2+-ATPase’ (SERCA) pump [48], induced gefitinib-sensitive phosphorylation of EGFR, ERK, and Akt (Figure 4C). Taken together, these data suggest that Ca2+ rather than PKC mediates the PGE2-induced transactivation of the EGFR in these cells.
Role of Ca2+and PKC in responses to PGE 2 in MH 1 C 1 cells. A) MH1C1 cells were pretreated for 30 min with the PKC inhibitor GF109203X (3.5 μM) before stimulation with PGE2 (100 μM) for 5 min. B) MH1C1 cells were pretreated for 30 min with the PKC inhibitor GF109203X (3.5 μM) before stimulation with PGE2 (100 μM) or TPA (1 μM) for 5 min. C) MH1C1 cells were treated with gefitinib (1µM) for 30 min before stimulation with either PGE2 (100 μM) or thapsigargin (1 μM) for 5 min. Cells were then harvested and subjected to immunoblot analysis as described in Materials and Methods. Representative blots of at least three experiments.
Role of Src and metalloproteinases in the transactivation of the EGFR
To further elucidate mechanisms involved in transactivation of the EGFR, we investigated the effects of Src inhibitors. As shown in Figure 5A, pretreatment of the cells with the Src inhibitor CGP77675 almost completely abolished the PGE2-induced phosphorylation of EGFR and the activation of ERK and Akt, but, in contrast, had little or no effect on the phosphorylation of these proteins elicited by EGF. The Src inhibitor PP2 similarly prevented the phosphorylation of ERK in response to PGE2, while the response to EGF was not significantly affected (Figure 5B). These results suggest an involvement of a Src family kinase in the PGE2-induced transactivation of EGFR in MH1C1 cells.
Effect of Src and MMP inhibitors on phosphorylation of EGFR and downstream targets. A) MH1C1 cells were pretreated for 90 min with the Src inhibitor CGP 77675 (10 μM). Cells were then stimulated with either PGE2 (100 μM) or EGF (10 nM) for 5 min before they were harvested and immunoblotting performed as described in Materials and Methods. Representative blots of at least three experiments. B) MH1C1 cells were pretreated for 30 min with the Src inhibitor PP2 (10 μM). Cells were then stimulated with either PGE2 (100 μM) or EGF (10 nM) for 5 min before they were harvested and immunoblotting performed as described in Materials and Methods. Representative blots of two experiments. C) MH1C1 cells were pretreated for 30 min with increasing concentrations of the metalloproteinase inhibitor GM6001. Cells were then stimulated with PGE2 (100 μM) for 5 min before they were harvested and immunoblotting performed as described in Materials and Methods. Representative blots of three experiments D) MH1C1 cells were pretreated for 30 min with the metalloproteinase inhibitor GM6001 (10 μM). Cells were then stimulated with either PGE2 (100 μM) or EGF (10 nM) for five minutes before they were harvested and immunoblotting performed as described in Materials and Methods. Representative blots of at least three experiments E) Same experiment as in D) performed in hepatocytes. Representative blots of at least three experiments.
Previous evidence has implicated proteinases of the ‘a-disintegrin-and-metalloproteinase’ (ADAM) family in EGFR transactivation by GPCRs in various cells [2, 49, 50]. To test the role of ADAMs in the PGE2-induced EGFR transactivation in MH1C1, we pretreated the cells with GM6001, which is a broad-spectrum metalloproteinase inhibitor [50]. This pretreatment resulted in complete inhibition of PGE2-induced phosphorylation of EGFR, ERK, and Akt, while the EGF-induced phosphorylation of these proteins was not affected (Fig 5C and D), indicating that the transactivation is dependent on mechanisms involving ADAM-mediated release of EGFR ligand(s). We also examined the effect of this inhibitor in the primary cultures of rat hepatocytes, and found neither inhibition of PGE2-induced phosphorylation of ERK and Akt in these cells nor any effect on EGF-induced phosphorylation of EGFR, ERK and Akt (Figure 5E).
Discussion
We have shown that in the MH1C1 hepatocarcinoma cells stimulation with PGE2 or PGF2α causes phosphorylation of the EGFR and an EGFR-dependent phosphorylation of ERK and Akt, indicating that these prostaglandins induced transactivation of EGFR. Further study of the PGE2 effect suggested that the transactivation was mediated by the Gq-coupled FP receptor and activation of PLCβ with downstream signalling by Ca2+ release, Src, and ADAM-mediated shedding of membrane-bound EGFR ligand precursors. In contrast, in primary hepatocytes, PGE2 did not phosphorylate the EGFR, and gefitinib did not prevent phosphorylation of Akt or ERK after PGE2-stimulation, which lends further support to our previous data suggesting that GPCR agonists do not transactivate the EGFR in normal rat hepatocytes, but rather signal via mechanisms that synergistically enhance the effects of EGF [34, 37, 38, 51, 52] (Figure 6).
Mechanisms by which PGE 2 interacts with EGFR-mediated signalling in hepatocytes and MH 1 C 1 hepatocarcinoma cells. A) In normal rat hepatocytes, PGE2 does not elicit transactivation of EGFR, but induces upregulation of the effectiveness in Ras/ERK and PI3K/Akt pathways downstream of EGFR, leading to an enhanced mitogenic response to EGF family growth factors [37, 38, 51]. Although not fully clarified, previous studies have indicated that this effect of PGE2 is mediated primarily through EP3 receptors and Gi proteins, requires several hours to develop, and is most likely a result of altered gene expression [34, 37, 38, 51, 52]. B) In MH1C1 rat hepatocarcinoma cells, PGE2 transactivates EGFR and thereby activates the Ras/ERK and PI3K/Akt signalling pathways. The results of the present study suggest that this effect is exerted via FP receptors, Gq proteins, PLCβ, intracellular Ca2+ (but not PKC), Src, and ADAM-mediated release of EGFR ligands.
Different receptors and pathways may be involved in mitogenic and tumour-promoting effects of prostaglandins [28]. qRT-PCR analysis showed that the prostaglandin receptors expressed in these cells are EP1, EP4, and FP. No significant increase in cAMP accumulation was detected, in accordance with previous results [53], suggesting either that the EP4 protein levels are low, or that these receptors are functionally uncoupled from adenylyl cyclase. In contrast, PGE2 stimulated accumulation of inositol phosphates. Pretreatment with the EP4 antagonist L161982 or the EP1 antagonist SC51322, had no effect on the PGE2-induced phosphorylation of EGFR, ERK, or Akt, while the phosphorylation of these proteins were markedly inhibited by the FP antagonist AL8810. PGF2α, which binds to FP receptors with high affinity, mimicked the effects of PGE2. Together, these results suggest that in contrast to the normal rat hepatocytes, where the effect of PGE2 seems to be mediated primarily through the EP3 receptor [37, 52, 54], the MH1C1 cells, which do not express EP3 receptors, respond to PGE2 through FP receptors, Gq, and PLCβ. It is of interest that expression of EP3 receptors has been found to be suppressed or absent in colon cancer in vivo and in vitro, as compared to normal mucosa [55].
PLCβ can regulate cellular functions via two distinct pathways, involving DAG-mediated activation of PKC and InsP3-induced release and elevation of cytosolic Ca2+, respectively. Our findings suggest that in the MH1C1 cells, the effect of PGE2 was mediated through Ca2+, since it was not mimicked by TPA and not inhibited by a PKC blocker, while thapsigargin, which elevates intracellular Ca2+, mimicked the PGE2 effect, inducing a gefitinib-sensitive phosphorylation of EGFR.
In other cells, both ligand-dependent and ligand-independent mechanisms have been found to mediate EGFR transactivation [5]. Ligand-dependent mechanisms involve the release of EGFR agonists by cleavage and shedding of membrane-associated precursors by proteinases of the ADAM family [2, 49]. Ligand-independent mechanisms have been suggested to involve intracellular molecules including Src family kinases and Pyk2 [1, 3, 56, 57]. Han et al. reported that in Hep3B cells, PGE2 induced phosphorylation of the EGFR through EP1 receptors and an intracellular mechanism involving Src [57]. Itabashi et al. demonstrated that in some hepatocarcinoma cell lines EGFR transactivation triggered by angiotensin II stimulation was mediated through release of EGFR ligand by members of the ADAM family [58]. In the MH1C1 cells, we observed that Src inhibitors abolished PGE2-stimulated phosphorylation of the EGFR, ERK, and Akt, but in contrast, only slightly affected the response to EGF, suggesting a role of Src in the transactivation in these cells. We also found evidence for the involvement of ligand shedding in the transactivation of EGFR after PGE2 stimulation, since pretreatment of the cells with the metalloproteinase inhibitor GM6001 almost completely prevented PGE2-induced, but not EGF-induced, phosphorylation of EGFR, Akt and ERK. GM6001 did not affect the effects of PGE2 in the normal hepatocytes. The lack of transactivation in response to PGE2 in these cells could be due to the low expression of metalloproteinases in hepatocytes as compared to hepatocarcinoma cells [59]. These results indicate that in the MH1C1 cells Src is involved in activating ADAMs rather than directly stimulating the EGFR, thus combining the two mechanisms. The involvement of both Src and ADAMs has been reported in normal gastrointestinal epithelial and colon cancer cell lines [60].
Several signalling pathways seem to be important in hepatocarcinomas [19], and there is evidence that both EGFR-mediated mechanisms and the COX/prostaglandin system may be involved in the pathobiology of these tumours [17, 18, 20, 35, 36]. The results of the present study suggest a functional interaction between the EGFR and the prostaglandins. It has been proposed that transactivation can explain the mitogenic effect of GPCR ligands in some cell systems [61] and that it represents a means of diversifying signalling in the cells, by linking the input from a large number of ligands stimulating GPCRs to the pleiotypic and potentially tumorigenic effects of the EGFR [62]. However, there seems to be great variation between cell types with respect to the different pathways involved in the signalling. We have recently shown that while neurotensin, a GPCR agonist, activates ERK and Akt in an EGFR-independent way in pancreatic cancer Panc-1 cells, as also found by others [63], and activates ERK and Akt via EGFR transactivation in the colon cancer cell line HT 29, neurotensin uses both EGFR-dependent and -independent pathways in the colon cancer cell line HCT 116 [12].
In the present study we have shown that PGE2 has different ways of stimulating the cells, acting by FP-mediated EGFR transactivation in the hepatocarcinoma cells, whereas the effect is mediated mainly via EP3 receptors without any involvement of the EGFR in the hepatocytes [37, 52]. This is further evidence of the diversity of intracellular cross-talk and underscores the importance of investigating such mechanisms in order to better understand the signalling in cancer cells.
Conclusion
The results indicate that in MH1C1 cells, unlike normal hepatocytes, PGE2 activates the MEK/ERK and PI3K/Akt pathways by transactivation of the EGFR, thus diversifying the GPCR-mediated signal. The data also suggest that the underlying mechanisms in these cells involve FP receptors, PLCβ, Ca2+, Src, and proteinase-mediated release of membrane-associated EGFR ligand(s).
Abbreviations
- ADAM:
-
A disintegrin and metalloproteinase
- DAG:
-
Diacylglycerol
- EGF:
-
Epidermal growth factor
- EGFR:
-
Epidermal growth factor receptor
- ERK:
-
Extracellular regulated kinase
- EP:
-
E prostaglandin
- FP:
-
F prostaglandin
- GPCR:
-
G protein-coupled receptor
- InsP3:
-
Inositol trisphosphate
- PG:
-
Prostaglandin
- PGE2:
-
Prostaglandin E2
- PGF2α:
-
Prostaglandin F2α
- PLCβ:
-
Phospholipase C-β
- PKC:
-
Protein kinase C
- RTK:
-
Receptor tyrosine kinase
- TPA:
-
Tetradecanoylphorbol acetate
- SERCA:
-
Sarco/endoplasmic reticulum Ca2+-ATPase.
References
Daub H, Weiss FU, Wallasch C, Ullrich A: Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature. 1996, 379 (6565): 557-560. 10.1038/379557a0.
Prenzel N, Zwick E, Daub H, Leserer M, Abraham R, Wallasch C, Ullrich A: EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature. 1999, 402 (6764): 884-888.
Buchanan FG, Wang D, Bargiacchi F, DuBois RN: Prostaglandin E2 regulates cell migration via the intracellular activation of the epidermal growth factor receptor. J Biol Chem. 2003, 278 (37): 35451-35457. 10.1074/jbc.M302474200.
Schafer B, Marg B, Gschwind A, Ullrich A: Distinct ADAM metalloproteinases regulate G protein-coupled receptor-induced cell proliferation and survival. J Biol Chem. 2004, 279 (46): 47929-47938. 10.1074/jbc.M400129200.
Bhola NE, Grandis JR: Crosstalk between G-protein-coupled receptors and epidermal growth factor receptor in cancer. Front Biosci. 2008, 13: 1857-1865. 10.2741/2805.
Carpenter G, Cohen S: Epidermal growth factor. J Biol Chem. 1990, 265 (14): 7709-7712.
Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW: Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003, 284 (1): 31-53. 10.1016/S0014-4827(02)00098-8.
Holbro T, Civenni G, Hynes NE: The ErbB receptors and their role in cancer progression. Exp Cell Res. 2003, 284 (1): 99-110. 10.1016/S0014-4827(02)00099-X.
Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, Carotenuto A, De Feo G, Caponigro F, Salomon DS: Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006, 366 (1): 2-16. 10.1016/j.gene.2005.10.018.
Christoffersen T, Guren TK, Spindler KL, Dahl O, Lonning PE, Gjertsen BT: Cancer therapy targeted at cellular signal transduction mechanisms: strategies, clinical results, and unresolved issues. Eur J Pharmacol. 2009, 625 (1–3): 6-22.
Ciardiello F, Tortora G: EGFR antagonists in cancer treatment. N Engl J Med. 2008, 358 (11): 1160-1174. 10.1056/NEJMra0707704.
Müller KM, Tveteraas IH, Aasrum M, Odegard J, Dawood M, Dajani O, Christoffersen T, Sandnes DL: Role of protein kinase C and epidermal growth factor receptor signalling in growth stimulation by neurotensin in colon carcinoma cells. BMC Cancer. 2011, 11: 421-10.1186/1471-2407-11-421.
Sand TE, Christoffersen T: Temporal requirement for epidermal growth factor and insulin in the stimulation of hepatocyte DNA synthesis. J Cell Physiol. 1987, 131 (2): 141-148. 10.1002/jcp.1041310202.
Mead JE, Fausto N: Transforming growth factor alpha may be a physiological regulator of liver regeneration by means of an autocrine mechanism. Proc Natl Acad Sci U S A. 1989, 86 (5): 1558-1562. 10.1073/pnas.86.5.1558.
Christoffersen T, Thoresen GH, Dajani OF, Melien Ø, Guren T, Refsnes M, Sandnes D: Mechanisms of hepatocyte growth regulation by hormones and growth factor. The hepatocyte review. edn. Edited by AM BMaE. 2000, Kluwer Academic Publishers, Dordrecht/Boston/London, 209-246.
Scheving LA, Stevenson MC, Taylormoore JM, Traxler P, Russell WE: Integral role of the EGF receptor in HGF-mediated hepatocyte proliferation. Biochem Biophys Res Commun. 2002, 290 (1): 197-203. 10.1006/bbrc.2001.6157.
Farazi PA, DePinho RA: Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006, 6 (9): 674-687. 10.1038/nrc1934.
Villanueva A, Newell P, Chiang DY, Friedman SL, Llovet JM: Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis. 2007, 27 (1): 55-76. 10.1055/s-2006-960171.
Whittaker S, Marais R, Zhu AX: The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010, 29 (36): 4989-5005. 10.1038/onc.2010.236.
Berasain C, Castillo J, Prieto J, Avila MA: New molecular targets for hepatocellular carcinoma: the ErbB1 signaling system. Liver Int. 2007, 27 (2): 174-185. 10.1111/j.1478-3231.2006.01424.x.
Jhappan C, Stahle C, Harkins RN, Fausto N, Smith GH, Merlino GT: TGF alpha overexpression in transgenic mice induces liver neoplasia and abnormal development of the mammary gland and pancreas. Cell. 1990, 61 (6): 1137-1146. 10.1016/0092-8674(90)90076-Q.
Sandgren EP, Luetteke NC, Qiu TH, Palmiter RD, Brinster RL, Lee DC: Transforming growth factor alpha dramatically enhances oncogene-induced carcinogenesis in transgenic mouse pancreas and liver. Mol Cell Biol. 1993, 13 (1): 320-330.
Ito Y, Takeda T, Sakon M, Tsujimoto M, Higashiyama S, Noda K, Miyoshi E, Monden M, Matsuura N: Expression and clinical significance of erb-B receptor family in hepatocellular carcinoma. Br J Cancer. 2001, 84 (10): 1377-1383. 10.1054/bjoc.2000.1580.
Hopfner M, Sutter AP, Huether A, Schuppan D, Zeitz M, Scherubl H: Targeting the epidermal growth factor receptor by gefitinib for treatment of hepatocellular carcinoma. J Hepatol. 2004, 41 (6): 1008-1016. 10.1016/j.jhep.2004.08.024.
Huether A, Hopfner M, Baradari V, Schuppan D, Scherubl H: EGFR blockade by cetuximab alone or as combination therapy for growth control of hepatocellular cancer. Biochem Pharmacol. 2005, 70 (11): 1568-1578. 10.1016/j.bcp.2005.09.007.
Villanueva A, Llovet JM: Targeted therapies for hepatocellular carcinoma. Gastroenterology. 2011, 140 (5): 1410-1426. 10.1053/j.gastro.2011.03.006.
Breyer RM, Bagdassarian CK, Myers SA, Breyer MD: Prostanoid receptors: subtypes and signaling. Annu Rev Pharmacol Toxicol. 2001, 41: 661-690. 10.1146/annurev.pharmtox.41.1.661.
Wang D, Dubois RN: Eicosanoids and cancer. Nat Rev Cancer. 2010, 10 (3): 181-193. 10.1038/nrc2809.
Dubois RN, Abramson SB, Crofford L, Gupta RA, Simon LS, Van De Putte LB, Lipsky PE: Cyclooxygenase in biology and disease. FASEB J. 1998, 12 (12): 1063-1073.
Gupta RA, Dubois RN: Colorectal cancer prevention and treatment by inhibition of cyclooxygenase-2. Nat Rev Cancer. 2001, 1 (1): 11-21. 10.1038/35094017.
Zha S, Yegnasubramanian V, Nelson WG, Isaacs WB, De Marzo AM: Cyclooxygenases in cancer: progress and perspective. Cancer Lett. 2004, 215 (1): 1-20. 10.1016/j.canlet.2004.06.014.
Andreis PG, Whitfield JF, Armato U: Stimulation of DNA synthesis and mitosis of hepatocytes in primary cultures of neonatal rat liver by arachidonic acid and prostaglandins. Exp Cell Res. 1981, 134 (2): 265-272. 10.1016/0014-4827(81)90425-0.
Refsnes M, Dajani OF, Sandnes D, Thoresen GH, Rottingen JA, Iversen JG, Christoffersen T: On the mechanisms of the growth-promoting effect of prostaglandins in hepatocytes: the relationship between stimulation of DNA synthesis and signaling mediated by adenylyl cyclase and phosphoinositide-specific phospholipase C. J Cell Physiol. 1995, 164 (3): 465-473. 10.1002/jcp.1041640304.
Refsnes M, Thoresen GH, Dajani OF, Christoffersen T: Stimulation of hepatocyte DNA synthesis by prostaglandin E2 and prostaglandin F2 alpha: additivity with the effect of norepinephrine, and synergism with epidermal growth factor. J Cell Physiol. 1994, 159 (1): 35-40. 10.1002/jcp.1041590106.
Koga H, Sakisaka S, Ohishi M, Kawaguchi T, Taniguchi E, Sasatomi K, Harada M, Kusaba T, Tanaka M, Kimura R, et al: Expression of cyclooxygenase-2 in human hepatocellular carcinoma: relevance to tumor dedifferentiation. Hepatology. 1999, 29 (3): 688-696. 10.1002/hep.510290355.
Tang TC, Poon RT, Lau CP, Xie D, Fan ST: Tumor cyclooxygenase-2 levels correlate with tumor invasiveness in human hepatocellular carcinoma. World J Gastroenterol. 2005, 11 (13): 1896-1902.
Dajani OF, Meisdalen K, Guren TK, Aasrum M, Tveteraas IH, Lilleby P, Thoresen GH, Sandnes D, Christoffersen T: Prostaglandin E2 upregulates EGF-stimulated signaling in mitogenic pathways involving Akt and ERK in hepatocytes. J Cell Physiol. 2008, 214 (2): 371-380. 10.1002/jcp.21205.
Nilssen LS, Odegard J, Thoresen GH, Molven A, Sandnes D, Christoffersen T: G protein-coupled receptor agonist-stimulated expression of ATF3/LRF-1 and c-myc and comitogenic effects in hepatocytes do not require EGF receptor transactivation. J Cell Physiol. 2004, 201 (3): 349-358. 10.1002/jcp.20075.
Richardson UI, Tashjian AH, Levine L: Establishment of a clonal strain of hepatoma cells which secrete albumin. J Cell Biol. 1969, 40 (1): 236-247. 10.1083/jcb.40.1.236.
Christoffersen T, Refsnes M, Bronstad GO, Ostby E, Huse J, Haffner F, Sand TE, Hunt NH, Sonne O: Changes in hormone responsiveness and cyclic AMP metabolism in rat hepatocytes during primary culture and effects of supplementing the medium with insulin and dexamethasone. Eur J Biochem. 1984, 138 (2): 217-226. 10.1111/j.1432-1033.1984.tb07904.x.
Bustin SA: Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000, 25 (2): 169-193. 10.1677/jme.0.0250169.
Skomedal T, Grynne B, Osnes JB, Sjetnan AE, Oye I: A radioimmunoassay for cyclic AMP (cAMP) obtained by acetylation of both unlabeled and labeled (3 H-cAMP) ligand, or of unlabeled ligand only. Acta Pharmacol Toxicol (Copenh). 1980, 46 (3): 200-204.
Sugimoto Y, Narumiya S: Prostaglandin E receptors. J Biol Chem. 2007, 282 (16): 11613-11617.
Ji R, Chou CL, Xu W, Chen XB, Woodward DF, Regan JW: EP1 prostanoid receptor coupling to G i/o up-regulates the expression of hypoxia-inducible factor-1 alpha through activation of a phosphoinositide-3 kinase signaling pathway. Mol Pharmacol. 2010, 77 (6): 1025-1036. 10.1124/mol.110.063933.
Griffin BW, Klimko P, Crider JY, Sharif NA: AL-8810: a novel prostaglandin F2 alpha analog with selective antagonist effects at the prostaglandin F2 alpha (FP) receptor. J Pharmacol Exp Ther. 1999, 290 (3): 1278-1284.
Machwate M, Harada S, Leu CT, Seedor G, Labelle M, Gallant M, Hutchins S, Lachance N, Sawyer N, Slipetz D, et al: Prostaglandin receptor EP(4) mediates the bone anabolic effects of PGE(2). Mol Pharmacol. 2001, 60 (1): 36-41.
Santiskulvong C, Rozengurt E: Protein kinase Calpha mediates feedback inhibition of EGF receptor transactivation induced by Gq-coupled receptor agonists. Cell Signal. 2007, 19 (6): 1348-1357. 10.1016/j.cellsig.2007.01.006.
Thastrup O, Cullen PJ, Drobak BK, Hanley MR, Dawson AP: Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc Natl Acad Sci U S A. 1990, 87 (7): 2466-2470. 10.1073/pnas.87.7.2466.
Ohtsu H, Dempsey PJ, Eguchi S: ADAMs as mediators of EGF receptor transactivation by G protein-coupled receptors. Am J Physiol Cell Physiol. 2006, 291 (1): C1-C10. 10.1152/ajpcell.00620.2005.
Santiskulvong C, Rozengurt E: Galardin (GM 6001), a broad-spectrum matrix metalloproteinase inhibitor, blocks bombesin- and LPA-induced EGF receptor transactivation and DNA synthesis in rat-1 cells. Exp Cell Res. 2003, 290 (2): 437-446. 10.1016/S0014-4827(03)00355-0.
Odegard J, Aasrum M, Tveteraas IH, Bharath SP, Sandnes D, Christoffersen T: Role of ErbB2 in the prostaglandin E(2)-induced enhancement of the mitogenic response to epidermal growth factor in cultured hepatocytes. Biochem Biophys Res Commun. 2012, 421 (2): 255-260. 10.1016/j.bbrc.2012.03.148.
Meisdalen K, Dajani OF, Christoffersen T, Sandnes D: Prostaglandins enhance epidermal growth factor-induced DNA synthesis in hepatocytes by stimulation of E prostanoid 3 and F prostanoid receptors. J Pharmacol Exp Ther. 2007, 322 (3): 1044-1050. 10.1124/jpet.107.121277.
Bronstad GO, Gladhaug IP, Haffner F, Rugstad HE, Christoffersen T: The regulation of cyclic AMP levels in cultured MH1C1 rat hepatoma cells and in solid tumours derived from MH1C1 cell inoculates. Anticancer Res. 1987, 7 (2): 155-160.
Hashimoto N, Watanabe T, Ikeda Y, Yamada H, Taniguchi S, Mitsui H, Kurokawa K: Prostaglandins induce proliferation of rat hepatocytes through a prostaglandin E2 receptor EP3 subtype. Am J Physiol. 1997, 272 (3 Pt 1): G597-G604.
Shoji Y, Takahashi M, Kitamura T, Watanabe K, Kawamori T, Maruyama T, Sugimoto Y, Negishi M, Narumiya S, Sugimura T, et al: Downregulation of prostaglandin E receptor subtype EP3 during colon cancer development. Gut. 2004, 53 (8): 1151-1158. 10.1136/gut.2003.028787.
Andreev J, Galisteo ML, Kranenburg O, Logan SK, Chiu ES, Okigaki M, Cary LA, Moolenaar WH, Schlessinger J: Src and Pyk2 mediate G-protein-coupled receptor activation of epidermal growth factor receptor (EGFR) but are not required for coupling to the mitogen-activated protein (MAP) kinase signaling cascade. J Biol Chem. 2001, 276 (23): 20130-20135. 10.1074/jbc.M102307200.
Han C, Michalopoulos GK, Wu T: Prostaglandin E2 receptor EP1 transactivates EGFR/MET receptor tyrosine kinases and enhances invasiveness in human hepatocellular carcinoma cells. J Cell Physiol. 2006, 207 (1): 261-270. 10.1002/jcp.20560.
Itabashi H, Maesawa C, Oikawa H, Kotani K, Sakurai E, Kato K, Komatsu H, Nitta H, Kawamura H, Wakabayashi G, et al: Angiotensin II and epidermal growth factor receptor cross-talk mediated by a disintegrin and metalloprotease accelerates tumor cell proliferation of hepatocellular carcinoma cell lines. Hepatol Res. 2008, 38 (6): 601-613. 10.1111/j.1872-034X.2007.00304.x.
Caja L, Ortiz C, Bertran E, Murillo MM, Miro-Obradors MJ, Palacios E, Fabregat I: Differential intracellular signalling induced by TGF-beta in rat adult hepatocytes and hepatoma cells: implications in liver carcinogenesis. Cell Signal. 2007, 19 (4): 683-694. 10.1016/j.cellsig.2006.09.002.
Pai R, Soreghan B, Szabo IL, Pavelka M, Baatar D, Tarnawski AS: Prostaglandin E2 transactivates EGF receptor: a novel mechanism for promoting colon cancer growth and gastrointestinal hypertrophy. Nat Med. 2002, 8 (3): 289-293. 10.1038/nm0302-289.
Daub H, Wallasch C, Lankenau A, Herrlich A, Ullrich A: Signal characteristics of G protein-transactivated EGF receptor. EMBO J. 1997, 16 (23): 7032-7044. 10.1093/emboj/16.23.7032.
Fischer OM, Hart S, Gschwind A, Ullrich A: EGFR signal transactivation in cancer cells. Biochem Soc Trans. 2003, 31 (Pt 6): 1203-1208.
Kisfalvi K, Guha S, Rozengurt E: Neurotensin and EGF induce synergistic stimulation of DNA synthesis by increasing the duration of ERK signaling in ductal pancreatic cancer cells. J Cell Physiol. 2005, 202 (3): 880-890. 10.1002/jcp.20187.
Acknowledgements
The work was supported by the Norwegian Cancer Society. We thank Eva Østby and Ellen Johanne Johansen for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
IHT participated in the design of the study, carried out immunoblotting experiments and drafted the manuscript. KMM carried out immunoblotting experiments, inositol phosphate experiments and helped revise the manuscript. MA helped revise the manuscript. JØ carried out qRT-PCR experiment and helped revise the manuscript. OD conceived of the study, carried out DNA synthesis and helped revise the manuscript. TG conceived of the study and helped revise the manuscript. DS conceived of the study, participated in the design of the study, carried out cAMP and inositol phosphate experiments and helped revise the manuscript. TC conceived of the study, participated in the design of the study and helped revise the manuscript. All authors read and approved of the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Tveteraas, I.H., Müller, K.M., Aasrum, M. et al. Mechanisms involved in PGE2-induced transactivation of the epidermal growth factor receptor in MH1C1 hepatocarcinoma cells. J Exp Clin Cancer Res 31, 72 (2012). https://doi.org/10.1186/1756-9966-31-72
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-9966-31-72