Abstract
Background
The most important means of identifying diseases before symptoms appear is through the discovery of disease-associated biomarkers. Recently, microRNAs (miRNAs) have become highly useful biomarkers of infectious, genetic and metabolic diseases in human but they have not been well studied in domestic animals. It is probable that many of the animal homologs of human disease-associated miRNAs may be involved in domestic animal diseases. Here we describe a computational biology study in which human disease miRNAs were utilized to predict orthologous miRNAs in cow, chicken, pig, horse, and dog.
Results
We identified 287 human disease-associated miRNAs which had at least one 100% identical animal homolog. The 287 miRNAs were associated with 359 human diseases referenced in 2,863 Pubmed articles. Multiple sequence analysis indicated that over 60% of known horse mature miRNAs found perfect matches in human disease-associated miRNAs, followed by dog (50%). As expected, chicken had the least number of perfect matches (5%). Phylogenetic analysis of miRNA precursors indicated that 85% of human disease pre-miRNAs were highly conserved in animals, showing less than 5% nucleotide substitution rates over evolutionary time. As an example we demonstrated conservation of human hsa-miR-143-3p which is associated with type 2 diabetes and targets AKT1 gene which is highly conserved in pig, horse and dog. Functional analysis of AKT1 gene using Gene Ontology (GO) showed that it is involved in glucose homeostasis, positive regulation of glucose import, positive regulation of glycogen biosynthetic process, glucose transport and response to food.
Conclusions
This data provides the animal and veterinary research community with a resource to assist in generating hypothesis-driven research for discovering animal disease-related miRNA from their datasets and expedite development of prophylactic and disease-treatment strategies and also influence research efforts to identify novel disease models in large animals. Integrated data is available for download at http://agbase.hpc.msstate.edu/cgi-bin/animal_mirna.cgi.
Similar content being viewed by others
Background
MicroRNAs (miRNAs) are naturally occurring single-stranded small RNA molecules that play important roles in post-transcriptional regulation of gene expression [1]. Studies have shown that miRNAs exert their regulatory role by partially binding their target (complementary) mRNAs at 3′ UTRs (untranslated regions) [2–5]. Phylogenetic studies indicate that animal miRNAs are highly conserved [6, 7]. Until recently, miRNAs were thought to be of little or no cellular significance [8–10]. The first miRNA shown to have a regulatory function was lin-4 from Caenorhabditis elegans[11, 12]. Lin-4 acted as a silencer of genes that regulate developmental timing, but it was considered a unique evolutionary adaptation as lin-4 homologs were not found in other species. The discovery of the regulatory miRNA let-7 in C. elegans in 2000 [10], with homologs in other species including humans, caused researchers to reconsider the idea that miRNAs may have a more widespread function within cells. We now know that many miRNAs play central roles in post-transcriptional gene regulation. Additionally, expression of specific miRNAs has been linked to various diseases [13–16]. Considerable research has been devoted to understanding regulatory roles of miRNAs in human diseases [17–26], and miRNAs are important biomarkers of several disease processes [27–32] including many cancers [33–43] and cardiovascular [18, 44–53], inflammation [54–57], and gastrointestinal diseases [58–60].
While databases for human disease-associated miRNAs are publicly available [61–70], there is no any integrated resource for disease-associated miRNAs in domestic animals. An integrated resource of animal disease-related miRNA data would provide the animal and veterinary research community with an invaluable resource for searching disease related miRNA subsets from their experimental data. Pubmed articles stand solely as the major reliable source of information for disease miRNA data. However, there are very few Pubmed articles currently (as of 12/31/2014) documenting disease-associated miRNAs in domestic animals compared to human, mouse or rat (Figure 1). Identification of miRNA/disease associations in domestic animals is critical for understanding miRNA involvement in the pathophysiology of these organisms.
Publication statistics of disease associated miRNAs as of 12/31/2013. Searches from NCBI-Pubmed titles or abstracts were conditioned to retrieve publications from the last ten years. The searches contained species names, miRNA abbreviations, disease(s) and duration (year). For example searching Pubmeds for dog disease associated miRNAs in the past 10 years were acquired using the following query statement: (dog OR canine OR (Canis familiaris) AND (microRNA OR microRNAs OR miRNA OR mirRNAs OR mir OR miRs) AND (cancer OR cancers OR disease OR diseases OR disorder OR disorders) AND ((“2004/01/01”[PDat] : “2013/12/31”[PDat])). Note that (A) and (B) are presented in different y-axis scale due to large difference in number of Pubmed articles.
The main objective of our study was to identify animal miRNA homologs of published human disease-associated miRNAs in cow, chicken, pig, horse and dog using phylogenetic techniques. Using the current available human and animal miRNA resources, we identified potential disease-related miRNAs in domestic animals based on integrated computational and manual approaches including assessing the sequence similarities and evolutionary relationships between human disease miRNAs and their animal orthologs. These predictions will serve as a resource to facilitate hypothesis-driven research in domestic animals, which upon verification in animals could suggest animal models for human diseases and strategies for developing therapeutic measures.
Methods
Human and domestic animal mature miRNA sizes
We compared the sizes of all known mature human miRNAs with the sizes of all known cow, chicken, pig, horse, and dog to establish their length diversity.
Extraction and verification of disease-associated human miRNAs
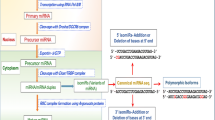
Briefly, we surveyed publicly available databases that link human miRNAs with diseases [65, 67, 71, 72] and selected the most up-to-date and comprehensive human disease miRNA database, i.e., the Human miRNA Disease Database (HMDD) version 2.0 [58], as a baseline for searching animal homologs. We then filled a request form available at http://202.38.126.151/hmdd/html/tools/hmdd_req.html to request all human disease-associated miRNA data from Dr. Qinghua Cui of department of Biomedical Informatics, Peking University Health Science Center. From this data we retrieved the miRNA IDs (identifiers) that were named according to miRBase [73] nomenclature standards, the disease(s) associated with each miRNA and the Pubmed articles from which the HMDD data was extracted. We then manually reviewed the Pubmed titles and abstracts to verify association of the miRNAs with human diseases.
Prediction of animal homologs of human disease-associated miRNAs
We used the IDs of human disease miRNAs (from HMDD) to extract corresponding mature (i.e., processed) miRNA nucleotide sequences from miRBase version 20 [73]. We also downloaded all sequences of mature and precursor (pre) miRNAs for cow, chicken, pig, horse and dog from miRBase version 20 and then used a Perl script to identify cow, chicken, pig, horse, and dog mature miRNA sequences that were 100% identical to sequences of human disease-associated miRNAs. The outputs of the Perl script were deemed “human disease miRNAs with animal counterparts” (HDMACs).
Phylogenetic analysis of HDMAC precursor sequences
The precursor miRNA sequences (pre-miRNAs) of HDMACs were compared using a multiple alignment and phylogenetic approach to detect conservation profiles and rapid sequence divergence in human and domestic animals. Briefly, we used Clustal Omega [74] tool for multiple alignment of pre-miRNA sequences and Clustalw2-Phylogeny tool [75] to generate the Neighbor-Joining (NJ) phylogenetic trees to determine nucleotide substitutions that have occurred over evolutionary time. Briefly, NJ method compares each sequence with each other, calculates distance matrices, then combines the least distant pair of sequences and construct phylogenetic tree. We displayed distances (divergence proportions) calculated from all pairs of sequences in the multiple alignments to facilitate evolutionary interpretation of phylograms. Divergence proportions less or equal to 5% (≤0.05) were considered to be highly conserved.
Annotation of human disease miRNA targets and their animal orthologs
We manually annotated the genes targeted by the human disease-associate miRNAs from the associated Pubmed articles and predict their animal orthologs using the Ensembl Biomart [76] tool. All human targets and their animal orthologs with one-to-one matches and reciprocal% identity >70 were integrated in the animal disease miRNA resource.
Integration of disease information with human-animal miRNA homologs
We integrated the information gathered from this study to form a computationally predicted animal disease miRNA resource which contained miRNA information including standardized miRBase identifiers of mature miRNAs, pre-miRNAs, and miRNA families linked to the associated human disease. Whenever applicable we used OMIM (Online Mendelian Inheritance in Man) [77], OMIA (Online Mendelian Inheritance in Animal) [78] and Disease Ontology (DO) [79] standardized names for disease phenotypes and BRENDA (BRaunschweig ENzyme DAtabase) Tissue Ontology (BTO) [80, 81] terms to standardize names of source tissues or cell types. Pubmed IDs were used as central literature references. Additional information regarding the genomic location(s) of miRNAs, type of experiment, and publication date was also integrated.
Results
Lengths of animal and human mature miRNAs
Generally, the distribution of mature miRNA lengths in all species followed same trend, with 22-nt sequences dominating in each species (Figure 2).
Dataset of human disease-associated miRNAs
After surveying various human disease-related miRNA databases, we found that the miRNAs in the Human miRNA Disease Database (HMDD) version 2.0 [71] (updated on 09/30/2013) were best suited for use as a training set (Figure 3). This database contained 578 published human miRNAs associated with 383 diseases referenced in 3,486 Pubmed articles. The human disease miRNA referenced in HMDD included 70% of the total 4,961 human disease miRNA articles we identified in Pubmed (Figure 1).
We identified 694 domestic animal mature miRNA sequences that showed 100% sequence identity with the 287 human disease miRNAs (Figure 4, Additional file 1). Over 60% of total horse mature miRNAs (216) showed perfect matches to human disease-associated miRNAs, followed by dog (50%). As expected, chicken had the least number of perfect matches (5%), most likely a result of the relative evolutionary distance between birds and mammals. We found that 14 human disease miRNAs were conserved in all animals, chicken included, while 41 were conserved only between the mammalian representatives (Table 1).
Mature human disease miRNA with animal counterparts (HDMACs) in key domestic animals. For a particular human-animal pair, blue bar indicates HDMACs in other animals but not in the selected animal, red bars indicates HDMACs in the select animal and blue & red bars together represent 287 HDMACs. Green sub-bars show non-HDMACs.
Conserved human disease pre-miRNAs in animals
In addition to producing multiple alignments of mature miRNAs, we established additional evolutionary relationship between pre-miRNAs of HDMACs, which included 265 and 649 human and animal pre-miRNA sequences, respectively. About 85% of human disease pre-miRNAs were found to be highly conserved in animals, showing less than 5% nucleotide substitution rates over evolutionary time (Figure 5; Additional file 2). We demonstrate phylogenetic analysis of pre-miRNA sequences of HDMACs that are clustered in the mir-154 family (Figure 6). In this family all pre-miRNAs had nucleotide substitution rate of <4%, among which, pre-miRNA sequences of eight animals; 4 in horse (eca-mir 409, 494, 377 & 382), 2 in dog (cfa-mir-494 & 377) and 2 in cow (bta-mir-377 & 382) had 100% percent identity with 4 of human disease pre-miRNAs sequences.
Nucleotide substitution rate of pre-miRNAs of HDMACs in key domestic animals. For a particular human-animal pair, blue indicates pre-miRNA sequences with zero nucleotide substitution rate; red bars indicates pre-miRNA sequences with <5% nucleotide substitution rate and green bars are pre-miRNA sequences with >5% nucleotide substitution rate.
Example of phylogenetic analysis of HDMACs pre-miRNAs in the mir-154 family. The branch labels are pre-miRNA IDs prefixed by abbreviation of species scientific name; hsa (Homo sapiens - human), bta (Bos Taurus - cow), gga (gallus gallus - chicken), ssc (Sus scrofa - pig), eca (Equine canibus - horse), and cfa (Canis familiaris - dog); followed by numbers that indicate the proportion of evolutionary distance in terms of nucleotide substitutions per site per unit time, which indicates the changes in sequences when they evolved from a common ancestral sequence. The alternating background colors (light pink, light green) facilitate visualization and comparison of miRNA species within the cluster.
Animal orthologs of human disease miRNA targets
Genes targeted by the human disease-associated miRNAs were utilized to predict their animal orthologs using the Ensembl Biomart [76] tool. Currently, over 45 targets conserved across human and domestic animals are included in the integrated resource available through AgBase [82, 83] at http://agbase.hpc.msstate.edu/cgi-bin/animal_mirna.cgi. Example of conserved disease miRNA targets linked to type 2 diabetes is included in this article (Figure 7, Table 2) and more targets are shown as Additional file 3.
Comparative structural analysis of human hsa-miR-143 and its animal orthologs. The multiple sequence alignment (A) shows the location of the mature miRNA on the 3′ arm of the pre-miRNA. The minimum free energy (MFE) secondary structure (B) of human mir-143 is compared to that of the animal orthologs and a normalized pre-miRNA consensus sequence. The species are abbreviated by their scientific names; hsa (Homo sapiens –human), ssc (Sus scrofa - pig), eca (Equine canibus - horse), and cfa (Canis familiaris - dog).
Data integration
In order to provide a unified view of data generated from this study we integrated all information to form a core resource of domestic animal disease-related miRNAs. The integrated data links all 694 animal mature miRNAs with 287 human miRNAs which are associated with 359 human diseases referenced in 2,863 Pubmed articles. This information is classified into five main categories including information for miRNA, associated disease, reference, genomic location and target (Table 3). The integrated resource is the main reference and preliminary data towards our efforts to develop an advanced farm and domestic animal disease-associated miRNA resource. The preliminary integrated resource is available at http://agbase.hpc.msstate.edu/cgi-bin/animal_mirna.cgi.
Application of the integrated resource
We have demonstrated how experimentally confirmed diabetes type 2-associated miRNA hsa-mir-143-3p can be used to identify related miRNAs in animals (Figure 7, Table 2) thus, providing a more focused hypothesis-driven investigations in animals. We found that the hsa-miR-143 which is located on the right arm (3′) of its pre-miRNA is highly conserved in pig, horse and dog. The hsa-miR-143-3p target gene is AKT1 [84] which has 97- 100% sequence identity with AKT1 found in pig, horse and dog. The biological processes annotated to AKT1 using Gene ontology (GO) [85, 86] indicate that this gene is involved in similar processes in all species including, glucose homeostasis, positive regulation of glucose import, positive regulation of glycogen biosynthetic process, response to food and glucose transport (Table 2).
Discussion
One means of identifying diseases before symptoms appear is through the discovery and utilization of disease-associated molecular biomarkers. Many biomarker techniques that have been widely applied in human and model organism studies have not been adequately implemented in the study of domestic animal diseases. It is now clear that miRNA play major regulatory role in various disease processes but financial investment is more committed to investigate miRNA involvement in human disease more than any other species (Figure 1). There are several miRNAs currently classified as biomarkers for human cancers [27, 42, 87–93], cardiovascular [45, 46, 94, 95], and inflammation [54–57] diseases. Although not experimentally found to be associated with miRNAs, some of these human diseases are also found in domestic animals [96–99]. Advances in knowledge about human disease-associated miRNAs warrant investigation of similar diseases in related species.
In this study we used homology approach to generate a resource that integrates animal miRNA data with human disease-associated miRNAs. As demonstrated using hsa-miR-143-3p which has been associated with diabetes type 2 [84] (Figure 7 and Table 2) it is logical that similar miRNAs perform comparable functions across related species, and therefore diseases correlated with miRNAs in one species may be correlated with homologous miRNA expression and disease in related species. The example of diabetes type 2-associated miRNA hsa-miR-143-3p gave a highlight on how to link disease-associated elements across species and develop hypothesis-driven investigation in animals. Integrating all data enabled us to identify some human disease miRNAs that are found in more than one animal species (Additional file 1), which indicates the likelihood of also sharing common diseases. Having miRNAs targeting similar genes across species provide clue of functional orthology. As indicated in this study one miRNA can be associated with multiple diseases. For example, hsa-miR-21-5p has been documented in nearly 400 Pubmed articles and is associated with 124 human disease phenotypes and has homologs in four animals including chicken.
However, not all human miRNA-related diseases may be relevant to all animals. Manual curation effort to continue building and updating the animal disease miRNA resource developed in this study is our long term process. The relevance of each human miRNA disease linked to each animal and targeted gene will continue to be established and integrated in the resource. Validation of the human disease miRNAs in the animal context will likely leverage the findings in human at the same time improve our understanding of their involvement in the pathogenesis, diagnosis, and prognosis of various animal diseases.
Conclusions
In this study we have shown that some human disease-associated miRNAs are well conserved across domestic animals. Also, human genes targeted by disease-associated miRNAs are highly conserved in animals. Conservation of both miRNAs and their target genes across human and domestic animals provides the likelihood of having functional orthology relationship which may also lead to similar diseases. Findings from this study are a step forward towards building an advanced animal disease miRNA resource, identifying miRNA-related diseases in animals and utilization of miRNA disease biomarkers in animal and veterinary research. In the long-term, validating these human disease miRNAs in domestic animals could identify new large animal models of diseases and most likely biomarkers to expedite development of therapeutic measures for human and animal diseases.
Availability of supporting data
The data supporting the results of this article is included within the article and its additional files. The integrated animal disease miRNA resource is freely available for download via AgBase at http://agbase.hpc.msstate.edu/cgi-bin/animal_mirna.cgi.
Ethical requirements
Our study used human and animal data from publicly available databases and did not require ethics approval from the Institutional Review Board for the Protection of Human Subjects in Research (IRB) or the Institutional Animal Care and Use Committee (IACUC).
Abbreviations
- BRENDA:
-
BRaunschweig ENzyme Database
- BTO:
-
BRENDA Tissue ontology
- DO:
-
Disease ontology
- GO:
-
Gene ontology
- HDMACs:
-
Human disease miRNAs with animal counterparts
- HMDD:
-
Human miRNA disease database
- IGBB:
-
Institute for genomics, biocomputing and biotechnology
- MFE:
-
Minimum free energy
- miRNAs:
-
MicroRNAs
- NJ:
-
Neighbor-joining
- OMIA:
-
Online mendelian inheritance in animal
- OMIM:
-
Online mendelian inheritance in man
- pre-miRNAs:
-
Precursor miRNAs
- UTRs:
-
Un-translated regions.
References
Winter J, Jung S, Keller S, Gregory RI, Diederichs S: Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009, 11 (3): 228-234.
Minguzzi S, Selcuklu SD, Spillane C, Parle-McDermott A: An NTD-Associated Polymorphism in the 3′ UTR of MTHFD1L can Affect Disease Risk by Altering miRNA Binding. Hum Mutat. 2014, 35 (1): 96-104.
Wang L, Liu W, Jiang W, Lin J, Jiang Y, Li B, Pang D: A miRNA binding site single-nucleotide polymorphism in the 3′-UTR region of the IL23R gene is associated with breast cancer. PLoS One. 2012, 7 (12): e49823-
Fang L, Du WW, Yang X, Chen K, Ghanekar A, Levy G, Yang W, Yee AJ, Lu WY, Xuan JW, Gao Z, Xie F, He C, Deng Z, Yang BB: Versican 3'-untranslated region (3'-UTR) functions as a ceRNA in inducing the development of hepatocellular carcinoma by regulating miRNA activity. FASEB J. 2013, 27 (3): 907-919.
Marin RM, Voellmy F, von Erlach T, Vanicek J: Analysis of the accessibility of CLIP bound sites reveals that nucleation of the miRNA: mRNA pairing occurs preferentially at the 3′-end of the seed match. RNA. 2012, 18 (10): 1760-1770.
Niwa R, Slack FJ: The evolution of animal microRNA function. Curr Opin Genet Dev. 2007, 17 (2): 145-150.
Christodoulou F, Raible F, Tomer R, Simakov O, Trachana K, Klaus S, Snyman H, Hannon GJ, Bork P, Arendt D: Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010, 463 (7284): 1084-1088.
Ma ZL, Yang HY, Tien P: Progress of miRNA and its functions in eukaryotes. Yi Chuan. 2003, 30 (7): 693-696.
Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G: Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000, 408 (6808): 86-89.
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G: The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000, 403 (6772): 901-906.
Lee RC, Feinbaum RL, Ambros V: The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993, 75 (5): 843-854.
Wightman B, Ha I, Ruvkun G: Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993, 75 (5): 855-862.
Lu TX, Sherrill JD, Wen T, Plassard AJ, Besse JA, Abonia JP, Franciosi JP, Putnam PE, Eby M, Martin LJ, Aronow BJ, Rothenberg ME: MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J Allergy Clin Immunol. 2012, 129 (4): 1064-1075 e1069.
Heneghan HM, Miller N, Kelly R, Newell J, Kerin MJ: Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 2010, 15 (7): 673-682.
Shi H, Xu J, Zhang G, Xu L, Li C, Wang L, Zhao Z, Jiang W, Guo Z, Li X: Walking the interactome to identify human miRNA-disease associations through the functional link between miRNA targets and disease genes. BMC Syst Biol. 2013, 7: 101-
Bhayani MK, Calin GA, Lai SY: Functional relevance of miRNA sequences in human disease. Mutat Res. 2012, 731 (1–2): 14-19.
Zheng Z, Zeng Y, Huang H, Xu F: MicroRNA-132 may play a role in coexistence of depression and cardiovascular disease: a hypothesis. Med Sci Monit. 2013, 19: 438-443.
Wu C, Gong Y, Sun A, Zhang Y, Zhang C, Zhang W, Zhao G, Zou Y, Ge J: The human MTHFR rs4846049 polymorphism increases coronary heart disease risk through modifying miRNA binding. Nutr Metab Cardiovasc Dis. 2013, 23 (7): 693-698.
Steinfeld I, Navon R, Ach R, Yakhini Z: miRNA target enrichment analysis reveals directly active miRNAs in health and disease. Nucleic Acids Res. 2013, 41 (3): e45-
Shrivastava S, Petrone J, Steele R, Lauer GM, Di Bisceglie AM, Ray RB: Up-regulation of circulating miR-20a is correlated with hepatitis C virus-mediated liver disease progression. Hepatology. 2013, 58 (3): 863-871.
Shimizu C, Kim J, Stepanowsky P, Trinh C, Lau HD, Akers JC, Chen C, Kanegaye JT, Tremoulet A, Ohno-Machado L, Burns JC: Differential expression of miR-145 in children with Kawasaki disease. PLoS One. 2013, 8 (3): e58159-
Shapshak P: Molecule of the month: miRNA and Human Prion brain disease. Bioinformation. 2013, 9 (13): 659-660.
Selth LA, Townley SL, Gillis JL, Tilley WD, Butler LM: Identification of prostate cancer-associated micrornas in circulation using a mouse model of disease. Methods Mol Biol. 2013, 1024: 235-246.
Qi J, Hou S, Zhang Q, Liao D, Wei L, Fang J, Zhou Y, Kijlstra A, Yang P: A functional variant of pre-miRNA-196a2 confers risk for Behcet’s disease but not for Vogt-Koyanagi-Harada syndrome or AAU in ankylosing spondylitis. Hum Genet. 2013, 132 (12): 1395-1404.
Qabaja A, Alshalalfa M, Bismar TA, Alhajj R: Protein network-based Lasso regression model for the construction of disease-miRNA functional interactions. EURASIP J Bioinform Syst Biol. 2013, 2013 (1): 3-
Patel V, Williams D, Hajarnis S, Hunter R, Pontoglio M, Somlo S, Igarashi P: miR-17~92 miRNA cluster promotes kidney cyst growth in polycystic kidney disease. Proc Natl Acad Sci U S A. 2013, 110 (26): 10765-10770.
Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A: Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg. 2014, 259 (4): 735-743.
Hussein F-K, Nizar B, Mehdi N, Philippe L, Mohammad F-K, Rabih B, Eva H, Ahmad D, Nader H, Rim ED, Fadwa B, Luc V, Arsène B, Philippe M, Redouane R, Bassam B: Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. J Transl Med. 2013, 11: 31-31.
Erener S, Mojibian M, Fox JK, Denroche HC, Kieffer TJ: Circulating miR-375 as a biomarker of β-cell death and diabetes in mice. Endocrinology. 2013, 154 (2): 603-608.
Zeng X, Xiang J, Wu M, Xiong W, Tang H, Deng M, Li X, Liao Q, Su B, Luo Z, Zhou Y, Zhou M, Zeng Z, Li X, Shen S, Shuai C, Li G, Fang J, Peng S: Circulating miR-17, miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. PLoS One. 2012, 7 (10): e46367-
Wang N, Zhou Y, Jiang L, Li D, Yang J, Zhang CY, Zen K: Urinary microRNA-10a and microRNA-30d serve as novel, sensitive and specific biomarkers for kidney injury. PLoS One. 2012, 7 (12): e51140-
Sun Y, Wang M, Lin G, Sun S, Li X, Qi J, Li J: Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS One. 2012, 7 (10): e47003-
Garofalo M, Romano G, Di Leva G, Nuovo G, Jeon Y-J, Ngankeu A, Sun J, Lovat F, Alder H, Condorelli G, Engelman JA, Ono M, Rho JK, Cascione L, Volinia S, Nephew KP, Croce CM: EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2012, 18 (1): 74-82.
Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, Okamoto K, Otsuji E: Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010, 102 (7): 1174-1179.
Zeng Z, Wang J, Zhao L, Hu P, Zhang H, Tang X, He D, Tang S, Zeng Z: Potential role of microRNA-21 in the diagnosis of gastric cancer: a meta-analysis. PLoS One. 2013, 8 (9): e73278-
Zeng R-C, Zhang W, Yan X-Q, Ye Z-Q, Chen E-D, Huang D-P, Zhang X-H, Huang G-L: Down-regulation of miRNA-30a in human plasma is a novel marker for breast cancer. Med Oncol. 2013, 30 (1): 477-
Yong FL, Law CW, Wang CW: Potentiality of a triple microRNA classifier: miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer. 2013, 13: 280-280.
Ye J, Wu X, Wu D, Wu P, Ni C, Zhang Z, Chen Z, Qiu F, Xu J, Huang J: miRNA-27b targets vascular endothelial growth factor C to inhibit tumor progression and angiogenesis in colorectal cancer. PLoS One. 2013, 8 (4): e60687-
Wu J, Yang T, Li X, Yang Q, Liu R, Huang J, Li Y, Yang C, Jiang Y: Alteration of serum miR-206 and miR-133b is associated with lung carcinogenesis induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Toxicol Appl Pharmacol. 2013, 267 (3): 238-246.
Yamada N, Nakagawa Y, Tsujimura N, Kumazaki M, Noguchi S, Mori T, Hirata I, Maruo K, Akao Y: Role of intracellular and extracellular MicroRNA-92a in colorectal cancer. Transl Oncol. 2013, 6 (4): 482-492.
Watahiki A, Macfarlane RJ, Gleave ME, Crea F, Wang Y, Helgason CD, Chi KN: Plasma miRNAs as biomarkers to identify patients with castration-resistant metastatic prostate cancer. Int J Mol Sci. 2013, 14 (4): 7757-7770.
Wang Z, Han J, Cui Y, Fan K, Zhou X: Circulating microRNA-21 as noninvasive predictive biomarker for response in cancer immunotherapy. Med Hypotheses. 2013, 81 (1): 41-43.
Wang JL, Hu Y, Kong X, Wang ZH, Chen HY, Xu J, Fang JY: Candidate microRNA biomarkers in human gastric cancer: a systematic review and validation study. PLoS One. 2013, 8 (9): e73683-
Gidlöf O, Smith JG, Miyazu K, Gilje P, Spencer A, Blomquist S, Erlinge D: Circulating cardio-enriched microRNAs are associated with long-term prognosis following myocardial infarction. BMC Cardiovasc Disord. 2013, 13: 12-12.
Zhu S, Cao L, Zhu J, Kong L, Jin J, Qian L, Zhu C, Hu X, Li M, Guo X, Han S, Yu Z: Identification of maternal serum microRNAs as novel non-invasive biomarkers for prenatal detection of fetal congenital heart defects. Clin Chim Acta. 2013, 424: 66-72.
Zhou X, Mao A, Wang X, Duan X, Yao Y, Zhang C: Urine and serum microRNA-1 as novel biomarkers for myocardial injury in open-heart surgeries with cardiopulmonary bypass. PLoS One. 2013, 8 (4): e62245-
Wang E, Nie Y, Zhao Q, Wang W, Huang J, Liao Z, Zhang H, Hu S, Zheng Z: Circulating miRNAs reflect early myocardial injury and recovery after heart transplantation. J Cardiothorac Surg. 2013, 8: 165-165.
Matsumoto S, Sakata Y, Suna S, Nakatani D, Usami M, Hara M, Kitamura T, Hamasaki T, Nanto S, Kawahara Y, Komuro I: Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ Res. 2013, 113 (3): 322-326.
Finn NA, Eapen D, Manocha P, Al Kassem H, Lassegue B, Ghasemzadeh N, Quyyumi A, Searles CD: Coronary heart disease alters intercellular communication by modifying microparticle-mediated microRNA transport. FEBS Lett. 2013, 587 (21): 3456-3463.
Dickinson BA, Semus HM, Montgomery RL, Stack C, Latimer PA, Lewton SM, Lynch JM, Hullinger TG, Seto AG, van Rooij E: Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur J Heart Fail. 2013, 15 (6): 650-659.
Wang H, Lu H-M, Yang W-H, Luo C, Lu S-H, Zhou Y, Lin Y-Z: The influence of statin therapy on circulating microRNA-92a expression in patients with coronary heart disease. Zhongguo Wei Zhong Bing Ji Jiu Yi Xue. 2012, 24 (4): 215-218.
Agarwal SM, Raghav D, Singh H, Raghava GP: CCDB: a curated database of genes involved in cervix cancer. Nucleic Acids Res. 2011, 39 (Database issue): D975-979.
Gong J, Tong Y, Zhang HM, Wang K, Hu T, Shan G, Sun J, Guo AY: Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum Mutat. 2012, 33 (1): 254-263.
Guo D, Liu J, Wang W, Hao F, Sun X, Wu X, Bu P, Zhang Y, Liu Y, Liu F, Zhang Q, Jiang F: Alteration in abundance and compartmentalization of inflammation-related miRNAs in plasma after intracerebral hemorrhage. Stroke. 2013, 44 (6): 1739-1742.
Wang H, Peng W, Shen X, Huang Y, Ouyang X, Dai Y: Circulating levels of inflammation-associated miR-155 and endothelial-enriched miR-126 in patients with end-stage renal disease. Braz J Med Biol Res. 2012, 45 (12): 1308-1314.
Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, Kodys K, Szabo G: Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 2012, 56 (5): 1946-1957.
Bhattacharyya S, Balakathiresan NS, Dalgard C, Gutti U, Armistead D, Jozwik C, Srivastava M, Pollard HB, Biswas R: Elevated miR-155 promotes inflammation in cystic fibrosis by driving hyperexpression of interleukin-8. J Biol Chem. 2011, 286 (13): 11604-11615.
Blanco-Calvo M, Calvo L, Figueroa A, Haz-Conde M, Antón-Aparicio L, Valladares-Ayerbes M: Circulating microRNAs: molecular microsensors in gastrointestinal cancer. Sensors (Basel). 2012, 12 (7): 9349-9362.
Barnard JA: Recent advances in pediatric gastroenterology, hepatology and nutrition. F1000Prime Rep. 2013, 5: 25-
Duttagupta R, DiRienzo S, Jiang R, Bowers J, Gollub J, Kao J, Kearney K, Rudolph D, Dawany NB, Showe MK, Stamato T, Getts RC, Jones KW: Genome-wide maps of circulating miRNA biomarkers for ulcerative colitis. PLoS One. 2012, 7 (2): e31241-
Li Y, Qiu C, Tu J, Geng B, Yang J, Jiang T, Cui Q: HMDD v2.0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014, 42 (Database issue): D1070-1074.
Bhattacharya A, Ziebarth JD, Cui Y: PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014, 42 (Database issue): D86-91.
Dong L, Luo M, Wang F, Zhang J, Li T, Yu J: TUMIR: an experimentally supported database of microRNA deregulation in various cancers. J Clin Bioinformatics. 2013, 3 (1): 7-
Bruno AE, Li L, Kalabus JL, Pan Y, Yu A, Hu Z: miRdSNP: a database of disease-associated SNPs and microRNA target sites on 3′UTRs of human genes. BMC Genomics. 2012, 13: 44-
Ruepp A, Kowarsch A, Theis F: PhenomiR: microRNAs in human diseases and biological processes. Methods Mol Biol. 2012, 822: 249-260.
Ziebarth JD, Bhattacharya A, Chen A, Cui Y: PolymiRTS Database 2.0: linking polymorphisms in microRNA target sites with human diseases and complex traits. Nucleic Acids Res. 2012, 40 (Database issue): D216-221.
Xie B, Ding Q, Han H, Wu D: miRCancer: a microRNA-cancer association database constructed by text mining on literature. Bioinformatics. 2013, 29 (5): 638-644.
Kiran C, Deepika P: Lung cancer: microRNA and target database. Zhongguo Fei Ai Za Zhi. 2012, 15 (7): 429-434.
Mitra S, Das S, Das S, Ghosal S, Chakrabarti J: HNOCDB: a comprehensive database of genes and miRNAs relevant to head and neck and oral cancer. Oral Oncol. 2012, 48 (2): 117-119.
Yang Z, Ren F, Liu C, He S, Sun G, Gao Q, Yao L, Zhang Y, Miao R, Cao Y, Zhao Y, Zhong Y, Zhao H: dbDEMC: a database of differentially expressed miRNAs in human cancers. BMC Genomics. 2010, 11 Suppl 4: S5-
Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, Cui Q: An analysis of human microRNA and disease associations. PLoS One. 2008, 3 (10): e3420-
Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y: miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009, 37 (Database issue): D98-104.
Kozomara A, Griffiths-Jones S: miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39 (Database issue): D152-157.
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG: Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011, 7: 539-
EMBL-EBI: ClustalW2 – Phylogeny: commonly used phylogenetic tree generation methods provided by the ClustalW2 program.http://www.ebi.ac.uk/Tools/services/web/toolform.ebi?tool=clustalw2_phylogeny,
Kasprzyk A: BioMart: driving a paradigm change in biological data management. Database (Oxford). 2011, 2011: bar049-
Baxevanis AD: Searching Online Mendelian Inheritance in Man (OMIM) for information on genetic loci involved in human disease. Curr Protoc Hum Genet. 2012, Chapter 9: Unit 9 13 11–10-
Lenffer J, Nicholas FW, Castle K, Rao A, Gregory S, Poidinger M, Mailman MD, Ranganathan S: OMIA (Online Mendelian Inheritance in Animals): an enhanced platform and integration into the Entrez search interface at NCBI. Nucleic Acids Res. 2006, 34 (Database issue): D599-601.
Schriml LM, Arze C, Nadendla S, Chang YW, Mazaitis M, Felix V, Feng G, Kibbe WA: Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res. 2012, 40 (Database issue): D940-946.
Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone SA, Scheuermann RH, Shah N, Whetzel PL, Lewis S, Consortium OBI: The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007, 25 (11): 1251-1255.
Gremse M, Chang A, Schomburg I, Grote A, Scheer M, Ebeling C, Schomburg D: The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 2011, 39 (Database issue): D507-513.
McCarthy FM, Gresham CR, Buza TJ, Chouvarine P, Pillai LR, Kumar R, Ozkan S, Wang H, Manda P, Arick T, Bridges SM, Burgess SC: AgBase: supporting functional modeling in agricultural organisms. Nucleic Acids Res. 2011, 39 (Database issue): D497-506.
McCarthy FM, Wang N, Magee GB, Nanduri B, Lawrence ML, Camon EB, Barrell DG, Hill DP, Dolan ME, Williams WP, Luthe DS, Bridges SM, Burgess SC: AgBase: a functional genomics resource for agriculture. BMC genomics. 2006, 7: 229-
Jordan SD, Kruger M, Willmes DM, Redemann N, Wunderlich FT, Bronneke HS, Merkwirth C, Kashkar H, Olkkonen VM, Bottger T, Braun T, Seibler J, Bruning JC: Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol. 2011, 13 (4): 434-446.
Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS: The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32 (Database issue): D258-261.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G: Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000, 25 (1): 25-29.
Zhao A, Li G, Peoc’h M, Genin C, Gigante M: Serum miR-210 as a novel biomarker for molecular diagnosis of clear cell renal cell carcinoma. Exp Mol Pathol. 2013, 94 (1): 115-120.
Yu X, Luo L, Wu Y, Yu X, Liu Y, Yu X, Zhao X, Zhang X, Cui L, Ye G, Le Y, Guo J: Gastric juice miR-129 as a potential biomarker for screening gastric cancer. Med Oncol. 2013, 30 (1): 365-
Yang IP, Tsai H-L, Huang C-W, Huang M-Y, Hou M-F, Juo S-HH, Wang J-Y: The functional significance of MicroRNA-29c in patients with colorectal cancer: a potential circulating biomarker for predicting early relapse. PLoS One. 2013, 8 (6):
Ting HJ, Messing J, Yasmin-Karim S, Lee YF: Identification of microRNA-98 as a therapeutic target inhibiting prostate cancer growth and a biomarker induced by vitamin D. J Biol Chem. 2013, 288 (1): 1-9.
Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H, Hu C: Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol. 2013, 139 (2): 223-229.
Mori F, Strano S, Blandino G: MicroRNA-181a/b: novel biomarkers to stratify breast cancer patients for PARPi treatment. Cell Cycle. 2013, 12 (12): 1823-1824.
Li C, Li JF, Cai Q, Qiu QQ, Yan M, Liu BY, Zhu ZG: MiRNA-199a-3p: A potential circulating diagnostic biomarker for early gastric cancer. J Surg Oncol. 2013, 108 (2): 89-92.
Endo K, Naito Y, Ji X, Nakanishi M, Noguchi T, Goto Y, Nonogi H, Ma X, Weng H, Hirokawa G, Asada T, Kakinoki S, Yamaoka T, Fukushima Y, Iwai N: MicroRNA 210 as a biomarker for congestive heart failure. Biol Pharm Bull. 2013, 36 (1): 48-54.
Bauters C, Kumarswamy R, Holzmann A, Bretthauer J, Anker SD, Pinet F, Thum T: Circulating miR-133a and miR-423-5p fail as biomarkers for left ventricular remodeling after myocardial infarction. Int J Cardiol. 2013, 168 (3): 1837-1840.
Parker HG, Ostrander EA: Cancer. Hiding in plain view--an ancient dog in the modern world. Science. 2014, 343 (6169): 376-378.
Rowell JL, McCarthy DO, Alvarez CE: Dog models of naturally occurring cancer. Trends Mol Med. 2011, 17 (7): 380-388.
Khanna C, Lindblad-Toh K, Vail D, London C, Bergman P, Barber L, Breen M, Kitchell B, McNeil E, Modiano JF, Niemi S, Comstock KE, Ostrander E, Westmoreland S, Withrow S: The dog as a cancer model. Nat Biotechnol. 2006, 24 (9): 1065-1066.
Davis JL, Gardner SY, Schwabenton B, Breuhaus BA: Congestive heart failure in horses: 14 cases (1984–2001). J Am Vet Med Assoc. 2002, 220 (10): 1512-1515.
Acknowledgements
This work was supported by the Office of Research and Graduate Studies, College of Veterinary Medicine and the Institute for Genomics, Biocomputing & Biotechnology. We thank the staff at the High Performance Computing Collaboratory (HPC2) for their technical support and routine backup of data generated from this study. We thank the reviewers for their valuable comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
TB generated data for this manuscript and performed data analysis and interpretation, MA provided computational data integration assistance, HW generated standardized disease phenotype names for the integrated resource, TB and DGP wrote the manuscript. All authors read, critiqued, edited and approved the final manuscript.
Electronic supplementary material
13104_2014_2942_MOESM1_ESM.xlsx
Additional file 1:Distribution of human disease microRNAs across animal counterparts. Multiple human diseases and locations are separated by a pipe (column 3). The (+) or (−) in columns 2–6 indicates presence or absence of homologous human disease miRNA (column 1) in the selected animal species. (XLSX 40 KB)
13104_2014_2942_MOESM2_ESM.xlsx
Additional file 2:Phylogenetic distances of pre-miRNAs of HDMACs. The data in column 1 & 3 are the pre-miRNA ID for animals and human, respectively prefixed with abbreviation of species scientific name i.e. bta (Bos Taurus - cow), gga (gallus gallus - chicken), ssc (Sus scrofa - pig), eca (Equine canibus - horse), cfa (Canis familiaris - dog), and hsa (Homo sapiens - human). Column 2 & 4 show the phylogenetic distances calculated based on nucleotide substitution rate within similar clusters. (XLSX 21 KB)
13104_2014_2942_MOESM3_ESM.xlsx
Additional file 3:Sample of disease miRNAs targets and their animal orthologs. Experimentally verified genes targeted by the disease miRNAs are manually curated from Pubmed articles and their % identity with an animal ortholog is generated via Ensembl Biomart [76]. (XLSX 20 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Buza, T., Arick, M., Wang, H. et al. Computational prediction of disease microRNAs in domestic animals. BMC Res Notes 7, 403 (2014). https://doi.org/10.1186/1756-0500-7-403
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-0500-7-403