Abstract
Background
The present study determines the feasibility of generating an average arterial input function (Avg-AIF) from a limited population of patients with neck nodal metastases to be used for pharmacokinetic modeling of dynamic contrast-enhanced MRI (DCE-MRI) data in clinical trials of larger populations.
Methods
Twenty patients (mean age 50 years [range 27–77 years]) with neck nodal metastases underwent pretreatment DCE-MRI studies with a temporal resolution of 3.75 to 7.5 sec on a 1.5T clinical MRI scanner. Eleven individual AIFs (Ind-AIFs) met the criteria of expected enhancement pattern and were used to generate Avg-AIF. Tofts model was used to calculate pharmacokinetic DCE-MRI parameters. Bland-Altman plots and paired Student t-tests were used to describe significant differences between the pharmacokinetic parameters obtained from individual and average AIFs.
Results
Ind-AIFs obtained from eleven patients were used to calculate the Avg-AIF. No overall significant difference (bias) was observed for the transfer constant (Ktrans) measured with Ind-AIFs compared to Avg-AIF (p = 0.20 for region-of-interest (ROI) analysis and p = 0.18 for histogram median analysis). Similarly, no overall significant difference was observed for interstitial fluid space volume fraction (ve) measured with Ind-AIFs compared to Avg-AIF (p = 0.48 for ROI analysis and p = 0.93 for histogram median analysis). However, the Bland-Altman plot suggests that as Ktrans increases, the Ind-AIF estimates tend to become proportionally higher than the Avg-AIF estimates.
Conclusion
We found no statistically significant overall bias in Ktrans or ve estimates derived from Avg-AIF, generated from a limited population, as compared with Ind-AIFs.
However, further study is needed to determine whether calibration is needed across the range of Ktrans. The Avg-AIF obtained from a limited population may be used for pharmacokinetic modeling of DCE-MRI data in larger population studies with neck nodal metastases. Further validation of the Avg-AIF approach with a larger population and in multiple regions is desirable.
Similar content being viewed by others
Background
A broad range of tumors [1–3] have been studied clinically by dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) which monitors the passage of intravenously administered Gadolinium (Gd) contrast agent through tumor tissue. The rate at which the agent passes from the intravascular space to the interstitial space depends on several factors including tumor perfusion, vascular density, and vascular permeability. MRI signal changes depend on these factors and also the leakage (interstitial) space. Tumors are considered to be leaky and to have high perfusion. Three different approaches have been reported for the analysis of the DCE-MRI time course data: a qualitative approach, or subjective assessment of the curve shape; semi-quantitative analysis of the signal time course, such as uptake slope and maximum signal intensity change; and quantitative analysis involving analytical pharmacokinetic modelling of the time course data. The qualitative and semi-quantitative approaches cannot be directly related to physiological parameters, although they do provide clinically useful data. The quantitative approach has proven to be the most sophisticated and provides physiological data [4–6]. Rijpkema et al [4] performed quantitative analyses of DCE-MRI data in eleven patients with tumors, of which six patients had tumor in the head and neck region. Their initial analysis presented the redistribution rate constant kep (= Ktrans/ve) maps while the Ktrans (transfer constant) and ve (interstitial fluid space volume fraction) maps could not be expressed in absolute quantitation values due to the scaling factor and were not shown. Kim et al [5] measured the AIF in nine individuals and found the effects of transcytolemmal water exchange to be an important factor in DCE-MRI data analysis. Use of standardized quantitative methods for analysis has been highly recommended so that DCE-MRI data may be compared from different sites, platforms and field strengths and be of widespread use, for example in clinical trials assessing tumor response to therapy [7].
Quantitative analysis of the DCE-MRI data requires knowledge of the arterial input function (AIF): the time-dependent contrast agent concentration in the arterial blood feeding the tissue of interest. The absolute accuracy of the pharmacokinetic parameters Ktrans and ve depends on the AIF accuracy [8]. The Ktrans and ve parameters are clinically relevant and have been used in oncological imaging for tumor detection and to evaluate response to therapy [3, 7]. DCE-MRI has become a useful tool also in evaluating head and neck cancers, differentiating tumor from non-tumor in the cervical lymph nodes or lymphoma from squamous cell carcinoma, assessing mandibular invasion, and predicting response to therapy [3, 4, 6, 9–13]. The need for reliable measurements of pharmacokinetic parameters has been the impetus for accurate measurement of AIFs in the carotid artery in head and neck cancer patients [4, 5]. However, obtaining individual AIFs for each DCE-MRI study may not be possible in all patients due to data acquisition constraints. This challenge can be overcome if a population-averaged AIF is used [14]. Recent studies in cancer [14, 15] have indicated that the use of a high-temporal-resolution population-based AIF allowed assessment of detailed physiological information with a good degree of precision even when individual AIF measurement was not possible. However, the use of Avg-AIF for quantitative analysis of DCE-MRI data in neck nodal metastases has not been tested so far. In the present study, we determined the feasibility of building an average AIF obtained from a limited population of head and neck cancer patients with neck nodal metastases for pharmacokinetic modeling of DCE-MRI data in studies of larger populations, e.g. in clinical trials to examine the effects of treatment on DCE-MRI parameters.
Methods
Patients
DCE-MRI was performed in twenty head and neck cancer patients (mean patient age 50 years [range 27–77 years]; 17 males, 3 females, with nodal disease [size >1 cc]; 2 patients stage III and 18 stage IV) before chemo-radiation therapy or surgery. The institutional review board granted exempt status for this retrospective study with a waiver of informed consent for two patients who underwent DCE-MRI between April and September 2005. From February, 2006 to October, 2007 DCE-MRI was performed as part of an ongoing National Institutes of Health (NIH) study investigating the use of MR imaging in patients with head and neck cancers; 18 patients gave informed consent for their participation in the institutional review board-approved NIH study. The study was also compliant with the Health Insurance Portability and Accountability Act. Although 20 patients were recruited, only 11 had AIFs that were included for averaging to produce a population AIF.
MRI Data acquisition
MRI data were acquired on a 1.5 Tesla GE Excite scanner (General Electric, Milwaukee, WI) with a 4 channel neurovascular phased-array coil (MRI Devices Corporation, Gainesville, FL) for signal reception and a body coil for transmission. The study consisted of MR imaging covering the entire neck. MRI data was acquired with spatial saturation to minimize the flow effects on the MR measurement in the arterial blood vessels. MR acquisition parameters for the neck survey were as follows: rapid scout images, multiplanar (axial, coronal and sagittal) T2-weighted, fat-suppressed, fast-spin echo images (TR = 5000 ms, TE = 102 ms, averages = 2, slice thickness = 5.0 mm and gap = 2.5 mm), multi-planar T1-weighted images (TR = 675 ms, TE = 8 ms, averages = 2, slice thickness = 5.0 mm and gap = 2.5 mm). Standard T1- and T2-weighted imaging were followed by proton density MRI acquired (for the purpose of determining the longitudinal relaxation rate constant R1 for each DCE-MRI data point) in the axial plane (TR = 350 ms, TE = 2 ms with a 30° flip angle (α), 2 excitation, 15.63 kHz receive bandwidth, field of view [FOV], slice location and thickness same as in the DCE-MRI scan [see below] and a 256 × 128 matrix), followed by an axial T1-weighted DCE-MRI scan and then post-contrast T1-weighted images in axial and coronal planes. At the beginning of the sixth image set (data point) of the DCE-MRI scan, a bolus of 0.1 mmol/kg GdDTPA contrast agent (Magnevist; Berlex Laboratories, Wayne, NJ, USA) was delivered through an antecubital vein catheter at 2 cc/sec, followed by a saline flush using an MR-compatible programmable power injector (Spectris; Medrad, Indianola, PA, USA). DCE-MRI data were acquired using a 2D-fast multi-phase spoiled gradient echo (FMSPGR) sequence. The entire node was covered contiguously with 5–7-mm thick slices, zero gap yielding 3–6 slices depending on the size of the node with 3.75–7.5 sec temporal resolution. Acquisition parameters included 9 ms TR, 2 ms TE, 30° flip angle (α), 15.63 kHz receive bandwidth, 18–20-cm field of view (FOV), 40–80 time course data points, and a 256 × 128 matrix sized zero filled to 256 × 256 during image reconstruction.
DCE-MRI Data Analysis
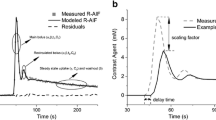
Data were exported to a Windows PC and the pharmacokinetic analysis based on the Toft's model was done using in-house software written to display and analyze data using IDL windows version 6.0 (Research Systems, Boulder, CO, USA) [16]. To perform the above pharmacokinetic analysis using the Tofts model [16], a reliable AIF (Cp(t)) is required. In all patients studied, the carotid artery was visible in most of the image slices. To minimize partial volume effects the image slice that contained the central portion of the artery was used. A region of interest (ROI) was placed within the carotid artery (Figure 1a) and this allowed the direct measurement of the arterial input function through monitoring of the changes in signal intensity (and converting the data into concentration) (Figure 1b). The signal intensity (S) of spoiled gradient echo with contrast agent bolus injection is modeled according to Equation 1, assuming TE << T2 so that the final exponential term can be taken as 1, [17]:
(a) A post-contrast axial MR image from a head and neck cancer patient extracted from the DCE-MRI scan showing bilateral nodal metastases (dashed white lines) in the neck and the right carotid artery (solid red line). (b) An arterial input function (AIF) plot (plasma Gd-DTPA concentration time course). The data points were measured from the region of interest placed within the right carotid artery. The AIF wash-out phase was fitted with a bi-exponential decay function.

where α is the flip angle, TR is the repetition time, S0 is a constant proportional to the proton density of the sample. Based on the method described by Parker et al [18], a calibration curve was generated of signal intensity ratio of T1-weighted image to proton density image as a function of T1. The details of the phantom experiment are reported by Wang et al [15]. The artery ROI values R1 for the DCE image series were obtained from the calibration curve and subsequently converted to Gd-DTPA concentrations. The apparent longitudinal relaxation rate constant, R1 (= 1/T1), is proportional to Cp (t) and is given by:

Where Cp(t) is the arterial plasma GdDTPA concentration at time t, r1 is the contrast agent relaxivity; taken to be 4.1 sec-1(mmol/L)-1 at 1.5T [19], and R10 is the pre-contrast R1. The derived Cp(t) time course was fitted with a bi-exponential decay function in the wash-out phase [20] to generate the AIF. The Ind-AIF's were inspected to determine whether they followed the "expected" contrast enhancement pattern: enhancement during the early phase followed by signal intensity attenuation. The residual (Sum of residual) and mean square error (MSE) were used as a reference as well as visual inspection done by a physicist. Ind-AIFs were excluded that clearly showed large deviations from the above enhancement pattern shown in Figure 1b (Sum of residual = 0.073; MSE = 0.325). By averaging the eleven individual AIFs (Ind-AIFs) obtained from the carotid artery that satisfied the above criteria, with peak height aligned, an average AIF (Avg-AIF) was obtained. The average AIF was calculated using the equation [16]:

Where D is the dose (mmole/kg), and amplitudes "a" are normalized for unit dose (so that Cp is then known for any size dose). For the parameters a1, a2, m1 and m2 Mean values are reported as Mean ± SE of the Mean in Table 1. The SE variation show that the changes are large during the initial spike (a) and then reduce to a small variation during the wash out phase (m). This is consistent with previous studies [14, 15].
Quantitative DCE-MRI analyses of the tumor tissue time course data was done in all the patients for the ROI as well as each pixel within the ROI using histogram analyses [15]. The latter calculates the pixel Ktrans and ve and the median values of these parameters.
For the tumor tissue time course data, ROI were manually drawn by an experienced neuro-radiologist outlining the contrast-enhanced tumor and were used for signal intensity measurements. All the slices containing tumor were outlined and analyzed. The model fitted the tissue contrast agent concentration, Ct(t), time course for the extraction of the Ktrans and ve parameters from the whole tumor (multiple slices), as shown in the following Kety-Schmidt equation:

The term that includes plasma volume fraction (vp), vpCp(t), was ignored on the right side of the equation. As shown by Li et al [21] there is sufficient contrast agent extravasation from plasma to interstitium tumor tissue such that Ktrans and veparameters are adequate for analyses. The exclusion of vp may lead to errors in situations when there is less contrast extravasation (such as Ktrans < 0.001 min-1). For each DCE-MRI data set, only the bi-exponential function-fitted AIF wash-out phase was used for Ct(t) curve fitting with time zero in Eq. [4] set as the time of AIF peak amplitude. This is due to the fact that the arrival of the bolus precedes the apparent rise of tumor tissue signal intensity, defined as signal intensity rising more than one standard deviation (SD) of the signal intensities of the five pre-contrast injection baseline data points.
A paired Student t test was used to evaluate differences in pharmacokinetic parameters resulting from the use of Ind- and Avg-AIFs. Bland-Altman plots [22] were used to explore possible trends across the range of observed ROI means for each parameter. An example of a single slice was plotted to show the pixel-by-pixel variation between the two estimates within the slice.
Results
In twenty patients ROI's were drawn in the carotid artery for calculation of the AIF and 55% (eleven patients) had AIF that met the expected pattern of enhancement. The data from these eleven patients was used to build the Average-AIF. The bi-exponential function-fitted Average-AIF with 2 cc/sec contrast agent injection rate is shown in Figure 2. Bland-Altman plots show the values of ROI Ktrans (Figure 3a) and ROI ve (Figure 3b) measured from pharmacokinetic modeling using both Ind-AIFs and the Avg-AIF for the eleven head and neck cancer patient's nodal tumor tissue. For each ROI, the difference between the two estimates is plotted against the mean of the two estimates. Reference lines show the average difference (bias) and limits of agreement (+/- two SD). There were no statistically significant bias between Ktrans values derived with Ind-AIFs and those derived with the Avg-AIF (Table 2; p = 0.20 for ROI Ktrans and 0.18 for histogram median Ktrans; paired t test). Similar results were obtained for the ve parameter (Table 2). In order to show how Avg-AIF performs on a pixel-by-pixel basis, Figure 4 displays the representative graph for a patient with pixel Ktrans values within the tumor tissue ROI obtained with the Avg-AIF plotted against those obtained with the Ind-AIF. The plot shows significant linear correlation (p < 0.0001) and a proportional divergence from the identity reference line. The slope value for linear regression through the origin was 1.35. Similar results were obtained from ROIs for the other 10 patients with reliable Ind-AIF measurements. This indicates that the use of Avg-AIF works equally well for both ROI- and pixel-wise analyses.
Bland-Altman plots showing variability of the differences between parameter estimates obtained by the two methods. Horizontal axes are means and vertical axes are the differences of the paired estimates. Dotted references lines show bias and limits of agreement (mean of differences +/- 2 SD of differences). (a) Ktrans Ind-AIF – KtransAvg-AIF (0.124 +/- .448); (b) ve Ind-AIF – veAvg-AIF (0.088 +/- .600).
Scatter plot of pixel K trans values obtained from single-image slice pharmacokinetic modeling analyses of a patient's DCE-MRI data. The Ktransextracted with the average AIF (Avg-AIF) approach are plotted against those extracted with the individually measured AIF (Ind-AIF) approach. Both identity line and regression line are shown. The slope of the linear regression is 1.35.
Discussion
DCE-MRI studies have shown great promise in several aspects of head and neck cancer management, including differential diagnosis and assessment of treatment response [3]. To date, the general methods of analysis of the data have ranged from qualitative to quantitative analysis [3–6, 9–12]. Quantitative modeling requires accurate AIF measurements [7]. Although it would be ideal to obtain AIFs from individual patients, which is often feasible, in many settings it is not possible to perform an AIF measurement reliably either due to data acquisition constraints or lack of a suitable artery within the imaging FOV from which to obtain an AIF [14, 15]. Parker et al suggested that in cases where a reliable AIF was not measured, a high-resolution population-averaged AIF can improve the reproducibility of parameters obtained using kinetic-modelling of DCE-MRI data and that in general only small changes in accuracy can be expected [14]. This study was performed on twenty three patients with cancer in the abdomen [14]. The authors commented that use of the Avg-AIF approach could be a useful alternative to the use of Ind-AIFs, especially for quantitative treatment studies where changes in microvascular properties are more important than the absolute values [14]. Our group has recently published the use of limited population based Avg-AIF for DCE-MRI data analysis in osteosarcomas [15]. In the present study we focus the use of the same principle on a different anatomic site with cancer i.e. head and neck. These preliminary studies can provide basis for large, validation studies and future application in clinical trials which use DCE-MRI parameters as non-invasive MR biomarkers. There is a need for widespread use of quantitative analysis of DCE-MRI data in order to compare and evaluate studies performed at different field strengths which would be independent of instrument platform and acquisition parameters.
Different groups have proposed various methods for analyzing DCE-MRI data for scenarios in which Ind- or Avg-AIFs cannot be obtained easily. These techniques may be an option for analysis of DCE-MRI data after appropriate validation or comparisons with standard methods. Riabkov et al [23] estimated the kinetic parameters without input functions using multichannel blind identification methods and iterative quadratic maximum-likelihood (IQML) gave the most accurate estimates. Yankeelov et al [24] and Walker-Samuel et al [25, 26] have proposed a method that compares the tissue of interest (TOI) curve shape to that of a reference region (RR), thereby eliminating the need for direct AIF measurements when a reliable AIF is not obtainable. Yang et al [27] proposed the double-reference-tissue method, which assumes that the AIFs of the two reference tissues have the same shape. The elegant simulations used in their study need to be tested in more complicated tissues, such as tumor tissue.
Clinical requirements for diagnosis often dictate large imaging spatial coverage and high image resolution, which result in poor temporal resolution for DCE-MRI acquisition. Roberts et al [28] showed that AIFs sampled at low temporal resolution introduced an unpredictable degree of error in the quantitative analysis. In such cases the use of Avg-AIF obtained from acquisitions with higher temporal resolution would be the method of choice. In the present study, the Avg-AIF was obtained from DCE-MR images of sufficient temporal resolution, and therefore it may be used to analyze DCE-MRI data that were acquired with poorer temporal resolution but otherwise with the same contrast injection set up, including dose, injection site and injection rate. In the present study ROIs were drawn on all arteries visible on the MR images but reliable AIF measurement was obtained only from the section that contained the central portion of the artery. This was because other sections showed small vessels containing only a small number of pixels, leading to a partial volume effect, or because other sections showed the bifurcation of the common carotid artery. Care was taken by the physicist during the selection process of Ind-AIFs so as to not pre-determine the outcome of the Avg-AIF, but rather to remove patient data that largely deviated from the expected contrast enhancement pattern.
A bi-exponential function was used for data analysis. Consistent use of this function for AIF curve fitting will not introduce systematic errors in longitudinal comparisons of changes in pharmacokinetic parameters caused by treatment. The use of an Avg-AIF implies that resulting parameter estimates will be higher than Ind-AIF estimates for some cases and lower for others. However, there is added benefit in the implicit gain in precision of the pooled or Avg-AIF and the possibility of parameter estimation when Ind-AIFs are not available. Although the sample size was small, other studies of cancers such as sarcoma have reported similar findings for Avg-AIF from small population [15]. Further validation of the Avg-AIF approach with a larger population and in multiple regions is desirable.
Conclusion
In conclusion, this study shows that parameter estimates derived from an Avg-AIF, generated from a limited population, yield similar results to estimates derived from Ind-AIFs. The Avg-AIF may be useful for pharmacokinetic modeling of DCE-MRI data in studies of larger populations of patients with neck nodal metastases where precise Ind-AIFs may not be available for all cases, e.g., in clinical trials to examine the effects of drugs on DCE-MRI parameters.
Abbreviations
- DCE-MRI:
-
Dynamic contrast-enhanced MRI
- AIF:
-
arterial input function
- Ktrans :
-
transfer constant
- ve :
-
interstitial fluid space volume fraction.
References
Dyke JP, Panicek DM, Healey JH, Meyers PA, Huvos AG, Schwartz LH, Thaler HT, Tofts PS, Gorlick R, Koutcher JA, Ballon D: Osteogenic and Ewing sarcomas: estimation of necrotic fraction during induction chemotherapy with dynamic contrast-enhanced MR imaging. Radiology. 2003, 228: 271-278. 10.1148/radiol.2281011651.
Knopp MV, Weiss E, Sinn HP, Mattern J, Junkermann H, Radeleff J, Magener A, Brix G, Delorme S, Zuna I, van Kaick G: Pathophysiologic basis of contrast enhancement in breast tumors. J Magn Reson Imaging. 1999, 10: 260-266. 10.1002/(SICI)1522-2586(199909)10:3<260::AID-JMRI6>3.0.CO;2-7.
Shah GV, Fischbein NJ, Gandhi D, Mukherji SK: Dynamic contrast-enhanced MR imaging. Top Magn Reson Imaging. 2004, 15: 71-77. 10.1097/01.ftd.0000136399.78067.dd.
Rijpkema M, Kaanders JH, Joosten FB, Kogel van der AJ, Heerschap A: Method for quantitative mapping of dynamic MRI contrast agent uptake in human tumors. J Magn Reson Imaging. 2001, 14: 457-463. 10.1002/jmri.1207.
Kim S, Quon H, Loevner LA, Rosen MA, Dougherty L, Kilger AM, Glickson JD, Poptani H: Transcytolemmal water exchange in pharmacokinetic analysis of dynamic contrast-enhanced MRI data in squamous cell carcinoma of the head and neck. J Magn Reson Imaging. 2007, 26: 1607-1617. 10.1002/jmri.21207.
Van Cann EM, Rijpkema M, Heerschap A, Bilt van der A, Koole R, Stoelinga PJ: Quantitative dynamic contrast-enhanced MRI for the assessment of mandibular invasion by squamous cell carcinoma. Oral Oncol. 2008, 44 (12): 1147-1154. 10.1016/j.oraloncology.2008.02.009.
Evelhoch J, Garwood M, Vigneron D, Knopp M, Sullivan D, Menkens A, Clarke L, Liu G: Expanding the use of magnetic resonance in the assessment of tumor response to therapy: workshop report. Cancer Res. 2005, 65: 7041-7044. 10.1158/0008-5472.CAN-05-0674.
Cheng HL: Investigation and optimization of parameter accuracy in dynamic contrast-enhanced MRI. J Magn Reson Imaging. 2008, 28: 736-743. 10.1002/jmri.21489.
Asaumi J, Yanagi Y, Konouchi H, Hisatomi M, Matsuzaki H, Kishi K: Application of dynamic contrast-enhanced MRI to differentiate malignant lymphoma from squamous cell carcinoma in the head and neck. Oral Oncol. 2004, 40: 579-584. 10.1016/j.oraloncology.2003.12.002.
Hoskin PJ, Saunders MI, Goodchild K, Powell ME, Taylor NJ, Baddeley H: Dynamic contrast enhanced magnetic resonance scanning as a predictor of response to accelerated radiotherapy for advanced head and neck cancer. Br J Radiol. 1999, 72: 1093-1098.
Konouchi H, Asaumi J, Yanagi Y, Shigehara H, Hisatomi M, Matsuzaki H, Kishi K: Evaluation of tumor proliferation using dynamic contrast enhanced-MRI of oral cavity and oropharyngeal squamous cell carcinoma. Oral Oncol. 2003, 39: 290-295. 10.1016/S1368-8375(02)00119-7.
Noworolski SM, Fischbein NJ, Kaplan MJ, Lu Y, Nelson SJ, Carvajal L, Henry RG: Challenges in dynamic contrast-enhanced MRI imaging of cervical lymph nodes to detect metastatic disease. J Magn Reson Imaging. 2003, 17: 455-462. 10.1002/jmri.10280.
Yanagi Y, Asaumi J, Konouchi H, Hisatomi M, Matsuzaki H, Murakami J, Maki Y, Unetsubo T, Kishi K: Enhancement effects of test injection with a small amount of MR contrast medium in the oral and maxillofacial region. Eur J Radiol. 2006, 59: 367-370. 10.1016/j.ejrad.2006.03.017.
Parker GJ, Roberts C, Macdonald A, Buonaccorsi GA, Cheung S, Buckley DL, Jackson A, Watson Y, Davies K, Jayson GC: Experimentally-derived functional form for a population-averaged high-temporal-resolution arterial input function for dynamic contrast-enhanced MRI. Magn Reson Med. 2006, 56: 993-1000. 10.1002/mrm.21066.
Wang Y, Huang W, Panicek DM, Schwartz LH, Koutcher JA: Feasibility of using limited population based arterial input function for pharmacokinetic modeling of osteosarcoma dynamic contrast enhanced MRI data. Magn Reson Med. 2008, 59: 1183-1189. 10.1002/mrm.21432.
Tofts PS: Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. J Magn Reson Imaging. 1997, 7: 91-101. 10.1002/jmri.1880070113.
Haase A, Snapshot FLASH MRI: Applications to T1, T2, and chemical-shift imaging. Magn Reson Med. 1990, 13: 77-89. 10.1002/mrm.1910130109.
Parker GJ, Suckling J, Tanner SF, Padhani AR, Revell PB, Husband JE, Leach MO: Probing tumor microvascularity by measurement, analysis and display of contrast agent uptake kinetics. J Magn Reson Imaging. 1997, 7: 564-574. 10.1002/jmri.1880070318.
Yankeelov TE, Rooney WD, Huang W, Dyke JP, Li X, Tudorica A, Lee JH, Koutcher JA, Springer CS: Evidence for shutter-speed variation in CR bolus-tracking studies of human pathology. NMR Biomed. 2005, 18: 173-185. 10.1002/nbm.938.
Weinmann HJ, Laniado M, Mutzel W: Pharmacokinetics of GdDTPA/dimeglumine after intravenous injection into healthy volunteers. Physiol Chem Phys Med NMR. 1984, 16: 167-172.
Li X, Rooney WD, Springer CS: A unified magnetic resonance imaging pharmacokinetic theory: intravascular and extracellular contrast reagents. Magn Reson Med. 2005, 54: 1351-1359. 10.1002/mrm.20684.
Bland JM, Altman : Statistical methods for assessing agreement between two methods of clinical measurements. Lancet. 1986, 1: 307-310.
Riabkov DY, Di Bella EV: Estimation of kinetic parameters without input functions: analysis of three methods for multichannel blind identification. IEEE Trans Biomed Eng. 2002, 49: 1318-1327. 10.1109/TBME.2002.804588.
Yankeelov TE, Luci JJ, Lepage M, Li R, Debusk L, Lin PC, Price RR, Gore JC: Quantitative pharmacokinetic analysis of DCE-MRI data without an arterial input function: a reference region model. Magn Reson Imaging. 2005, 23: 519-529. 10.1016/j.mri.2005.02.013.
Walker-Samuel S, Leach MO, Collins DJ: Reference tissue quantification of DCE-MRI data without a contrast agent calibration. Phys Med Biol. 2007, 52: 589-601. 10.1088/0031-9155/52/3/004.
Walker-Samuel S, Parker CC, Leach MO, Collins DJ: Reproducibility of reference tissue quantification of dynamic contrast-enhanced data: comparison with a fixed vascular input function. Phys Med Biol. 2007, 52: 75-89. 10.1088/0031-9155/52/1/006.
Yang C, Karczmar GS, Medved M, Stadler WM: Estimating the arterial input function using two reference tissues in dynamic contrast-enhanced MRI studies: fundamental concepts and simulations. Magn Reson Med. 2004, 52: 1110-1117. 10.1002/mrm.20243.
Roberts C, Buckley DL, Parker GJ: Comparison of errors associated with single- and multi-bolus injection protocols in low-temporal-resolution dynamic contrast-enhanced tracer kinetic analysis. Magn Reson Med. 2006, 56: 611-619. 10.1002/mrm.20971.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1756-6649/9/4/prepub
Acknowledgements
We thank Ms. Maayan Korenblit for managing the patient data base, Dr Yousef Mazaheri for helping with the phantom study and Dr Jacobus F.A. Jansen for helpful suggestions. This study was conducted with support from National Cancer Institute/National Institutes of Health (grant number 1 R01 CA115895).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
1) All authors (ASD, NL, HS, YW, WH, SGP, JPS and JAK) have made contributions to conceptions and design of this study. NL, SGP, JPS referred patients for the study. ASD, YW, HS, WH, JAK made contribution in acquisition of data, and interpretation of data; 2) ASD, WH and JAK have been involved in drafting the manuscript or revising it critically for important intellectual content; and 3) All authors have given final approval of the version to be submitted to BMC Medical Physics.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Shukla-Dave, A., Lee, N., Stambuk, H. et al. Average arterial input function for quantitative dynamic contrast enhanced magnetic resonance imaging of neck nodal metastases. BMC Med Phys 9, 4 (2009). https://doi.org/10.1186/1756-6649-9-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-6649-9-4