Abstract
Background
Dirofilaria immitis and Dirofilaria repens are transmitted by bloodsucking culicid mosquitoes belonging to Culex, Aedes, Ochlerotatus, Anopheles and Mansonia genera.
The detection of filarioids in mosquitoes for assessing distribution of vectors and/or of pathogens in a given area (also known as “xenomonitoring”), when based on individual dissection of wild-caught female mosquitoes is time consuming and hardly applicable in large epidemiological surveys.
Our study aimed to evaluate the recently developed duplex real-time PCR for screening large number of culicids and to assess their positivity for D. immitis and D. repens in an area where both species are endemic.
Methods
A duplex real-time PCR was used to detect and differentiate D. immitis and D. repens in mosquitoes collected in six provinces of the Veneto region using 43 carbon dioxide-baited traps under the frame of an entomological surveillance program to monitor the vectors of West Nile disease. From early May till October 2010, unfed female mosquitoes (n = 40,892) were captured in 20 selected sites.
Results
Mosquitoes identified as Culex pipiens, Ochlerotatus caspius, Aedes vexans and Culex modestus were grouped into 995 pools according to species, day and site of collection (from minimum of 1 to maximum of 57). Out of 955 pools, 23 (2.41 %) scored positive for Dirofilaria spp. of which, 21 (2.2 %) for D. immitis and two (0.21 %) for D. repens. An overall Estimated Rate of Infection (ERI) of 0.06 % was recorded, being higher in Och. caspius and Ae. vexans (i.e., 0.18 % and 0.14 %, respectively). At least one mosquito pool was positive for Dirofilaria spp. in each province with the highest ERI recorded in Vicenza and Padova provinces (i.e., 0.42% and 0.16 %, respectively). Mosquitoes collected in all provinces were positive for D. immitis whereas, only two (i.e., Padova and Rovigo) provinces scored positive for D. repens. All mosquito species, except for Cx. modestus, were positive for D. immitis, whereas D. repens was only found in Cx. pipiens.
Conclusions
The results suggest that both Dirofilaria species are endemic and may occur in sympatry in the examined area. The molecular approach herein used represents a powerful tool for surveillance programs of D. immitis and D. repens in the culicid vectors towards a better understanding of the epidemiology of the infections they cause and their seasonal transmission patterns.
Similar content being viewed by others
Background
Dirofilaria immitis and Dirofilaria repens (Spirurida, Onchocercidae) are transmitted by bloodsucking culicid mosquitoes belonging to Culex, Aedes, Ochlerotatus Anopheles and Mansonia genera [1–5]. Dirofilaria immitis causes severe cardiopulmonary disease in dogs and it is of major veterinary importance compared to D. repens, which causes a low pathogenic subcutaneous infestation. Nonetheless, both filarial nematodes are of zoonotic concern worldwide being agents of human dirofilariosis [6–13].
In Europe, canine filariosis caused by D. immitis has been diagnosed for a long time mostly in southern regions [14], with the highest endemic area (i.e., prevalence rates as high as 80 %) along the Po River Valley of northern Italy [15, 16]. Although D. repens remains the species most common in central and southern regions of Italy [17, 18], recent reports have suggested that a change in the distribution of this parasite is occurring throughout the Italian territory [19]. Indeed, over the last decades, a high number of cases of canine dirofilariosis caused by D. immitis and D. repens occurred in areas previously regarded as non-endemic, as a consequence of the occurrence of simultaneous infections of the two species in both animal and vector populations [19–21]. In spite of the large amount of information available on the distribution of canine dirofilariosis in Europe [20], field data on vector species of D. immitis and D. repens and on the vector infection rate are exiguous [22, 23].
The detection of filarioids in mosquitoes for assessing distribution of vectors and/or of pathogens in a given area (also known as “xenomonitoring”), when based on individual dissection of wild-caught female mosquitoes is time consuming and hardly applicable in large epidemiological surveys [24]. Furthermore, the morphological identification of the retrieved larval stages of Dirofilaria spp. is challenging and requires specialised parasitological skills, impairing a reliable, prompt diagnosis [10, 25]. Over the past decades, several molecular PCR-based assays have been shown to provide rapid, sensitive, and species-specific methods for the detection and delineation of D. immitis and D. repens DNA in invertebrate hosts [22, 23, 26–30]. Some molecular tools have been applied especially for the entomological monitoring of human filariasis in endemic areas [24, 31–33].
Nevertheless, none of these methods were used on a large scale due to inherent limitations (i.e., double species specific PCRs, low sensitivity). Recently, a sensitive SsoFast™ EvaGreen® based duplex real-time polymerase chain reaction (dqPCR) assay coupled with melting-curve analysis targeting on partial cytochrome c oxidase 1 (cox 1) mitochondrial DNA and on second internal transcribed spacer (ITS-2) of nuclear ribosomal DNA [34] showed its powerfulness in sensitivity and specificity for the detection of small amounts of D. immitis and D. repens genomic DNA from dog blood and mosquito vectors.
The aims of this study were: to evaluate (i) the positivity of field collected culicids for D. immitis and D. repens in an area of north-eastern Italy (Veneto region) where both species are endemic; (ii) the usefulness of the recently developed dqPCR for screening large numbers of culicids; and (iii) the association among mosquito species captured and their positivity for D. immitis and D. repens.
Methods
Sampling area and mosquitoes collection
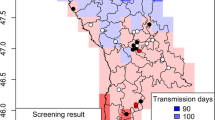
From May to October 2010, 43 carbon dioxide-baited traps were placed for the entomological monitoring of West Nile virus, introduced in this area since 2008 [35], in six provinces of Veneto region (north-eastern Italy). Samples herein examined came from 20 sites (Figure 1) where one or more dogs were present in the surroundings of the trap, hence restricting the survey to areas where culicid species were effectively attracted by proper hosts of Dirofilaria spp. The selected sites were located in rural areas, in lowland (altitude ranging from 2 to 178 m above sea level) devoted mainly to agriculture. Traps were activated every 15 days for one night from sunset to the following morning (i.e., 10.00 am). Mosquito collection was performed until two consecutive captures were negative. Females captured were visually discriminated as fed and/or unfed during identification under the stereomicroscope by size and red-brown color of the abdomen. All specimens were maintained refrigerated until being counted and identified using standard morphological keys [36].
Nine hundred and fifty-five pooled (from minimum of 1 to maximum of 57) female specimens were prepared according to species, date and site of collection. Additionally, Cx. pipiens naturally infested with D. repens (n = 2) and D. immitis (n = 10) were used as positive vector controls. Genomic DNA from Aedes albopictus and from microfilariae of both Dirofilaria species was mixed to obtain an artificially co-infested vector. Specimens of Ae. albopictus (n = 10) raised from eggs collected in the Apulia region (southern Italy), were used as vector negative control samples [34]. Genomic DNA from all mosquito samples was extracted as described in Sangioni and colleagues [37].
Duplex real-time PCR
The duplex real-time PCR for the detection of both D. immitis and D. repens was previously assessed using two species-specific primer sets targeting cox 1 and ITS-2 [34]. Reactions were carried out in a final volume of 20 μl, consisting of 7 μl of SsoFastTM EvaGreen® Supermix (Bio-Rad Laboratories, Hercules CA, USA), 5 μl of Di-Ethyl Pyro-Carbonate (DEPC) treated pyrogen-free DNase/RNase–free water (Invitrogen, Carlsbad, CA, USA), 4 μl of template DNA (except no-template controls), 5 pmol and 100 pmol of each DI COI-F1/DI COI-R1 and Dr ITS2-F/Dr ITS2-R primer pairs, respectively. The run consisted of a hot-start at 95°C for 10 min, and 40 cycles of denaturation (95°C for 15 sec) and annealing-extension (60°C for 1 min). The melting curve was obtained by heating the product at 95°C for 20 sec, cooling it to 55°C for 20 sec and then slowly heating it at 95°C in increments of 0.5°C. The real-time PCR was performed in a CFX96TM Real-Time System (Bio-Rad Laboratories, Inc., Hercules CA, USA). The increase in the fluorescent signal was registered during the extension step of the reaction and the data were analysed by the CFX ManagerTM Software Version 2.1 (Bio-Rad).
The specificity of the duplex real-time PCR assay was established by melting-curve analysis, and PCR products were detected by electrophoresis in 2 % ethidium bromide-stained agarose gels (Gellyphor, EuroClone, Milan, Italy), band sizes being compared with those of an appropriate molecular marker (Gene RulerTM 100-bp DNA Ladder, MBI Fermentas, Vilnius, Lithuania). Furthermore, all the real-time PCR products were purified using Ultrafree-DA columns (Millipore; Bedford, USA) and then sequenced directly using Taq Dye Deoxy Terminator Cycle Sequencing Kit (v.2, Applied Biosystems Inc.) in an automated sequencer (ABI-PRISM 377; Applied Biosystems Inc.), from both strands, using the same sets of primers used in the real-time PCR. All sequences generated were compared to sequences available in GenBank using Basic Local Alignment Search Tool (BLAST) [38].
Statistical analysis
The rate of infection in mosquitoes was adjusted for pooled samples, calculating the Estimated Rate of Infection (ERI) using the following formula: ERI = 1 − (1- x/m)1/k[39], where x is the number of positive pools; m the number of examined pools and k the average number of specimens in each pool. Differences in mosquito rates of infection among species and locations were tested using Yates’ chi-squared test.
Results
Among the mosquito species collected, Cx. pipiens was the most represented (n = 37,865, 92.6 %) followed by Och. caspius (n = 2,264, 5.5 %), Ae. vexans (n = 720, 1.8 %) and Cx. modestus (n = 43, 0.1 %). At least one pool positive for Dirofilaria spp. was detected in each of the examined provinces with the highest mosquito rate of infection (ERI = 0.42 %) found in Vicenza province, where two sites were significantly more infested than all other provinces, but not Padova (Table 1). Out of the 955 mosquito pools representative for 40,892 unfed female individuals examined through the 20 sites monitored, 23 (2.41 %) scored positive for Dirofilaria spp. over 13 (65 %) sites (Tables 1 and 2, Figure 1). In particular, 21 pools (2.2 %) were positive for D. immitis and two (0.21 %) for D. repens by melting-curve analysis (Figure 2, Table 3). The results for positive mosquito-pool samples showed two melting peaks (i.e., mean Tm = 75 and 69°C) corresponding to species-specific Tm range of D. immitis (mean ± SD = 75.7 ± 0.3°C) and D. repens (mean ± SD = 70 ± 0.7°C) positive controls, respectively (Figure 2). No melting peaks generated simultaneously were displayed, except for the Ae. albopictus artificially co-infested vector, as well as no fluorescence signal was detected for any of the negative controls (Figure 2). Dirofilaria immitis was found in all mosquito species except for Cx. modestus, which was poorly represented, whereas D. repens was detected only in Cx. pipiens (Table 3). The overall ERI was 0.06 %, being about one mosquito infested every 1,800 (Table 2). In particular, Och. caspius and Ae. vexans showed the highest rate of infestation (0.18 % and 0.14 %, respectively). Overall, Cx. pipiens was about four times less infested (0.05 %) than other species and significantly less infested than Och. caspius (p <0.05; Table 2). However, considering single sites, Cx. pipiens reached the highest rate of infestation (i.e., 1.1 %) whereas the infestation rates for D. immitis and D. repens were comparable in positive sites (Table 3).
Representative melting curve analyses.
Melt peak of Dirofilaria immitis (green), Dirofilaria repens (blue), and constructed positive sample with a simultaneous amplification of cox 1 and ITS2 (black), pools of mosquito positive to Dirofilaria immitis (yellow) and Dirofilaria repens (red), respectively. Negative (Aedes albopictus) and No template controls (NTC) (grey).
All real-time PCR products were confirmed by detection on 2 % ethidium bromide-stained agarose gels showing a single band of 200 and 300 bp for D. immitis and D. repens, respectively (Figure 3). The sequences derived from the amplicons matched (99-100 % identity) appropriate reference sequences of Dirofilaria species (accession numbers JF461464 and AY693808). Positive pools were found each month throughout the sampling period from the first week of May (start of monitoring) till October, without any apparent seasonality (Table 3).
Discussion
This study reports data on the first survey employing a duplex real-time PCR to assess the occurrence of D. immitis and D. repens in culicids collected in a large-scale entomological monitoring programme and provides information about the relative infestation rate for both filarial nematodes in mosquito species of north-eastern Italy. Collecting mosquitoes under the frame of a regional surveillance programme for the West Nile virus vectors allowed optimizing samples and collection efforts, thus saving economic resources.
Results indicate that Dirofilaria spp. were evenly detected in mosquito pools throughout the Veneto region with the highest rate of infestation found in Vicenza and Padova (i.e., ERI = 0.42% and 0.16 %, respectively). In particular, while D. immitis was detected in mosquito pools collected in all provinces, D. repens was found only in Padova and Rovigo. The results above confirm the occurrence of D. repens in north-eastern Italy, where the species was reported more than 20 years ago [40], it also expands current knowledge on the distribution of this parasite, which was previously detected in western-regions of the country (Piedmont and Lombardy) [15, 41, 42]. Interestingly, the high D. immitis infestation rate in Och. caspius (i.e., overall ERI = 0.18 % and local ERI = 0.7 %) indicates the role of this species as a putative vector, as previously suggested, based on the retrieval of infective larvae at the dissection [43]. Conversely, a molecular study based on classic PCR failed to detect D. immitis in Och. caspius, indicating that this mosquito species was refractory to filarial infestation [44]. Aedes vexans was found positive for D. immitis only once in a small number of mosquitoes, corresponding to a considerable infestation rate (i.e., ERI = 0.14 %). A similar result was already reported in a study from Turkey in which this mosquito species was suggested as the main vector of D. immitis[45]. However, this finding needs to be confirmed in a larger collection of Ae. vexans, poorly represented in our study likely because of its diurnal activity. Previous studies carried out in central and northern Italy failed to detect D. immitis in Ae. vexans, both by insect dissection and classic PCR testing [44, 46]. Hence, our findings might be a result of either the higher sensitivity of our PCR protocol in detecting genomic D. immitis DNA (detection limit of the assay as low as 2.5 pg/μl of DNA) [34] or of the effective presence of this nematode in our sampled Ae. vexans population. Culex modestus scored negative for D. immitis in contrast to previous reports indicating this species as a competent vector [47, 48]. This result may be due to the small number of mosquitoes specimens collected, due to its host preference. Indeed, Cx. modestus is considered unwilling to feed on dogs [41, 49–52], differently from Och. caspius and Cx. pipiens. The latter species was highly positive for D. immits and D. repens as already demonstrated by insect dissection and/or PCR processing [22, 53].
Even if the present assay does not allow differentiation between infested and infective mosquitoes (abdomen and thorax-head have not been analyzed separately), data herein presented clearly indicate the usefulness of the molecular diagnosis assay for a broader field application in order to gain more information on the epidemiology of the mosquitoes acting as vectors of Dirofilaria spp.
Moreover, an effective surveillance system is pivotal considering the expansion of certain vectors of Dirofilaria spp. throughout areas previously regarded as non-endemic. This is the case of Ae. albopictus, a mosquito species highly adapted to many ecological niches in southern Europe [54] which, in turn, is also a more competent vector of D. immitis than native Cx. pipiens populations [23, 28]. Importantly, continuous epidemiological surveillance systems need to be implemented by local authorities in order to test autochthonous vector populations as well as recently introduced potential vectors (e.g., Aedes koreicus) [55]. A continuous monitoring of infestation rates for mosquito vector populations is pivotal to evaluate the success of an anti-filarial campaign in endemic areas, thus providing evidence on whether drug administration in a given area can be terminated or not, in order to reduce risk for drug resistance in filarioid populations. The advantages of using a highly sensitive molecular diagnostic tool for xenomonitoring was shown to be pivotal for assessing the efficacy and progress of eradication programmes of lymphatic human filariasis based on mass drug administration [56, 57].
Conclusions
The results presented here suggest a high sensitivity and specificity of the assay herein used in field-collected samples, clearly representing an alternative to classic microscopic methods and to PCR-based assays for the xenomonitoring of D. repens and D. immitis, mainly in areas where these species are endemic and/or occur in sympatry [4, 58]. Moreover, it represents a non-invasive method to assess the presence of both Dirofilaria species in an area. Field information on vector distribution and their rates of infestation will contribute to implementing warning among veterinarians, physicians and health authorities, also considering that human dirofilariasis is reported to be increasing in the Old World [20].
References
Coluzzi M: Osservazioni sperimentali sul comportamento di Dirofilaria repens in diversi gruppi di artropodi vettori. Parassitologia. 1964, 4: 57-62.
Sauerman DM: A survey for natural potential vectors of Dirofilaria immitis in Vero beach. Mosq News. 1985, 43: 222-225.
Cancrini G, Pietrobelli M, Frangipane Di Regalbono A, Della Torre A: Development of Dirofilaria and Setaria nematodes in Aedes albopictus. Parassitologia. 1995, 37: 141-145.
Cancrini G, Frangipane di Regalbono A, Ricci I, Tessarin C, Gabriella S, Pietrobelli M: Aedes albopictus is a natural vector of Dirofilaria immitis in Italy. Vet Parasitol. 2003, 118: 195-202. 10.1016/j.vetpar.2003.10.011.
Cancrini G, Gabrielli S: Vectors of Dirofilaria nematods: biology behaviour and host/parasites relationships. Dirofilaria immitis and D. repens in dog and cat and human infections. Edited by Genchi C, Rinaldi L, Cringoli G. Napoli: Rolando; 2007:49–58.
Orihel TC, Ash LR: Parasites in human tissues. 1995, American Society of Clinical Pathology Press, Chicago, 386-
Pampiglione S, Canestri Trotti G, Rivasi F: Human dirofilariasis due to Dirofilaria (Nochtiella) repens in Italy: a review of word literature. Parassitologia. 1995, 37: 149-193.
Pampiglione S, Rivasi F, Gustinelli A: Dirofilarial human cases in the Old World, attributed to Dirofilaria immitis: a critical analysis. Histopathology. 2009, 54: 192-204. 10.1111/j.1365-2559.2008.03197_a.x.
Orihel TC, Eberhard ML: Zoonotic filariasis. Clin Microbiol Rev. 1998, 11: 366-381.
McCall JW, Genchi C, Kramer LH, Guerrero J, Venco L: Heartworm disease in animals and humans. Adv Parasitol. 2008, 66: 193-285.
Pampiglione S, Rivasi F: Human dirofilariasis due to Dirofilaria (Nochtiella) repens: an update of world literature from 1995 to 2000. Parassitologia. 2000, 42: 231-254.
Simón F, Morchón R, González-Miguel J, Marcos-Atxutegi C, Siles-Lucas M: What is new about animal and human dirofilariosis?. Trends Parasitol. 2009, 25: 404-409. 10.1016/j.pt.2009.06.003.
Otranto D, Eberhard ML: Zoonotic helminths affecting the human eye. Parasit Vectors. 2011, 4: 41-10.1186/1756-3305-4-41.
Genchi C, Rinaldi L, Cascone C, Mortarino M, Cringoli G: Is heartworm disease really spreading in Europe?. Vet Parasitol. 2005, 133: 137-148. 10.1016/j.vetpar.2005.04.009.
Rossi L, Pollono F, Meneguz PG, Gribaudo L, Balbo T: An epidemiological study of canine filarioses in north-west Italy: what has changed in 25 years?. Vet Res Commun. 1996, 20: 308-315. 10.1007/BF00366536.
Genchi C, Kramer LH, Prieto G: Epidemiology of canine and feline Dirofilariosis: a global view. Heartworm infection in humans and animals. Edited by Simon F, Genchi C. Salamanca; 2001:121–133.
Marconcini A, Magi M: Incidence of canine dirofilariasis in “Maremma” (Grosseto-Italy). Ann Fac Med Vet. 1991, 44: 153-156.
Giannetto S, Pampiglione S, Santoro V, Virga A: Research of canine filariasis in Trapani province (Western Sicily). Morphology on SEM of male Dirofilaria repens. Parassitologia. 1997, 39: 403-405.
Otranto D, Capelli G, Genchi C: Changing distribution patterns of canine vector borne diseases in Italy: leishmaniosis vs. dirofilariosis. Parasit Vectors. 2009, 2 (Suppl 1): S2-10.1186/1756-3305-2-S1-S2.
Genchi C, Kramer LH, Rivasi F: Dirofilarial Infections in Europe. Vector Borne Zoonotic Dis. 2011, 11: 1307-1317. 10.1089/vbz.2010.0247.
Colwell DD, Dantas-Torres F, Otranto D: Vector-borne parasitic zoonoses: emerging scenarios and new perspectives. Vet Parasitol. 2011, 182: 14-21. 10.1016/j.vetpar.2011.07.012.
Cancrini G, Kramer LH: Insect vectors of Dirofilaria spp.:. Heartworm infection in humans and animals. Edited by Simon F and Genchi C. Universidad Salamanca; 2001:63–82.
Cancrini G, Romi R, Gabriella S, Toma L, Di Paolo M, Scaramozzino P: First finding of Dirofilaria repens in a natural population of Aedes albopictus. Med Vet Entomol. 2003, 17: 448-451. 10.1111/j.1365-2915.2003.00463.x.
Bockarie MJ: Molecular xenomonitoring of lymphatic filariasis. Am J Trop Med Hyg. 2007, 77: 591-592.
Traversa D, Di Cesare A, Conboy G: Canine and feline cardiopulmonary parasitic nematodes in Europe: emerging and underestimated. Parasit Vectors. 2010, 3: 62-10.1186/1756-3305-3-62.
Favia G, Lanfrancotti A, Della Torre A, Cancrini G, Coluzzi M: Polymerase chain reaction-identification of Dirofilaria repens and Dirofilaria immitis. Parasitology. 1996, 113: 567-571. 10.1017/S0031182000067615.
Mar PH, Yang IC, Chang GN, Fei AC: Specific polymerase chain reaction for differential diagnosis of Dirofilaria immitis and Dipetalonema reconditum using primers derived from internal transcribed spacer region 2 (ITS2). Vet Parasitol. 2002, 106: 243-252. 10.1016/S0304-4017(02)00032-8.
Cancrini G, Scaramozzino P, Gabriella S, Di Paolo M, Toma L, Romi R: Aedes albopictus and Culex pipiens implicated as natural vectors of Dirofilaria repens in central Italy. J Med Entomol. 2007, 44: 1064-1066. 10.1603/0022-2585(2007)44[1064:AAACPI]2.0.CO;2.
Lee SE, Kim HC, Chong ST, Klein TA, Lee WJ: Molecular survey of Dirofilaria immitis and Dirofilaria repens by direct PCR for wild caught mosquitoes in the Republic of Korea. Vet Parasitol. 2007, 148: 149-155. 10.1016/j.vetpar.2007.04.015.
Thanchomnang T, Intapan PM, Lulitanond V, Sangmaneedet S, Chungpivat S, Taweethavonsawat P, Choochote W, Maleewong W: Rapid detection of Dirofilaria immitis in mosquito vectors and dogs using a real-time fluorescence resonance energy transfer PCR and melting curve analysis. Vet Parasitol. 2010, 168: 255-260. 10.1016/j.vetpar.2009.12.002.
Mishra K, Raj DK, Hazra RK, Dash AP, Supakar PC: The development and evaluation of a single step multiplex PCR method for simultaneous detection of Brugia malayi and Wuchereria bancrofti. Mol Cell Probes. 2007, 21: 355-362. 10.1016/j.mcp.2007.05.001.
Chambers EW, McClintock SK, Avery MF, King JD, Bradley MH, Schmaedick MA, Lammie PJ, Burkot TR: Xenomonitoring of Wuchereria bancrofti and Dirofilaria immitis infections in mosquitoes from American Samoa: trapping considerations and a comparison of polymerase chain reaction assays with dissection. Am J Trop Med Hyg. 2009, 80: 774-781.
Intapan PM, Thanchomnang T, Lulitanond V, Maleewong W: Rapid detection of Wuchereria bancrofti and Brugia malayi in mosquito vectors (Diptera: Culicidae) using a real-time fluorescence resonance energy transfer multiplex PCR and melting curve analysis. J Med Entomol. 2009, 46: 158-164. 10.1603/033.046.0119.
Latrofa MS, Dantas-Torres F, Annoscia G, Genchi M, Traversa D, Otranto D: A duplex real-time polymerase chain reaction assay for the detection of and differentiation between Dirofilaria immitis and Dirofilaria repens in dogs and mosquitoes. Vet Parasitol. 2012, 185: 181-185. 10.1016/j.vetpar.2011.10.038.
Busani L, Capelli G, Cecchinato M, Lorenzetto M, Savini G, Terregino C, Vio P, Bonfanti L, Pozza MD, Marangon S: West Nile virus circulation in Veneto region in 2008–2009. Epidemiol Infect. 2010, 30: 1-8.
Severini F, Toma L, Di Luca M, Romi R: Le zanzare italiane: generalità e identificazione degli adulti (Diptera, Culicidae). Fragmenta Entomologica. 2009, 41: 213-372.
Sangioni LA, Horta MC, Vianna MC, Gennari SM, Soares RM, Galvão MA, Schumaker TT, Ferreira F, Vidotto O, Labruna MB: Rickettsial infection in animals and Brazilian spotted fever endemicity. Emerg Infect Dis. 2005, 11: 265-270. 10.3201/eid1102.040656.
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25: 3389-3402. 10.1093/nar/25.17.3389.
Cowling DW, Gardner IA, Johnson WO: Comparison of methods for estimation of individual-level prevalence based on pooled samples. Prev Vet Med. 1999, 39: 211-225. 10.1016/S0167-5877(98)00131-7.
Martini M, Poglayen G, Capelli G, Roda R: Diagnosis of canine filariosis: relative sensitivity and specificity of some haematological techniques. Angew Parasitol. 1991, 32: 133-136.
Pollono F, Rossi L, Cancrini G: Research on Culicidae attracted to dog bait in Piedmont. Parassitologia. 1998, 40: 439-445.
Petruschke G, Rossi L, Pollono F, Genchi C: Sulle filariasi canine in provincia di Varese e nel basso Canton Ticino. Parassitologia. 1998, 40: 133-
Rossi L, Pollono F, Meneguez PG, Cancrini G: Quattro specie di culicidi come possibili vettori di Dirofilaria immitis nella risaia piemontese. Parassitologia. 1999, 41: 537-542.
Cancrini G, Magi M, Gabrielli S, Arispici M, Tolari F, Dell’Omodarme M, Prati MC: Natural vectors of dirofilariasis in rural and urban areas of the Tuscan region, central Italy. J Med Entomol. 2006, 43: 574-579. 10.1603/0022-2585(2006)43[574:NVODIR]2.0.CO;2.
Yildirim A, Inci A, Duzlu O, Biskin Z, Ica A, Sahin I: Aedes vexans and Culex pipiens as the potential vectors of Dirofilaria immitis in Central Turkey. Vet Parasitol. 2011, 178: 143-147. 10.1016/j.vetpar.2010.12.023.
Petruschke G, Rossi L, Genchi C, Pollono F: Canine dirofilariasis in the canton of Ticino and in the neighboring areas of northern Italy. Schweiz Arch Tierheilkd. 2001, 143: 141-147.
Rossi L, Favia G, Ricci I, Cancrini G: Looking for the vectors of canine heartworm infection in the north of Italy. Parassitologia. 2002, 44: 160-
Cancrini G, Gabrielli S, Frangipane di Regalbono A, Magi M, Scaramozzino P, Toma L: Advances in the identification of natural vectors of zoonotic filariae by PCR-based approaches [abstracts]. IX EMOP 2004:400
Iori A, Cancrini G, Vezzoni A, Del Ninno G, Tassi P, Genchi C, Della Torre A, Coluzzi M: Osservazioni sul ruolo di Culex pipiens nella trasmissione della filariasi del cane in Italia. Parassitologia. 1990, 32: 151-152.
Di Sacco B, Cancrini G, Genchi C: Studio del tropismo nei riguardi del cane e del gatto da parte dei ditteri potenziali vettori delle filariasi in provincia di Pavia. Parassitologia. 1992, 34: 11-12.
Di Sacco B, Del Zoppo B, Genchi C: Survey of dog Dirofilaria immitis mosquito vectors in Milan and vicinity. Parassitologia. 1994, 36: 50-
Genchi C, Di Sacco B, Cancrini G: Epizoology of canine and feline heartworm infection in Northern Italy: possible mosquito vectors. Wilmington: American Heartworm Society. 1992, 1992: 39-46.
Cancrini G, Della Torre A, Coluzzi M: Different probabilities of transmission of Dirofilaria immitis and D. repens by Culex pipiens. Proceedings of 6th European Multicolloquium of Parasitology. 1992, Hague, Netherlands, 90-
Straetemans M: Vector-related risk mapping of the introduction and establishment of Aedes albopictus in Europe. ECDC consultation group on vector-related risk for chikungunya virus transmission in Europe. Euro Surveill. 2008, 13: 8040-
Capelli G, Drago A, Martini S, Montarsi F, Soppelsa M, Delai N, Ravagnan S, Mazzon L, Schaffner F, Mathis A, Di Luca M, Romi R, Russo F: First report in Italy of the exotic mosquito species Aedes (Finlaya) koreicus, a potential vector of arboviruses and filariae. Parasit Vectors. 2011, 4: 188-10.1186/1756-3305-4-188.
Farid HA, Morsy ZS, Helmy H, Ramzy RM, El Setouhy M, Weil GJ: A critical appraisal of molecular xenomonitoring as a tool for assessing progress toward elimination of Lymphatic Filariasis. Am J Trop Med Hyg. 2007, 77: 593-600.
Boakye DA, Baidoo HA, Glah E, Brown C, Appawu M, Wilson MD: Monitoring lymphatic filariasis interventions: Adult mosquito sampling, and improved PCR – based pool screening method for Wuchereria bancrofti infection in Anopheles mosquitoes. Filaria J. 2007, 6: 13-10.1186/1475-2883-6-13.
Otranto D, Dantas-Torres F: Canine and feline vector-borne diseases in Italy: current situation and perspectives. Parasit Vectors. 2010, 3: 2-10.1186/1756-3305-3-2.
Acknowledgments
Authors wish to thank Matteo Mazzucato (CREV-GIS office, IZSVe) for map drawing and Bayer Animal Health GmbH for supporting the publication expenses.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
FM, SC, and SR collected and identified the mosquito specimens. MSL and GA run the real-time PCRs. GC, LMS and DO contributed in writing the first draft of the manuscript. LMS, DO, FDT and GC equally contributed with data analysis and interpretation and revision of the manuscript. All authors read and approved the final version of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Latrofa, M.S., Montarsi, F., Ciocchetta, S. et al. Molecular xenomonitoring of Dirofilaria immitis and Dirofilaria repens in mosquitoes from north-eastern Italy by real-time PCR coupled with melting curve analysis. Parasites Vectors 5, 76 (2012). https://doi.org/10.1186/1756-3305-5-76
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-3305-5-76