Abstract
Background
The long noncoding RNA MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) is described as a potential biomarker for NSCLC (non-small cell lung cancer). Diagnostic biomarkers need to be detectable in easily accessible body fluids, should be characterized by high specificity, sufficient sensitivity, and robustness against influencing factors. The aim of this study was to evaluate the performance of MALAT1 as a blood based biomarker for NSCLC.
Results
MALAT1 was shown to be detectable in the cellular fraction of peripheral human blood, showing different expression levels between cancer patients and cancer-free controls. For the discrimination of NSCLC patients from cancer-free controls a sensitivity of 56% was calculated conditional on a high specificity of 96%. No impact of tumor stage, age, gender, and smoking status on MALAT1 levels could be observed, but results based on small numbers.
Conclusions
The results of this study indicate that MALAT1 complies with key characteristics of diagnostic biomarkers, i.e., minimal invasiveness, high specificity, and robustness. Due to its relatively low sensitivity MALAT1 might not be feasible as a single biomarker for the diagnosis of NSCLC in the cellular fraction of blood. Alternatively, MALAT1 might be applicable as a complementary biomarker within a panel in order to improve the entire diagnostic performance.
Similar content being viewed by others
Background
Lung cancer is the leading cause of cancer death worldwide[1] with NSCLC (non-small cell lung cancer) as the most prominent subgroup accounting for approximately 80% of all lung cancer cases. Commonly, the disease is detected in late stages resulting in short survival rates, whereas for patients with early-stage lung cancer longer survival rates could be observed[2]. Thus, the detection of lung cancer in early stages when clinical symptoms have not yet occurred appears to be a promising opportunity to decrease mortality, because in more cases a curative therapy might become possible.
In principal, biomarkers should be feasible for the detection of cancer in early stages. Thus, a major aim in cancer research is the identification of proper biomarkers. Key characteristics of diagnostic biomarkers among others are: (i) minimally-invasive to measure the biomarker in easily accessible body fluids, (ii) high specificity to avoid false-positive results in cancer-free individuals, (iii) sufficient sensitivity to detect the tumors, and (iv) robustness against potential influencing factors.
In recent years biomarker research focused on noncoding RNAs (ncRNAs), in particular microRNAs (miRNAs). MiRNAs are small RNA molecules with a length of ~ 22 nucleotides (nt), playing a central role in the regulation of gene expression[3] and acting as tumor suppressors or oncogenes in cancer[4]. Several studies show the feasibility of using miRNAs as biomarkers in body fluids for the diagnosis of lung cancer[5–8]. However, there is a lack of consistent results between studies focused on the identification of miRNAs as biomarkers[9]. Thus, the discovery of alternative or complementing biomarkers is essential.
In addition to miRNAs, long noncoding RNAs (lncRNAs) are a promising alternative within the group of ncRNAs. LncRNAs are commonly described as RNA molecules with a length > 200 nt, playing regulatory and structural roles in biological processes. As lncRNAs are implicated as tumor suppressors and oncogenes[10], they might be feasible as diagnostic biomarkers[11]. Currently, only few lncRNAs have been described as candidate biomarkers in human body fluids[10]. HULC (highly up-regulated in liver cancer) is highly expressed in hepatocellular carcinoma patients and detectable in human blood[12]. PCA3 (prostate cancer gene 3) is detectable in urine of prostate cancer patients, showing high accuracy[13]. In addition, MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) might be a candidate biomarker for NSCLC[14]. MALAT1 is a well-described lncRNA widely expressed in normal tissues[15]. In several human carcinomas MALAT1 was shown to be upregulated[16], particularly in early-stage metastasizing NSCLC.
The aim of this study was the evaluation of MALAT1 as a blood-based biomarker for NSCLC. The expression of MALAT1 was measured in the cellular fraction of peripheral human blood and the expression levels of NSCLC patients and cancer-free controls of the general population were compared.
Methods
Study population
The study was designed according to rules guarding patient privacy and with the approval from the ethics committee of the Ruhr-Universität Bochum (No. 3217–08). All participants provided written informed consent.
The cancer group of 45 NSCLC patients consisted of 21 patients with AdCa (adenocarcinoma) and 24 patients with SqCC (squamous cell carcinoma). Participants were recruited at the HELIOS Clinic Emil von Behring, Berlin, Germany. Tumor staging was performed according to the TNM classification of malignant tumors[17]. Cancer patients had not been treated by surgery, chemotherapy, or radiation therapy before blood collection. The control group of 25 cancer-free subjects was drawn from the Heinz Nixdorf Recall study, a population-based cohort of elderly subjects[18]. Characteristics of the study groups are summarized in Table 1. Detailed subject characteristics are listed in Additional file1.
RNA isolation
Peripheral blood samples were collected from each participant in 9.0 ml S-Monovette EDTA gel tubes (Sarstedt, Nümbrecht, Germany) and centrifuged (2000 x g for 10 minutes) within 30 minutes after collection. The cellular fraction was separated from plasma and stored frozen until RNA isolation.
Samples were thawed at room temperature and RNA isolation including DNase I treatment was performed from 0.5 ml of the cellular fraction using the RiboPure Blood Kit according to the manufacturers’ instructions (Life Technologies, Darmstadt, Germany).
Quantitative real-time PCR (qRT-PCR)
TaqMan assays (Life Technologies) were used for quantitative expression analyses of MALAT1 (Hs00273907_s1) as potential biomarker and of GAPDH (Hs99999905_m1), HPRT1 (Hs02800696_m1), and RPLP0 (Hs99999902_m1) as potential reference genes for normalization. Quantitative real-time PCR (qRT-PCR) was performed using a 7900 HT Fast Real-Time PCR System (Life Technologies). For the reverse transcription reaction 12 μl RNA and for the PCR reaction 5 μl cDNA were used as templates. Samples were analyzed in duplicate and non-template controls were included. For cycle threshold (Ct) estimation a fixed threshold of 0.2 was used. Ct values > 35 were considered to be under the detection limit[19] and marked as 35 for analysis[20]. Raw Ct values are presented in Additional file1.
The performance of potential references was analyzed utilizing RefFinder[21], a web-based comprehensive tool (http://www.leonxie.com/referencegene.php), including the four commonly used algorithms geNorm[22], NormFinder[23], BestKeeper[24], and comparative ΔCt method[25], to evaluate the most stable reference across study groups. As the geometric mean (GM) of several reference genes is more reasonable than a single reference gene[22], the GM of potential references was calculated. Normalized MALAT1 levels were expressed as ΔCt, with ΔCt = Ct(MALAT1) - Ct(Reference).
Statistical analysis
Median and inter-quartile range (IQR) were used to describe the distribution of MALAT1 levels. Groups were compared using the non-parametric Kruskal-Wallis test for continuous variables. Sensitivity and specificity of MALAT1 were determined from receiver operating characteristic (ROC) curves illustrating the performance of MALAT1 to discriminate the studied groups. In brief, NSCLC vs. controls, AdCa vs. controls, SqCC vs. controls, and AdCa vs. SqCC were analyzed. The bootstrap procedure (1000 runs) was used for internal validation of the estimates in the ROC analyses.
Potential factors influencing MALAT1 levels were evaluated using a linear regression model. Estimates were given as β with 95% confidence intervals (CI) and p values. Here, values of β > 0 indicate a negative association between the influencing factor and MALAT1 levels, values of β < 0 a positive association. Logistic regression modeling was performed to estimate the odds ratio (OR) with 95% CI for detecting NSCLC as a function of normalized MALAT1 levels.
Statistical analyses were performed using SAS/STAT and SAS/IML software, version 9.3 (SAS Institute Inc., Cary, NC).
Results
Expression stability of candidate references
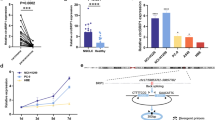
The potential reference genes GAPDH, HPRT1, and RPLP0 were measured in all samples from NSCLC patients and cancer-free controls. Using raw Ct values no significant differences between NSCLC patients and controls could be observed for GAPDH and HPRT1 in contrast to RPLP0 (p = 0.0002), (Figure 1). Thus, RPLP0 was excluded from further evaluation as reference gene.
In order to identify the most stable reference across the study groups RefFinder was used to rank the analyzed references. Lowest rank represents the most stable reference and highest rank represents the least stable reference (Table 2). The GM of GAPDH and HPRT1 was identified as the most stable reference and used for normalization of MALAT1.
Distribution of MALAT1 in the study groups
Table 3 depicts the distribution of normalized MALAT1 levels in the study groups. The median normalized MALAT1 value for NSCLC was -0.35 (IQR -1.34; 1.00), for AdCa -0.59 (IQR -1.34; -0.25), and for SqCC -0.23 (IQR -1.31; 1.47), whereas for controls the median normalized MALAT1 level was -2.07 (IQR -2.53; -0.83).
Differences of normalized MALAT1 levels between cancer patients and cancer-free subjects were significant for NSCLC vs. controls (p < 0.0001), AdCa vs. controls (p = 0.0043), and SqCC vs. controls (p = 0.0001), whereas the difference between AdCa and SqCC was not significant (Figure 2).
Scatter dot plots of MALAT1 levels in NSCLC (N = 45), AdCa (N = 21), SqCC (N = 24), and cancer-free controls (N = 25). Expression values were normalized by the geometric mean of GAPDH and HPRT1 and expressed as ΔCt. Horizontal bars represent median and IQR. Groups were compared using the Kruskal-Wallis test. NSCLC: non-small cell lung cancer, AdCa: adenocarcinoma, SqCC: squamous cell carcinoma.
MALAT1 as biomarker of NSCLC
Using ROC analyses, for NSCLC patients and controls an area under the curve (AUC) of 0.79 (95% CI 0.68 – 0.89), (Figure 3A), for AdCa patients and controls an AUC of 0.75 (95% CI 0.59 – 0.90), (Figure 3B), for SqCC patients and controls an AUC of 0.82 (0.70 – 0.94), (Figure 3C), and for AdCa vs. SqCC an AUC of 0.58 (95% CI 0.42 – 0.76), (Figure 3D), were calculated.
Receiver operating characteristics (ROC) curves of normalized MALAT1 . The area under curve (AUC) was determined for MALAT1 of (A) NSCLC (non-small cell lung cancer) patients and controls, (B) AdCa (adenocarcinoma) patients and controls, (C) SqCC (squamous cell carcinoma) patients and controls, and (D) AdCa patients and SqCC patients.
Sensitivity and specificity of normalized MALAT1 are shown in Table 4, calculated due false-positive rates (FPR) of 0% (no false-positive test), 4% (one false-positive test), and 8% (two false-positive tests), and to the maximum Youden’s Index (YI = sensitivity + specificity -1), respectively. A FPR of 4%, representing 96% specificity, resulted in 56% sensitivity for the discrimination of NSCLC from controls. The sensitivity to discriminate SqCC from controls is higher (63%) than the sensitivity to discriminate AdCa from controls (48%). A FPR of 8% (92% specificity) did not lead to any increase in sensitivities, whereas a FPR of 0% (100% specificity) resulted in lowest sensitivities for the discrimination of controls from patients with NSCLC (47%), AdCa (38%), or SqCC (54%). Use of the maximum YI leads to an increase in sensitivity to 81% only for AdCa vs. controls, but specificity decreased to 64%. For the discrimination of AdCa from SqCC a FPR of 5% (95% specificity, one false-positive test) resulted in 8% sensitivity and a FPR of 10% (90% specificity, two false-positive tests) resulted in 21% sensitivity, whereas using the maximum YI resulted in 33% sensitivity and 86% specificity.
ROC analyses on 1000 bootstrap samples resulted in similar cutoffs, sensitivities, and specificities of MALAT1 in comparison to the original analyses. The calculated 95% CI regarding NSCLC vs. controls and SqCC vs. controls indicate a good precision of this assessment, whereas AdCa vs. controls the 95% CI shows a less precision (Additional file2).
The application of logistic regression models revealed a two-fold increased risk of detecting NSCLC per normalized MALAT1 unit. The OR of MALAT1 was 1.88 (95% CI 1.26 - 2.83) without adjustment and 2.03 (95% CI 1.30-3.16) with adjustment for gender, age and smoking status.
Potential factors influencing MALAT1
The influence of tumor characteristics on MALAT1 levels is shown in Table 5. MALAT1 is not affected by tumor size, metastasis status or lymph node status.
The impact of potential influencing factors on the expression levels of MALAT1 are shown in Table 6. NSCLC showed a significant 1.63-fold (95% CI 0.75 – 2.51) decrease of MALAT1 (p = 0.0003), whereas the factors gender, age, and smoking status showed no impact on the MALAT1 levels in human blood.
Discussion
NSCLC is commonly detected in late stages of the disease. Biomarkers have the potential to detect cancer at early stages, facilitating an earlier and therefore more curative therapy that ideally results in decreased mortality. In NSCLC, Gutschner et al. showed that MALAT1 regulates the expression of several metastasis-associated genes, e.g. CDCP1 (CUB domain containing protein 1) and GPC6 (glypican 6), indicating a major role of MALAT1 in disease progression[26]. Additionally, it was suggested that MALAT1 might also regulate other important cellular processes in lung cancer[26]. Thus, MALAT1 is a candidate biomarker for NSCLC[14].
For quantitative expression analysis of messengerRNAs (mRNAs) and miRNAs qRT-PCR is considered to be the gold standard[27] and the same might be true for lncRNAs. However, to produce reliable data in qRT-PCR assays the use of appropriate reference genes for normalization is an important issue[28] and candidate reference genes need to be tested prior to application[29]. As no information regarding lncRNAs as references were accessible, mRNAs were selected as potential references. HPRT1 and RPLP0 are well-described reference genes for analyses in NSCLC tissues[30] and GAPDH was already applied for normalization of MALAT1[15]. However, in this study RPLP0 seems to be no feasible reference which is in agreement with Falkenberg et al., showing that RPLP0 is not appropriate as reference gene in human blood samples[31]. In this study, GAPDH and HPRT1 were suitable reference genes, particularly the GM of GAPDH and HPRT1 showed the best reference performance. This is in accordance with Ulivi et al., using GAPDH and HPRT1 for normalization of mRNAs in blood samples of NSCLC patients and controls[32].
One key characteristic of proper diagnostic biomarkers is the need to be detectable in easily accessible body fluids like peripheral blood. In this study MALAT1 was measured in the cellular fraction of human blood, showing that this matrix is in principle appropriate for the analysis of lncRNAs. Comparable results for the usability of the cellular blood fraction were shown for miRNAs[33, 34]. Commonly, the cellular fraction obtained during plasma preparation is discarded, but it might be reasonable to collect this matrix in biobanks for subsequent biomarker discovery.
In this study, a significant downregulation of MALAT1 in NSCLC patients in comparison to cancer-free controls was shown. Comparable results were achieved by Zhang et al., showing a downregulation of MALAT1 in patients with hepatocellular carcinomas[35]. However, MALAT1 was implicated to play an oncogenic role[10] and upregulation of MALAT1 was observed in several other cancers, e.g. of the breast and prostate[16]. Such differences might be caused by the paradigm that in fact MALAT1 is expressed ubiquitously but fulfills tissue-specific functions depending on the cellular environment[36]. Commonly, MALAT1 is analyzed in tissues[14–16], whereas in this study and the study of Zhang et al.[35]MALAT1 was detected in blood. Because MALAT1 was detected in the cellular fraction of blood, it is unlikely to be directly produced by the tumor tissue. Its downregulation in blood cells may be an indirect effect of the tumor, e.g., on the immune system. The source of MALAT1 in the cellular fraction of blood remains unclear. Theoretically, it might originate from leucocytes altered by the tumor. However, further analyses are needed to evaluate the origin of MALAT1 in human blood. Very recently, it was shown that MALAT1 was detectable in plasma of patients with gastric or prostate cancer[37, 38]. Thus, it would be reasonable to analyze MALAT1 in plasma of NSCLC patients instead of the cellular fraction, because the presence of MALAT1 in plasma might be a direct effect of the tumor, e.g., release of lncRNA-containing extracellular vesicles[39].
Tani et al. showed that the stability of MALAT1 varied in various cell types and indicated that the half-life of MALAT1 is shorter than the median half-life of mRNA[40]. Such decay of MALAT1 might also prevail in blood cells. It is well known that systems like PAXgene, Tempus, and RNAlater stabilize mRNAs and miRNAs in whole blood samples[41–43]. Thus, the performance of the assay was additionally tested in a few available blood samples stabilized by PAXgene or RNAlater. In the stabilized blood samples MALAT1 is detectable at lower Ct values corresponding to larger quantities (data not shown). The results implicate that the use of stabilization systems might be meaningful for lncRNA analyses in blood. However, this assumption needs to be verified in more detail.
Regarding the key characteristics of an obligatory high specificity and a sufficiently high sensitivity of diagnostic biomarkers, the candidate biomarker MALAT1 does not fulfill both criteria. Generally, in screening cohorts a high specificity is needed to avoid an unacceptably high number of false-positive tests that would result in psychological pressure and needless intervention for the patients. Thus, the sensitivity of candidate biomarkers should be calculated at a fixed high specificity level[44]. In regard to the relatively small study group the specificity of 96% is quite high, particularly as this corresponds to only one single false-positively tested control. On the other hand, the calculated sensitivity is too low (56%) for the use of MALAT1 as a single biomarker for the diagnosis of NSCLC, particularly for the subtype AdCa (48%). However, lower sensitivity could be balanced by the use of several biomarkers in a panel. Theoretically, in an optimal panel every single biomarker is characterized by sufficiently high sensitivity and the necessary high specificity, perfectly complement each other in order to obtain superior diagnostic performance[45]. Thus, it might be reasonable to verify MALAT1 in combination with other biomarkers in larger study groups to improve the entire diagnostic performance of the biomarker panel. However, for the discrimination of AdCa and SqCC, a sensitivity of only 8% precludes MALAT1 as a biomarker for the differential diagnosis of NSCLC subtypes.
Bootstrap analysis showed that the calculated cutoffs, sensitivities, and specificities remain stable, indicating that the calculated values are appropriate for the discrimination of patients and controls.
Regarding the fourth key characteristic of diagnostic biomarkers, the results indicate that MALAT1 values in blood are not correlated with tumor size, metastasis status, or lymph node status. However, more cases of early-stage metastasizing NSCLC need to be analyzed in subsequent studies because this study comprises only three cases with tumor stage T1 or T2. Additionally, MALAT1 seems to be relatively independent from common influencing factors like age, gender, and smoking status, indicating the robustness of the candidate biomarker. These observations are in agreement with MALAT1 expression in tissue[46]. However, it has to be clarified if other potential influencing factors from the multitude of biological, preanalytical, and analytical factors show an impact on MALAT1 levels in human blood.
Conclusions
MALAT1 could be detected in peripheral blood, showing different expression levels between NSCLC patients and cancer-free controls. It was shown that MALAT1 complies with key characteristics of diagnostic biomarkers, being minimally-invasive, exhibiting high specificity, and robustness. On the contrary, the observed sensitivity is too low for the use of MALAT1 as a single biomarker for the diagnosis of NSCLC using the cellular fraction of peripheral blood. However, it might be reasonable to verify the performance of MALAT1 as a complementary biomarker within a panel in larger studies including more cases of early-stage metastasizing NSCLC.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D: Global cancer statistics. CA Cancer J Clin. 2011, 61: 69-90. 10.3322/caac.20107.
Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V, Sobin L: International Association for the Study of Lung Cancer International Staging C, Participating I: the IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007, 2: 706-714. 10.1097/JTO.0b013e31812f3c1a.
Ambros V: The functions of animal microRNAs. Nature. 2004, 431: 350-355. 10.1038/nature02871.
Kent OA, Mendell JT: A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006, 25: 6188-6196. 10.1038/sj.onc.1209913.
Foss KM, Sima C, Ugolini D, Neri M, Allen KE, Weiss GJ: miR-1254 and miR-574-5p: Serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol. 2011, 6: 482-488. 10.1097/JTO.0b013e318208c785.
Zheng D, Haddadin S, Wang Y, Gu LQ, Perry MC, Freter CE, Wang MX: Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011, 4: 575-586.
Bianchi F, Nicassio F, Marzi M, Belloni E, Dall'olio V, Bernard L, Pelosi G, Maisonneuve P, Veronesi G, Di Fiore PP: A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med. 2011, 3: 495-503. 10.1002/emmm.201100154.
Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, Calabro E, Croce CM, Pastorino U, Sozzi G: MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011, 108: 3713-3718. 10.1073/pnas.1100048108.
Pass HI, Beer DG, Joseph S, Massion P: Biomarkers and molecular testing for early detection, diagnosis, and therapeutic prediction of lung cancer. Thorac Surg Clin. 2013, 23: 211-224. 10.1016/j.thorsurg.2013.01.002.
Reis EM, Verjovski-Almeida S: Perspectives of long non-coding RNAs in cancer diagnostics. Front Genet. 2012, 3: 32-
Tano K, Akimitsu N: Long non-coding RNAs in cancer progression. Front Genet. 2012, 3: 219-
Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K: Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007, 132: 330-342. 10.1053/j.gastro.2006.08.026.
Fradet Y, Saad F, Aprikian A, Dessureault J, Elhilali M, Trudel C, Masse B, Piche L, Chypre C: uPM3, a new molecular urine test for the detection of prostate cancer. Urology. 2004, 64: 311-315. 10.1016/j.urology.2004.03.052. discussion 315–316
Schmidt LH, Spieker T, Koschmieder S, Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, et al: The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011, 6: 1984-1992. 10.1097/JTO.0b013e3182307eac.
Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003, 22: 8031-8041. 10.1038/sj.onc.1206928.
Lin R, Maeda S, Liu C, Karin M, Edgington TS: A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007, 26: 851-858. 10.1038/sj.onc.1209846.
Sobin LH, Gospodarowicz MK, Wittekind C: International Union against Cancer.: TNM classification of malignant tumours. 7th edn. Chichester, West Sussex, UK. 2010, Wiley-Blackwell: Hoboken, NJ
Schmermund A, Möhlenkamp S, Stang A, Grönemeyer D, Seibel R, Hirche H, Mann K, Siffert W, Lauterbach K, Siegrist J, et al: Assessment of clinically silent atherosclerotic disease and established and novel risk factors for predicting myocardial infarction and cardiac death in healthy middle-aged subjects: rationale and design of the Heinz Nixdorf RECALL study. Risk factors, evaluation of coronary calcium and lifestyle. Am Heart J. 2002, 144: 212-218. 10.1067/mhj.2002.123579.
Guthrie JL, Seah C, Brown S, Tang P, Jamieson F, Drews SJ: Use of Bordetella pertussis BP3385 to establish a cutoff value for an IS481-targeted real-time PCR assay. J Clin Microbiol. 2008, 46: 3798-3799. 10.1128/JCM.01551-08.
Ning B, Dial S, Sun Y, Wang J, Yang J, Guo L: Systematic and simultaneous gene profiling of 84 drug-metabolizing genes in primary human hepatocytes. J Biomol Screen. 2008, 13: 194-201. 10.1177/1087057108315513.
Xie F, Xiao P, Chen D, Xu L, Zhang B: miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol. 2012, 80: 75-84. 10.1007/s11103-012-9885-2.
Vandesompele J, De Preter K, Pattyn F, Poppe B, van Roy N, De Paepe A, Speleman F: Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3: RESEARCH0034.1-0034.11-
Andersen CL, Jensen JL, Orntoft TF: Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64: 5245-5250. 10.1158/0008-5472.CAN-04-0496.
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP: Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004, 26: 509-515.
Silver N, Best S, Jiang J, Thein SL: Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006, 7: 33-10.1186/1471-2199-7-33.
Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al: The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013, 73: 1180-1189. 10.1158/0008-5472.CAN-12-2850.
Becker C, Hammerle-Fickinger A, Riedmaier I, Pfaffl MW: mRNA and microRNA quality control for RT-qPCR analysis. Methods. 2010, 50: 237-243. 10.1016/j.ymeth.2010.01.010.
Davoren PA, McNeill RE, Lowery AJ, Kerin MJ, Miller N: Identification of suitable endogenous control genes for microRNA gene expression analysis in human breast cancer. BMC Mol Biol. 2008, 9: 76-10.1186/1471-2199-9-76.
Schmittgen TD, Zakrajsek BA: Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000, 46: 69-81. 10.1016/S0165-022X(00)00129-9.
Gresner P, Gromadzinska J, Wasowicz W: Reference genes for gene expression studies on non-small cell lung cancer. Acta Biochim Pol. 2009, 56: 307-316.
Falkenberg VR, Whistler T, Murray JR, Unger ER, Rajeevan MS: Identification of Phosphoglycerate Kinase 1 (PGK1) as a reference gene for quantitative gene expression measurements in human blood RNA. BMC Res Notes. 2011, 4: 324-10.1186/1756-0500-4-324.
Ulivi P, Mercatali L, Casoni GL, Scarpi E, Bucchi L, Silvestrini R, Sanna S, Monteverde M, Amadori D, Poletti V, Zoli W: Multiple marker detection in peripheral blood for NSCLC diagnosis. PLoS One. 2013, 8: e57401-10.1371/journal.pone.0057401.
Weber DG, Johnen G, Bryk O, Jöckel KH, Brüning T: Identification of miRNA-103 in the cellular fraction of human peripheral blood as a potential biomarker for malignant Mesothelioma - a pilot study. PLoS One. 2012, 7: e30221-10.1371/journal.pone.0030221.
Häusler SF, Keller A, Chandran PA, Ziegler K, Zipp K, Heuer S, Krockenberger M, Engel JB, Hönig A, Scheffler M, et al: Whole blood-derived miRNA profiles as potential new tools for ovarian cancer screening. Br J Cancer. 2010, 103: 693-700. 10.1038/sj.bjc.6605833.
Zhang PJ, Wei R, Wen XY, Ping L, Wang CB, Dong ZN, Deng XX, Bo W, Bin C, Tian YP: Genes expression profiling of peripheral blood cells of patients with hepatocellular carcinoma. Cell Biol Int. 2012, 36: 803-809. 10.1042/CBI20100920.
Eissmann M, Gutschner T, Hämmerle M, Günther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun T, Zörnig M, Diederichs S: Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012, 9: 1076-1087. 10.4161/rna.21089.
Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T, et al: Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013, 33: 3185-3193.
Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J, Wei M, Xu C, Wu C, Zhang Z, et al: Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013, 49: 2949-2959. 10.1016/j.ejca.2013.04.026.
Kogure T, Yan IK, Lin WL, Patel T: Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: a mechanism of intercellular signaling in human hepatocellular cancer. Genes & Cancer. 2013, 4: 261-272. 10.1177/1947601913499020.
Tani H, Nakamura Y, Ijiri K, Akimitsu N: Stability of MALAT-1, a nuclear long non-coding RNA in mammalian cells, varies in various cancer cells. Drug Discoveries & Therapeutics. 2010, 4: 235-239.
Weber DG, Casjens S, Rozynek P, Lehnert M, Zilch-Schöneweis S, Bryk O, Taeger D, Gomolka M, Kreuzer M, Otten H, et al: Assessment of mRNA and microRNA stabilization in peripheral human blood for multicenter studies and biobanks. Biomark Insights. 2010, 5: 95-102.
Matheson LA, Duong TT, Rosenberg AM, Yeung RS: Assessment of sample collection and storage methods for multicenter immunologic research in children. J Immunol Methods. 2008, 339: 82-89. 10.1016/j.jim.2008.08.003.
Kruhoffer M, Dyrskjot L, Voss T, Lindberg RL, Wyrich R, Thykjaer T, Orntoft TF: Isolation of microarray-grade total RNA, microRNA, and DNA from a single PAXgene blood RNA tube. J Mol Diagn. 2007, 9: 452-458. 10.2353/jmoldx.2007.060175.
Zhu CS, Pinsky PF, Cramer DW, Ransohoff DF, Hartge P, Pfeiffer RM, Urban N, Mor G, Bast RC, Moore LE, et al: A framework for evaluating biomarkers for early detection: validation of biomarker panels for ovarian cancer. Cancer Prev Res . 2011, 4: 375-383. 10.1158/1940-6207.CAPR-10-0193.
Mai PL, Wentzensen N, Greene MH: Challenges related to developing serum-based biomarkers for early ovarian cancer detection. Cancer Prev Res . 2011, 4: 303-306. 10.1158/1940-6207.CAPR-11-0053.
Gutschner T, Hämmerle M, Diederichs S: MALAT1 - a paradigm for long noncoding RNA function in cancer. J Mol Med. 2013, 91: 791-801. 10.1007/s00109-013-1028-y.
Acknowledgement
We thank Prof. Dr. T. Bauer and B. Krone for collaborating in obtaining human blood samples.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DGW conceived of the study, participated in its design and coordination, and drafted the manuscript. GJ participated in study design and coordination and helped to draft the manuscript. SC performed the statistical analyses and helped to draft the manuscript. OB performed the experiments and helped to draft the manuscript. BP and KHJ participated in the statistical analysis and helped to draft the manuscript. JK participated in study design and helped to draft the manuscript. TB participated in study coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Daniel Gilbert Weber, Georg Johnen contributed equally to this work.
Electronic supplementary material
13104_2013_2527_MOESM2_ESM.xlsx
Additional file 2:Marker cutoffs with 95% CI for NSCLC (non-small cell lung cancer) vs. controls, AdCa (adenocarcinoma) vs. controls, SqCC (squamous cell carcinoma) vs. controls, and AdCa vs. SqCC after bootstrap analysis with 1000 random samples, according to false positive rates (FPR) corresponding to none, one, and two false-positive tests and maximum Youden’s Index (YI).(XLSX 42 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Weber, D.G., Johnen, G., Casjens, S. et al. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes 6, 518 (2013). https://doi.org/10.1186/1756-0500-6-518
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-0500-6-518