Abstract
Background
The Leishmaniases are a group of clinically diverse diseases caused by parasites of the genus Leishmania. To distinguish between species is crucial for correct diagnosis and prognosis as well as for treatment decisions. Recently, sequencing of the HSP70 coding region has been applied in phylogenetic studies and for identifying of Leishmania species with excellent results.
Methods
In the present study, we analyzed the 3’-untranslated region (UTR) of Leishmania HSP70-type I gene from 24 strains representing eleven Leishmania species in the belief that this non-coding region would have a better discriminatory capacity for species typing than coding regions.
Results
It was observed that there was a remarkable degree of sequence conservation in this region, even between species of the subgenus Leishmania and Viannia. In addition, the presence of many microsatellites was a common feature of the 3´-UTR of HSP70-I genes in the Leishmania genus. Finally, we constructed dendrograms based on global sequence alignments of the analyzed Leishmania species and strains, the results indicated that this particular region of HSP70 genes might be useful for species (or species complex) typing, improving for particular species the discrimination capacity of phylogenetic trees based on HSP70 coding sequences. Given the large size variation of the analyzed region between the Leishmania and Viannia subgenera, direct visualization of the PCR amplification product would allow discrimination between subgenera, and a Hae III-PCR-RFLP analysis might be used for differentiating some species within each subgenera.
Conclusions
Sequence and phylogenetic analyses indicated that this region, which is readily amplified using a single pair of primers from both Old and New World Leishmania species, might be useful as a molecular marker for species discrimination.
Similar content being viewed by others
Background
Protozoan parasites of Leishmania genus cause a severe disease, Leishmaniasis, which threatens 350 million people worldwide and some 2 million new cases occur yearly. Furthermore, mortality and morbidity from Leishmaniasis worldwide show a worrying increasing trend [1]. Infection by the parasite Leishmania can result in a broad spectrum of pathological outcomes in the human host, ranging from simple self-healing cutaneous lesions (cutaneous Leishmaniasis, CL) to acute visceral Leishmaniasis (VL), commonly referred to as kala-azar. The different pathologies usually correlate with infection by different species [2]. Thus, Leishmania donovani and Leishmania infantum usually cause VL, whereas the rest of Leishmania species generally cause cutaneous Leishmaniasis (CL). Nevertheless, exceptional cases have been described, such as visceral outcomes in individuals infected with L. tropica. After self-cure or successful chemotherapy, many patients remain asymptomatically infected; however, such persistence often gives rise to disease reactivation, including post-kala-azar dermal Leishmaniasis after cure of VL caused by L. donovani, diffuse cutaneous Leishmaniasis (DCL) lesions in L. mexicana-infected people, or the development of destructive mucocutaneous Leishmaniasis (espundia) months or years after healing of a localized cutaneous ulcer caused by L. braziliensis[1].
On the other hand, treatment outcome in patients can vary with the infecting Leishmania species because of different sensitivity of the parasite species both to the standard drugs, pentavalent antimonials and miltefosine, and those on clinical trial, paromomycin [3]. Thus, correct diagnosis of the infecting Leishmania species is crucial for making decisions regarding prognosis and treatments, and also for epidemiological monitoring of the parasite spread [4].
To date, around 20 Leishmania species are known to infect humans, even though the species status is under discussion for some of them [1, 5]. In fact, a major conceptual problem in epidemiology of Leishmaniasis is the taxonomy of the etiological agent as Leishmania is a morphologically uniform genus. At present, multilocus enzyme electrophoresis (MLEE), an isoenzyme analysis that is based on 13 enzymes, is the reference method for Leishmania typing [1]. However, MLEE has important limitations: strains with the same enzyme phenotype may have distinct amino acid sequences, the degree of relationship between different phenotypes is not known, and putative heterozygous phenotypes are difficult to interpret [6, 7]. Furthermore, this technique is time consuming, requires mass parasite culturing, and it should be done by one of the few reference centres [1]. Therefore, its usefulness for taxonomic purposes is under dispute [8]. In conclusion, new methods for species identification are needed to ensure proper identification and therapy.
Molecular biology techniques, such as PCR and DNA sequencing, are replacing traditional methods of taxonomy, as they tend to be more specific and easier to perform. Thus, sequencing of genes such as DNA polymerase α [9], RNA polymerase II [10], gp63 [11], cytochrome oxidase II [12, 13], N-acetylglucosamine-1-phosphate transferase [14], cystein protease b [15], cytochrome b [16, 17], SSU rDNA [18], and 7SL RNA [19] has been carried out for phylogenetic studies. In addition, sequencing of ribosomal DNA internal transcribed spacers [20, 21] and kinetoplast DNA minicircles [22] is being used for taxonomic purposes within the Leishmania genus. Remarkably, in recent studies, coding regions of genes for most metabolic enzymes used in the MLEE typing method have been determined and used for Leishmania typing [6, 23, 24]. As expected, these studies have demonstrated that DNA sequencing has greater discriminatory power than MLEE typing. Accordingly, this approach, based on sequence analysis of selected genes, has been dubbed MLST (for “multilocus sequence typing”) [6].
The HSP70 gene is being extensively used as a target for PCR-RFLP assays for Leishmania species discrimination [25, 26]. In fact, Montalvo and co-workers [27] have designed a PCR-RFLP method allowing identification of most medically relevant Old and New World Leishmania species on the basis of HSP70 PCR amplification. More recently, sequence analysis of the 1380-bp fragment of the HSP70 coding region, commonly used in the PCR-RFLP method, has been directly used for phylogenetic and taxonomic purposes [26, 28]. The phylogenetic trees obtained by da Silva et al.[26] and Fraga et al.[28] demonstrate that DNA sequencing of HSP70 coding regions represent a valuable technique for Leishmania species identification. As a remarkable result from these studies, Fraga and coworkers [28] defined eight, highly supported, monophyletic groups within the genus Leishmania: (1) L. donovani/L. infantum, (2) L. tropica/L. aethiopica, (3) L. major, (4) L. mexicana/L. amazonensis, (5) L. naiffi, (6) L. braziliensis/L. peruviana, (7) L. guyanensis/L. panamensis, and (8) L. lainsoni. Nevertheless, the HSP70 protein and the coding region of its gene are among the best conserved sequences along the evolutionary tree of life [29]. As untranslated regions (UTR) of genes are expected to have a lower evolutionary constriction than coding regions (CDR), it is conceivable that UTR sequences may be a better target than CDR for typing of closely related organisms [30]. In the present study, we sequenced most of the 3’-untranslated region (UTR) of Leishmania HSP70-type I (HSP70-I) gene from different species and strains with the belief that this region might be useful for species typing within the genus, having hopefully a more discriminating capacity than the HSP70 coding region.
Methods
DNA extraction, amplification, and cloning
The Leishmania isolates used in this study are listed in Table 1. Promastigotes were cultivated in RPMI medium supplemented with 10% heat-inactivated fetal bovine serum at 26°C. Total genomic DNA was extracted from promastigotes using a standard sodium dodecyl sulfate-proteinase K procedure as previously described [31]. For amplification, the primers 70-IR-D (5’- CCAAGGTCGA GGAGGTCGAC TA- 3’) and 70-IR-M (5’- ACGGGTAGGG GGAGGAAAGA -3’) were used (Figure 1). Amplifications were done with the Maxime PCR PreMix Kit (i-Taq; Intron Biotechnology), using 20–40 ng of genomic DNA, dNTPs (2.5 mM each) and 0.5 μM of each primer. The PCR profile was 95°C for 2 min followed by 30 cycles of 95°C for 30 sec, 62.5°C for 30 sec, 72°C for 1 min and 20 sec, and then a final elongation step at 72°C for 5 min. The PCR products were checked by agarose gel electrophoresis and purified using a commercial DNA purification kit (FavorPrep™ Gel/PCR Purification Kit; Favorgen), following the manufacturer’s instructions.
Schematic map of the Leishmania HSP70 locus (fragment), showing the region amplified in this study. The map was drawn to scale [39]. PCR primer annealing sites are indicated by arrows. CDR, protein coding region; 5’- and 3’-UTR, untranslated regions; IR, intergenic region.
Microsatellite distribution in the 3’-UTR of HSP70-I genes in several Leishmania species. The repeated motifs correspond to those found in the sense strand. Note that the drawing scale is not proportional for the different Leishmania species; however, for a given sequence, the relative positions of the microsatellites are scaled.
For cloning, the purified PCR products were ligated into the pCR2.1 vector using the TA Cloning Kit (Invitrogen), and the ligation product used to transform XL1-blue competent bacteria [35]. DNA sequencing was carried out in the facilities of the Servicio de Genómica (Parque Tecnológico de Madrid-UAM), using Big Dye Terminators v3.1 kit (Applied Biosystem).
Sequence analysis
The chromatograms were visualized using the BioEdit Sequence Alignment Editor [36]. For all the clones, sequencing of both strands was carried out.
For all analyses, the priming sites were trimmed from both ends of all sequences. A search for microsatellite sequences was performed using the “Microsatellite repeats finder” program [37]. Parameter settings were: length of repeated sequence (minimum, 2; maximum, 6); minimum number of repeats, 3; minimum length of tandem repeat, 6; and allowed percentage of mismatches, 10%.
All sequences were multiple aligned with the default option using ClustalW2.1 at The European Bioinformatics Institute [33]. For phylogenetic studies, the multiple sequence alignment was analyzed using the software MEGA4 [32]. The evolutionary history was inferred using the Neighbor-Joining and Minimum Evolution methods [34], and the bootstrap consensus tree inferred from 1000 replicates. The evolutionary distances were computed using the Maximum Composite Likelihood method [38] and the units represent the number of base substitutions per site. All positions containing gaps and missing data were eliminated from the dataset.
Sequence data
Sequences generated in this study have been deposited in the EMBL-EBI database under accession numbers HE575325-HE575343 (Table 1).
Results and discussion
PCR amplification of non-coding sequences of Leishmania HSP70-I genes
The HSP70 locus in Leishmania consists of several HSP70 gene copies arranged in a head-to-tail manner [39, 40]. All the genes are highly conserved in sequence with the exception of the gene located at the 3’ end of the cluster, which presents a 3’-UTR different to the rest of genes. Hence, the latter gene is referred to as HSP70-II gene and the other genes as HSP70-I genes. Given the high sequence conservation of HSP70 CDR among the different Leishmania species [41], we designed oligonucleotides at the 5’ and 3’ extremities of the CDR with the purpose of PCR amplify the entire intercoding region of HSP70-I genes. The intercoding region (3’-UTR + intergenic region (IR) + 5’-UTR) in the L. infantum HSP70 locus is 1600 bp in length [39]. However, the task turned out to be complicated, as it was only possible to amplify and clone the expected region for a few Leishmania species. In order to overcome this difficulty, we searched for conserved sequence blocks within the 3’-UTR of HSP70-I genes from different Leishmania species, whose sequences were available in the databases. Finally, we succeeded and designed an oligonucleotide pair able to efficiently amplify a 3’-UTR sequence from all the Leishmania DNA samples, irrespective of the subgenus (Leishmania or Viannia) to which they belonged. See Figure 1 for details on the location of amplification primers 70-IR-D and 70-IR-M in the HSP70 locus.
The UTR-I fragment for 20 strains of different Leishmania species were PCR-amplified and cloned. However, it was not possible to obtain a complete sequence for the strain MHOM/GR/80/GR-L35 of L. tropica, even after using alternative sequencing protocols in which DMSO or betaine was added [42]. As a result, this strain was not included in subsequent studies. For many of the other sequences, the sequence chromatogram was not uniform, showing a drastic decrease in intensity after passing GC-rich regions. This fact may be an indication that the UTR-I fragment contains regions with strong secondary structures that were hindering (or totally blocking in one case, as mentioned above) the polymerase advance during sequencing reaction.
In addition to the 19 sequences obtained in this work, we retrieved 5 sequences from databases, amounting to a total of 24 sequences (Table 1). It should be noted that sequence information of this region is not yet available in L. mexicana[43], L. donovani[43], and L. tarentolae[44] genome sequencing projects, which are ongoing. The HSP70-UTR-I amplified region varied in size from 516 (in L. peruviana) to 733 bp (in L. infantum). Thus, the region is clearly shorter in Leishmania species of the subgenus Viannia than in species of the subgenus Leishmania, and this fact might be exploited as a PCR-based method for quick discrimination between both Leishmania subgenera. Within a given species, the length of this region shows a very low inter-strain variation; for example, the size of the region in four different L. donovani strains varies from 711 to 718, in L. infantum (four strains) ranges from 729 to 733, in L. major (3 strains) ranges from 715 to 718, and in L. aethiopica (3 strains) ranges from 696 to 701. Within a given Leishmania species, size variants are a consequence of variations in the repeat number of microsatellite sequences (see Figure 2).
The HSP70 UTR-I contains a remarkable abundance of microsatellites
A search for structural motifs along the sequenced UTR-I fragment in the different Leishmania species was carried out. The study showed a significant richness in microsatellites sequences (Figure 2). Microsatellites are tandemly repeated stretches of short nucleotide motives of 1–8 bp ubiquitously distributed in eukaryotic genomes [45]. The Leishmania genome is not an exception, and multilocus microsatellite typing (MLMT) is being used as a powerful tool for population genetic and epidemiological studies of Leishmania[46]. In the L. infantum, L. major and L. braziliensis genomes, the CA repeats represent by far the most common microsatellite [47]. Indeed, CA-microsatellites (read as TG or GT on the complementary strand) were the more abundant class in the UTR-I of all Leishmania species, showing most of them have an antisense orientation (Figure 2). TGC-microsatellites, another abundant repeat in Leishmania genomes [47], were found in the Leishmania species of the subgenus Leishmania but absent in the species of the subgenus Viannia. However, the analyzed HSP70 UTR-I fragment contains many other microsatellites (C, GC, GTG, GGC, GCG) that may be categorized as uncommon for Leishmania.
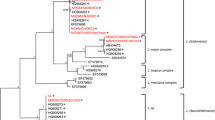
Phylogeny of Leishmania according to the Neighbor-joining (panel A) and Minimum Evolution (panel B) methods based on the multiple sequence alignment of 3’UTR of HSP70-I gene. Numbers above the branch represent percent recovery in 1,000 bootstrap pseudoreplicates analysis (only values above 90 are shown). All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). There were a total of 493 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 [32]. Distance represents the number of base substitutions per site. Species are abbreviated as follows: Lae, L. aethiopica; Lam, L. amazonensis; Lb, L. braziliensis; Ld, L. donovani; Lg, L. guyanensis; Li, L. infantum; Ll, L. lainsoni; Lmj, L. major; Lp, L. peruviana; Lpa, L. panamensis; Lt, L. tropica. See Table 1 for additional information of the strains.
The HSP70 locus in T. cruzi, T. brucei and Leishmania spp. has a similar genomic organization, i.e. it contains several genes that are head-to-tail tandemly organized. However, the UTRs of HSP70 genes are extremely divergent between Trypanosoma and Leishmania genera. Thus, the 3’UTR of HSP70 genes in T. brucei and T. cruzi are only 186 and 213 nucleotides in length, respectively [48, 49]. In contrast, the 3’-UTR of Leishmania HSP70 genes are higher than 1000 nucleotides in length for species of the Leishmania subgenus [39], and 936 nucleotides for the 3’-UTR-I in Leishmania braziliensis[30]. These findings suggest that UTRs of HSP70 genes have evolved towards opposite ends regarding sequence complexity in Trypanosoma and Leishmania. On the other hand, from the analysis of Figure 2, it became clear that the analyzed region of the HSP70 3’UTR has experienced also a notable evolution after separation of subgenus Leishmania and Viannia. In the species of the subgenus Leishmania, this region has grown both in size and microsatellite complexity. Interestingly, the regulation of HSP70-I expression seems to be more complex in species of the subgenus Leishmania than in L. braziliensis[40]. Thus, it would be interesting to analyze the contribution of the different microsatellites to the specific regulation of HSP70-I genes in Leishmania.
Phylogenetic relationships of Leishmania species based on sequences of the HSP70-I UTR
From the structural analysis of this region in the different Leishmania species, it became evident that this fragment of HSP70-I genes has greatly diverged between the subgenus Leishmania and Viannia (Figure 2). In order to study the usefulness of these sequences to discriminate at the level of species within each subgenus, phylogenetic analyses were carried out (Figure 3). The inferred phylogenetic tree based on either the Neighbor-Joining or Minimum Evolution methods indicated that the sequenced region may be useful for clustering Leishmania strains at the level of species or complex of species (99 to 100% bootstrap support). Not surprisingly, the subgenera Leishmania and Viannia constitute two distinct monophyletic lines. In addition, our phylogenetic analyses gave a robust support for the following Leishmania species or clusters: L. amazonensis (which forms a separated branch from the Old World Leishmania species), L. infantum/L. donovani (L. donovani complex), L. major L. tropica and L. aethiopica, within the Leishmania subgenus; and L. braziliensis/L. peruviana (L. braziliensis complex), L. guyanensis/L. panamensis (L. guyanensis complex), and L. lainsoni, within the Viannia subgenus. With few differences, our phylogenetic tree was coincident with that obtained by Fraga et al.[28] using sequences of the Leishmania HSP70 CDR. Nevertheless, our phylogenetic results support a separation between L. tropica and L. aethiopica (99% of bootstrap value; Figure 3), whereas they cannot be distinguished based on HSP70 coding sequences.
Separation between L. donovani and L. infantum strains remains as an issue to be resolved, even though several comprehensive studies have been conducted [20, 46, 50–53]. In our phylogenetic tree (Figure 3), separation of L. donovani and L. infantum strains was observed, even though the bootstrap value (50-52%, depending on the phylogenetic method) was not statistically significant. In the future, we plan to analyze a large number of L. donovani and L. infantum strains in order to define signatures within the 3’-UTRI that could allow discriminating specifically among both species (or subspecies).
Although sequencing is becoming a cost effective methodology, it is still not feasible for routine diagnosis, being more attractive other methodologies such as restriction fragment length polymorphisms of PCR amplicons (PCR-RFLP). This methodology has been found to be valuable for Leishmania species discrimination, using amplicons from the HSP70 coding region [26, 27]. We analyzed the sequence of the UTR-I amplicons described in this study searching for restriction enzyme cleavage sites that could be used for PCR-RFLP-based identification of Leishmania species. On the basis of digestion with Hae III restriction enzyme, our in silico analyses predicted seven different patterns (Table 2) that would allow the separation of the following species of groups of species: L. aethiopica /L. tropica, L. amazonensis, L. braziliensis/L. peruviana, L. donovani/L. infantum, L. guyanensis/L. panamensis, L. lainsoni and L. major.
Conclusions
Identification of the exact parasite species isolated from human and animal hosts, as well as from sand fly vectors, gains increasing importance for diagnostics, epidemiological surveillance, and clinical studies. As sequencing is becoming a cost-effective methodology, it is possible to envision the use of a battery of genes as genomic markers for phylogenetic analyses of Leishmania species towards the goal of both a molecular taxonomy and a definitive diagnostic method. Our data show the potential of the UTR of HSP70-I genes as a molecular marker for Leishmania species typing. In addition, this study has shown the existence of complex arrays of microsatellites in the UTR of HSP70-I, whose involvement in regulatory mechanisms of gene expression will be analyzed in future studies.
Abbreviations
- MLEE:
-
Multilocus enzyme electrophoresis
- UTR:
-
Untranslated region
- CDR:
-
Coding region.
References
WHO Expert Committee: Control of the Leishmaniases. 2010, WHO Press, Geneva, Switzerland
Bañuls AL, Hide M, Prugnolle F: Leishmania and the Leishmaniases: a parasite genetic update and advances in taxonomy, epidemiology and pathogenicity in humans. Adv Parasitol. 2007, 64: 1-109.
Croft SL, Sundar S, Fairlamb AH: Drug resistance in Leishmaniasis. Clin Microbiol Rev. 2006, 19 (1): 111-126. 10.1128/CMR.19.1.111-126.2006.
Mugasa CM, Laurent T, Schoone GJ, Basiye FL, Saad AA, El Safi S, Kager PA, Schallig HDFH: Simplified molecular detection of Leishmania parasites in various clinical samples from patients with Leishmaniasis. Parasit Vectors. 2010, 3 (1): 13-10.1186/1756-3305-3-13.
Schonian G, Mauricio I, Cupolillo E: Is it time to revise the nomenclature of Leishmania?. Trends Parasitol. 2010, 26 (10): 466-469. 10.1016/j.pt.2010.06.013.
Mauricio IL, Yeo M, Baghaei M, Doto D, Pratlong F, Zemanova E, Dedet JP, Lukes J, Miles MA: Towards multilocus sequence typing of the Leishmania donovani complex: resolving genotypes and haplotypes for five polymorphic metabolic enzymes (ASAT, GPI, NH1, NH2, PGD). Int J Parasitol. 2006, 36 (7): 757-769. 10.1016/j.ijpara.2006.03.006.
Schonian G, Kuhls K, Mauricio IL: Molecular approaches for a better understanding of the epidemiology and population genetics of Leishmania. Parasitology. 2011, 138 (4): 405-425. 10.1017/S0031182010001538.
Van der Auwera G, Fraga J, Montalvo AM, Dujardin JC: Leishmania taxonomy up for promotion?. Trends Parasitol. 2011, 27 (2): 49-50. 10.1016/j.pt.2010.11.007.
Croan DG, Morrison DA, Ellis JT: Evolution of the genus Leishmania revealed by comparison of DNA and RNA polymerase gene sequences. Mol Biochem Parasitol. 1997, 89 (2): 149-159. 10.1016/S0166-6851(97)00111-4.
Croan D, Ellis J: Phylogenetic relationships between Leishmania, Viannia and SauroLeishmania inferred from comparison of a variable domain within the RNA polymerase II largest subunit gene. Mol Biochem Parasitol. 1996, 79 (1): 97-102. 10.1016/0166-6851(96)02629-1.
Mauricio IL, Howard MK, Stothard JR, Miles MA: Genomic diversity in the Leishmania donovani complex. Parasitology. 1999, 119 (Pt 3): 237-246.
Ibrahim ME, Barker DC: The origin and evolution of the Leishmania donovani complex as inferred from a mitochondrial cytochrome oxidase II gene sequence. Infect Genet Evol. 2001, 1 (1): 61-68. 10.1016/S1567-1348(01)00009-0.
Cao DP, Guo XG, Chen DL, Chen JP: Species delimitation and phylogenetic relationships of Chinese Leishmania isolates reexamined using kinetoplast cytochrome oxidase II gene sequences. Parasitol Res. 2011, 109 (1): 163-173. 10.1007/s00436-010-2239-6.
Waki K, Dutta S, Ray D, Kolli BK, Akman L, Kawazu S, Lin CP, Chang KP: Transmembrane molecules for phylogenetic analyses of pathogenic protists: Leishmania-specific informative sites in hydrophilic loops of trans- endoplasmic reticulum N-acetylglucosamine-1-phosphate transferase. Eukaryot Cell. 2007, 6 (2): 198-210. 10.1128/EC.00282-06.
Hide M, Bras-Goncalves R, Bañuls AL: Specific cpb copies within the Leishmania donovani complex: evolutionary interpretations and potential clinical implications in humans. Parasitology. 2007, 134 (Pt 3): 379-389.
Asato Y, Oshiro M, Myint CK, Yamamoto Y, Kato H, Marco JD, Mimori T, Gomez EAL, Hashiguchi Y, Uezato H: Phylogenic analysis of the genus Leishmania by cytochrome b gene sequencing. Exp Parasitol. 2009, 121 (4): 352-361. 10.1016/j.exppara.2008.12.013.
Martinez LP, Rebollo JA, Luna AL, Cochero S, Bejarano EE: Molecular identification of the parasites causing cutaneous Leishmaniasis on the Caribbean coast of Colombia. Parasitol Res. 2010, 106 (3): 647-652. 10.1007/s00436-009-1712-6.
de Lima ACS, Zampieri RA, Tomokane TY, Laurenti MD, Silveira FT, Corbett CEP, Floeter-Winter LM, Gomes CMC: Leishmania sp. identification by PCR associated with sequencing of target SSU rDNA in paraffin-embedded skin samples stored for more than 30 years. Parasitol Res. 2011, 108 (6): 1525-1531. 10.1007/s00436-010-2208-0.
Stevenson LG, Fedorko DP, Zelazny AM: An enhanced method for the identification of Leishmania spp. using real-time polymerase chain reaction and sequence analysis of the 7SL RNA gene region. Diagn Microbiol Infect Dis. 2010, 66 (4): 432-435. 10.1016/j.diagmicrobio.2009.11.005.
Kuhls K, Mauricio IL, Pratlong F, Presber W, Schonian G: Analysis of ribosomal DNA internal transcribed spacer sequences of the Leishmania donovani complex. Microbes Infect. 2005, 7 (11–12): 1224-1234.
Odiwuor SOC, Saad AA, De Doncker S, Maes I, Laurent T, El Safi S, Mbuchi M, Buscher P, Dujardin JC, Van der Auwera G: Universal PCR assays for the differential detection of all Old World Leishmania species. Eur J Clin Microbiol Infect Dis. 2011, 30 (2): 209-218. 10.1007/s10096-010-1071-3.
Pita-Pereira D, Lins R, Oliveira MP, Lima RB, Pereira BA, Moreira OC, Brazil RP, Britto C: SYBR Green-based real-time PCR targeting kinetoplast DNA can be used to discriminate between the main etiologic agents of Brazilian cutaneous and visceral Leishmaniases. Parasit Vectors. 2012, 5: 15-10.1186/1756-3305-5-15.
Zemanova E, Jirku M, Mauricio IL, Horak A, Miles MA, Lukes J: The Leishmania donovani complex: genotypes of five metabolic enzymes (ICD, ME, MPI, G6PDH, and FH), new targets for multilocus sequence typing. Int J Parasitol. 2007, 37 (2): 149-160. 10.1016/j.ijpara.2006.08.008.
Tsukayama P, Lucas C, Bacon DJ: Typing of four genetic loci discriminates among closely related species of New World Leishmania. Int J Parasitol. 2009, 39 (3): 355-362. 10.1016/j.ijpara.2008.08.004.
Garcia L, Kindt A, Bermudez H, Llanos-Cuentas A, De Doncker S, Arevalo J, Wilber Quispe Tintaya K, Dujardin JC: Culture-independent species typing of neotropical Leishmania for clinical validation of a PCR-based assay targeting heat shock protein 70 genes. J Clin Microbiol. 2004, 42 (5): 2294-2297. 10.1128/JCM.42.5.2294-2297.2004.
da Silva LA, Santos de Sousa CD, da Graca GC, Porrozzi R, Cupolillo E: Sequence analysis and PCR-RFLP profiling of the hsp70 gene as a valuable tool for identifying Leishmania species associated with human Leishmaniasis in Brazil. Infect Genet Evol. 2010, 10 (1): 77-83. 10.1016/j.meegid.2009.11.001.
Montalvo AM, Fraga J, Monzote L, Montano I, De Doncker S, Dujardin JC, Van der Auwera G: Heat-shock protein 70 PCR-RFLP: a universal simple tool for Leishmania species discrimination in the New and Old World. Parasitology. 2010, 137 (8): 1159-1168. 10.1017/S0031182010000089.
Fraga J, Montalvo AM, De Doncker S, Dujardin JC, Van der Auwera G: Phylogeny of Leishmania species based on the heat-shock protein 70 gene. Infect Genet Evol. 2010, 10 (2): 238-245. 10.1016/j.meegid.2009.11.007.
Folgueira C, Requena JM: A postgenomic view of the heat shock proteins in kinetoplastids. FEMS Microbiol Rev. 2007, 31 (4): 359-377. 10.1111/j.1574-6976.2007.00069.x.
Ramirez CA, Requena JM, Puerta CJ: Identification of the HSP70-II gene in Leishmania braziliensis HSP70 locus: genomic organization and UTRs characterization. Parasit Vectors. 2011, 4: 166-10.1186/1756-3305-4-166.
Requena JM, Lopez MC, Jimenez-Ruiz A, de la Torre JC, Alonso C: A head-to-tail tandem organization of hsp70 genes in Trypanosoma cruzi. Nucleic Acids Res. 1988, 16 (4): 1393-1406. 10.1093/nar/16.4.1393.
Tamura K, Dudley J, Nei M, Kumar S: MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007, 24 (8): 1596-1599. 10.1093/molbev/msm092.
European Bioinformatics Institute: [http://www.ebi.ac.uk/]
Saitou N, Nei M: The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987, 4 (4): 406-425.
Inoue H, Nojima H, Okayama H: High efficiency transformation of Escherichia coli with plasmids. Gene. 1990, 96 (1): 23-28. 10.1016/0378-1119(90)90336-P.
BioEdit Sequence Alignment Editor: [http://www.mbio.ncsu.edu/BioEdit/bioedit.html]
Microsatellite repeats finder : [http://www.biophp.org/minitools/microsatellite_repeats_finder/demo.php]
Tamura K, Nei M, Kumar S: Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004, 101 (30): 11030-11035. 10.1073/pnas.0404206101.
Quijada L, Soto M, Alonso C, Requena JM: Analysis of post-transcriptional regulation operating on transcription products of the tandemly linked Leishmania infantum hsp70 genes. J Biol Chem. 1997, 272 (7): 4493-4499. 10.1074/jbc.272.7.4493.
Folgueira C, Cañavate C, Chicharro C, Requena JM: Genomic organization and expression of the HSP70 locus in New and Old World Leishmania species. Parasitology. 2007, 134 (3): 369-377. 10.1017/S0031182006001570.
Quijada L, Requena JM, Soto M, Alonso C: During canine viscero-cutaneous Leishmaniasis the anti-Hsp70 antibodies are specifically elicited by the parasite protein. Parasitology. 1996, 112 (Pt 3): 277-284.
Kieleczawa J, Mazaika E: Optimization of protocol for sequencing of difficult templates. J Biomol Tech. 2010, 21 (2): 97-102.
Wellcome Trust Sanger Institute, Pathogen Sequencing Unit. GeneDB : [http://www.genedb.org/]
Raymond F, Boisvert S, Roy G, Ritt JF, Legare D, Isnard A, Stanke M, Olivier M, Tremblay MJ, Papadopoulou B, Ouellette M, Corbeil J: Genome sequencing of the lizard parasite Leishmania tarentolae reveals loss of genes associated to the intracellular stage of human pathogenic species. Nucleic Acids Res. 2012, 40 (3): 1131-1147. 10.1093/nar/gkr834.
Richard GF, Kerrest A, Dujon B: Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. 2008, 72 (4): 686-727. 10.1128/MMBR.00011-08.
Kuhls K, Keilonat L, Ochsenreither S, Schaar M, Schweynoch C, Presber W, Schonian G: Multilocus microsatellite typing (MLMT) reveals genetically isolated populations between and within the main endemic regions of visceral Leishmaniasis. Microbes Infect. 2007, 9 (3): 334-343. 10.1016/j.micinf.2006.12.009.
Fakhar M, Motazedian MH, Daly D, Lowe CD, Kemp SJ, Noyes HA: An integrated pipeline for the development of novel panels of mapped microsatellite markers for Leishmania donovani complex, Leishmania braziliensis and Leishmania major. Parasitology. 2008, 135 (5): 567-574.
Lee MG: The 3' untranslated region of the hsp 70 genes maintains the level of steady state mRNA in Trypanosoma brucei upon heat shock. Nucleic Acids Res. 1998, 26 (17): 4025-4033. 10.1093/nar/26.17.4025.
Rodrigues DC, Silva R, Rondinelli E, Urmenyi TP: Trypanosoma cruzi: modulation of HSP70 mRNA stability by untranslated regions during heat shock. Exp Parasitol. 2010, 126 (2): 245-253. 10.1016/j.exppara.2010.05.009.
Quispe-Tintaya KW, Laurent T, Decuypere S, Hide M, Banuls AL, De Doncker S, Rijal S, Cañavate C, Campino L, Dujardin JC: Fluorogenic assay for molecular typing of the Leishmania donovani complex: taxonomic and clinical applications. J Infect Dis. 2005, 192 (4): 685-692. 10.1086/432077.
Botilde Y, Laurent T, Quispe Tintaya W, Chicharro C, Canavate C, Cruz I, Kuhls K, Schonian G, Dujardin JC: Comparison of molecular markers for strain typing of Leishmania infantum. Infect Genet Evol. 2006, 6 (6): 440-446. 10.1016/j.meegid.2006.02.003.
Lukes J, Mauricio IL, Schonian G, Dujardin JC, Soteriadou K, Dedet JP, Kuhls K, Tintaya KWQ, Jirku M, Chocholova E: Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci U S A. 2007, 104 (22): 9375-9380. 10.1073/pnas.0703678104.
Haralambous C, Antoniou M, Pratlong F, Dedet JP, Soteriadou K: Development of a molecular assay specific for the Leishmania donovani complex that discriminates L. donovani/Leishmania infantum zymodemes: a useful tool for typing MON-1. Diagn Microbiol Infect Dis. 2008, 60 (1): 33-42. 10.1016/j.diagmicrobio.2007.07.019.
Acknowledgements
The authors would like to thank J. P. Dedet and Francine Pratlong (Centre National de Référence des Leishmania, Montpellier, France) who kindly donated some of the strains used in this study. We gratefully acknowledge assistance from our technician (Virginia Franco) and a Master’s student (Marta Truchado), who contributed to the cloning step. The careful revision of two anonymous reviewers and their comments and suggestions are also appreciated. This work was supported by grants from the Ministerio de Ciencia y Tecnología (BFU2009-08986), the Fondo de Investigaciones Sanitarias (ISCIII-RETIC RD06/0021/0008-FEDER and ISCIII-RETIC RD06/0021/0009-FEDER), and Agencia Española de Cooperación Internacional para el Desarrollo (AECID, A/024740/09). Also, an institutional grant from Fundación Ramón Areces is acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that non-financial competing interests exist.
Authors' contributions
CC and JMR conceived and designed the experiments. CCh and JMR carried out the molecular genetic studies. LG, RP and CJP contributed DNA samples and analysis tools. JMR performed the sequence data analyses and drafted the manuscript. CC helped to finalize the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Requena, J.M., Chicharro, C., García, L. et al. Sequence analysis of the 3’-untranslated region of HSP70 (type I) genes in the genus Leishmania: its usefulness as a molecular marker for species identification. Parasites Vectors 5, 87 (2012). https://doi.org/10.1186/1756-3305-5-87
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-3305-5-87