Abstract
Background
Methicillin-resistant staphylococci can colonize and cause diseases in companion animals. Unfortunately, few molecular studies have been carried out in Brazil and other countries with the aim of characterizing these isolates. Consequently, little is known about the potential role of companion animals in transmitting these resistant bacteria to humans. In this work we searched for mecA gene among Staphylococcus isolates obtained from nasal microbiota of 130 healthy dogs and cats attended in a veterinary clinic located in the west region of Rio de Janeiro. The isolates recovered were identified to the species level and characterized using molecular tools.
Results
A community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) isolate related to USA1100 (Southwest Pacific clone) and susceptible to all non-β-lactams was detected in a cat (1.7%, 1/60). Another coagulase-positive isolate harboring mecA was recovered from a dog (1.4%, 1/70) and identified as Staphylococcus pseudintermedius (MRSP) related to the European clone (ST71). The two isolates of Staphylococcus conhii subsp. urealyticus (1.4%, 1/70 dogs and 1.7%, 1/60 cats), similarly to the MRSP isolate, also presented high-level multiresistance. The majority of the methicillin-resistant coagulase-negative staphylococci recovered were Staphylococcus saprophyticus (5.7%, 4/70 dogs and 6.7%, 4/60 cats) and all clustered into the same PFGE type.
Conclusions
This work demonstrates that mecA-harboring Staphylococcus isolates are common members of the nasal microbiota of the healthy companion animals studied (9.2%, 12/130 animals), including some high-level multiresistant isolates of S. pseudintermedius and S. conhii subsp. urealyticus. The detection, for the first time in South America, of USA1100-related CA-MRSA and of ST71 MRSP (European clone), colonizing companion animals, is of concern. Both S. pseudintermedius and S. aureus are important agents of infections for animals. The USA1100 CA-MRSA is a causative of severe and disseminated diseases in healthy children and adults. Additionally, MRSP is a nosocomial pathogen in veterinarian settings. It had already been demonstrated that the virulent ST71 MRSP is geographically spread over Europe and USA, with potential for zoonotic infections.
Similar content being viewed by others
Background
The end of the 1980 decade was marked by the emergency of community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) among human populations from a remote area in Australia [1]. The first strain reported was named Western-Australia 1 (WA-1) and later genotyped as ST1-SCCmec IV. WA-1 isolates did not frequently carry the genes lukSF, encoding for the two subunits (S and F) of the Panton-Valentine leukocidin (PVL) [1, 2]. Soon after, CA-MRSA clones spread over different countries [3]. In the USA, three main PVL-positive CA-MRSA clones have been prevailing: USA 300 (ST8-SCCmec IV), USA 400 (ST1-SCCmec IV) and USA1100 (ST30-SCCmec IV), with USA 300 isolates (ST8-SCCmec IV, PVL+) ranking first among the causatives of skin/soft skin infections in healthy individuals from the community [4–6]. In Brazil, there are few reports on CA-MRSA infections and most of the isolates are related to USA 1100, which have been detected causing infections ranging from simple and localized to severe and invasive diseases, in previously healthy children and adults [7]. More recently, MRSA isolates have been found colonizing and infecting both companion and livestock animals with clear economical and public health implications [8–10]. The detection of MRSA in animals has raised concerns among scientists over their role as potential reservoirs or vectors for CA-MRSA infections in humans. Besides MRSA, other methicillin-resistant staphylococci (MRS) have recently been detected among companion animals such as methicillin-resistant Staphylococcus pseudintermedius (MRSP). S. pseudintermedius is a major cause of purulent and opportunistic infections in dogs, such as dermatitis, otitis and wound infections. Indeed, MRSP are frequently multidrug resistant pathogens resembling typical hospital isolates [11, 12].
Methicillin-resistant coagulase negative staphylococci (MRCoNS) have also been isolated in low frequencies from small companion animals and horses [13, 14]. Staphylococcus saprophyticus, for example, is a common agent of uncomplicated urinary tract infection (UTI) in young women [15]. Although the report of mecA-harboring S. saprophyticus is very rare, these isolates have recently been detected from human patients presented with UTI [16, 17]. Besides, MRCoNS are recognized as opportunistic pathogens for immunocompromized patients, including high-risk human neonates [18]. Of concern is the fact that MRCoNS are considered a potential reservoir of resistance determinants, including staphylococcal cassette mec[17]. In the study presented here we searched for mecA-harboring Staphylococcus isolates in the nasal microbiota of 130 companion animals attended in a veterinary clinic in Rio de Janeiro. The isolates recovered were identified at subspecies level, and the antimicrobial susceptibility and genotypic profiles determined.
Results and discussion
The mecA gene was confirmed in the 12 staphylococci that grew in the enrichment broth (9.2%, 12/130 animals). From these methicillin-resistant streptococci (MRS) isolates 10 were MRCoNS (7.7%, 10/130) and 2 coagulase-positive MRS (1.5%, 2/130). The majority of MRCoNS were identified as Staphylococcus saprophyticus (80%, 8/10) and the remaining as Staphylococcus conhii ssp. urealyticus (20%, 2/10). One coagulase-positive isolate (BMBSA87), collected from a cat, harbored mecA and was identified as MRSA (8.3%, 1/12) and the other isolate (BMBSP02), obtained from a dog, was initially identified as Staphylococcus intermedius by the automated method (8.3%, 1/12). Because all dog isolates identified thus far by routine phenotypic tests as S. intermedius clustered into the newly identified species S. pseudintermedius[19], additional PCR identification was carried out yielding an expected single band of 926 bp, confirming the classification of this isolate as S. pseudintermedius.
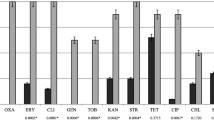
The MRSA isolate was susceptible to all drugs tested except β-lactams. All isolates identified as S. saprophyticus (except one that was also resistant to rifampicin; 7/8; 87.5%) displayed antimicrobial resistance only to erythromycin and clindamycin, in addition to β-lactams. Despite that, the MRSP isolate was resistant to ciprofloxacin, clindamycin, erythromycin, gentamicin and trimethoprim sulphamethoxazole, in addition to β-lactams. Multiresistance was also detected in 2 isolates of S. conhii ssp. urealyticus (Table 1).
The prevalence of MRS among dogs was as follows: 1.4% (1/70) carried MRSP, 1.4% (1/70) methicillin-resistant S. conhii-urea (MRSC) and 5.7% (4/70) methicillin-resistant S. saprophyticus (MRSS), and among cats was: 1.7% (1/60) carried MRSA, 1.7% (1/60) MRSC and 6.7% (4/60) MRSS. The MRSA isolate BMBSA87 was typed as SCCmec IV and displayed a PFGE pattern indistinguishable to that of CA-MRSA isolate WB49, a representative of the USA1100 clone (ST30-SCCmec IV; Figure 1A, Lanes 5–6), previously detected in Porto Alegre, RS, Brazil, from human skin/soft skin infection [20]. In addition, isolate BMBSA87 displayed the multilocus sequencing type (MLST) allelic profile: arcc = 2, aroe = 2, glpf = 2, gmk_ = 2, pta_ = 6, tpi_ = 3, yqil = 2, corresponding to ST30. However, amplification for the genes lukSF (encoding for PVL) could not be detected when total DNA of this isolate was tested with two different, specific, set of primers. PVL is commonly produced by ST30-SCCmec IV isolates [21, 22]. Despite that, cases of skin/soft tissue infections (SSTI) associated with PVL-negative CA-MRSA isolates have been reported [22, 23]. In addition, the MRSA isolate detected in the present study harbored pmsα3 encoding for the phenol soluble moduline α3, which has been associated with the pathogenesis of SSTI [24]. Our data showed that this MRSA isolate also carried the enterotoxin genes seg, sei, seo of the egc cluster, a highly prevalent operon of enterotoxin genes. It was suggested that the apparent redundancy of these superantigens provides selective advantage towards bacterial colonization and/or spread of the host and not only for toxemia [25]. The rate of MRSA colonization in dogs (1.4%, 1/70) was similar to those obtained in other studies (about 1- 2%) carried out in Hong Kong (2.1%, 15/704) and UK (0.7%, 6/850) [26, 27]. Indeed, small number of MRSA was detected as cause of urinary tract infections in dogs in Brazil (0.9%, 3/348) [13]. This incidence is in accordance with previous data reporting low percentage (1.2%) of CA-MRSA carriers among human infant in Brazil [28].
Pulsed-field gel electrophoresis (PFGE) of the Sma I-fragmented genomic DNA of the methicillin-resistant Staphylococcus isolates obtained from nasal swabs of companion animals. A, lanes 1–4: Representives of the methicillin-resistant S. saprophyticus ; 5: MRSA from cat (isolate 87); 6: Human isolate of CA-MRSA belonging to USA 1100 (ST30-SCCmec IV); 7: Lambda-ladder molecular size marker. B, lane 1: S. pseudintermedius from dog (isolate 02); 2: Representative isolate of the S. conhii-urea (isolate 28) and 3: Lambda-ladder molecular size marker.
S. pseudintermedius is an important opportunistic pathogen of companion animals. It is the most frequent Staphylococcus isolates collected from infected dogs. Since 2006 there has been a significant emergence of MRSP [12]. Many of MRSP isolates from Europe and USA (about 54%) were classified as ST71-SCCmec II, III or NT (NT: nontypable) [11, 12]. The MRSP recovered here from dog (isolate BMBSP02) was typed as SCCme c NT. Additionally, the lambda-ladder molecular size marker was used as reference to calculate the sizes of the PFGE bands (Figure 1B) of the isolate BMBSP02. These values were compared with those obtained from PFGE images of the MRSP isolates published by Ruscher et al. [12] and Soedarmanto et al. [29], and it was found that the banding patterns were quite similar. Thus, MLST analysis was then carried out and confirmed the presence of this international virulent MRSP clone (European clone) in our country, since the BMBSP02 clustered into ST71, corresponding to the MLST allelic profile ack = 3, cpn60 = 9, fdh = 1, pta = 2, sar = 2, tuf = 1. In Brazil, oxacillin resistance had already been described among S. pseudintermedius isolates causing urinary tract infections (8.6%, 6/70 dogs) and external otitis (14.3%, 13/91 dogs), on the basis of oxacillin 1 μg disk. However, these isolates were not fully molecular characterized [13, 30]. In a large study, carried out in two distinct localities in South China, with 785 infected or colonized companion animals (612 dogs and 173 cats), it was found a total of 8.8% (69/785) of MRSP, which represented 47.9% (69/144) of the total S. pseudintermedius isolates [31]. It is impressive the high level of multiresistance showed by MRSP isolates. In fact, increased antibiotic resistance has recently been a tendency among MRSP isolates of the international clonal complex [12] and cross-transmission dog-owner of a multiresistant MRSP has already been reported [29].
All eight MRSS isolates, from both dogs and cats, clustered into the same PFGE clone type (Figure 1A; lanes 1–4) and generated amplification products for both DCS and mecI primers described in 2007 by Milheiriço et al. [32] (Figure 2). These data indicated that the genes around mec complex were similar to those of SCCmec II and III (type II-III). Interestingly, eight MRSS isolates detected in Japan, which were nontypeable by the current classification for MRSA or by SCCmec sequencing, had the novel mec type II-III [16]. Dispite the fact that MRSS isolates were the most frequent MRS colonizing both cats and dogs in our study, S. saprophyticus have been isolated in low frequency from infected cats and dog [33]. It is well known that in young women, S. saprophyticus is a common agent of urinary tract infections [15], but still its relative importance is low compared with E. coli[34]. In a study conducted during one year in Sweden, with women and men of all ages, growth of S. saprophyticus was a quite uncommon finding [34]. The majority of individuals with S. saprophyticus were women between 15–29 years old (63.8%). However it is important to mention that, in this group, S. saprophyticus constituted 12.5% of all urinary tract pathogens [34]. It is also of significance that although still rare, isolates of MRSS have already been reported infecting humans in few countries as Canada [35], USA [36], Japan [16], Sweden [17], and also Brazil [37].
The PFGE type of a representative of S. conhii ssp. urealyticus (S. conhii-urea) is presented in Figure 1B (lane 2). Similarly to the MRSP isolate, S. conhii-urea isolates could not be typed for SCCmec with the type system used. Although very rare and generally considered a commensal bacteria, the presence of mecA has already been described in S. conhii either from human or animal origins [38].
Conclusions
This work demonstrates that mecA-harboring Staphylococcus isolates are common members of the nasal microbiota of the companion animals examined. Some bacterial species recovered are involved not only in animals but also in human infections. Moreover, the presence of USA1100-related MRSA colonizing animals is of concern. ST30 isolates are important CA-MRSA pathogens worldwide spread that has been associated to serious invasive diseases in adult and children in this country [39]. S. pseudintermedius is the most prevalent pathogenic streptococci in companion animals. To the best of our knowledge, that is the first report recognizing the prevalence of the European clone of MRSP (ST71) and of the USA1100 clone of MRSA (ST30) colonizing animals in Brazil. Both CA-MRSA and MRSP have already been incriminated in potential zoonotic diseases, reflecting the recent emergency of these important multidrug-resistant pathogens in animals.
The fact that most of mecA-positive isolates were S. saprophyticus also poses an important question concerning potential transfer of MRSS between animals and humans. Finally, our data indicate that continued surveillance studies involving fully identification and genotyping of Staphylococcus isolates should be carried out in this and other countries for monitoring the spread of MRS among companion animals.
Methods
Sample collection
Hundred and thirty nasal swabs were obtained from 70 dogs and 60 cats which were visiting a veterinarian clinic, located in Rio de Janeiro city, RJ, for pet grooming, during the year of 2010. The animals included in this study were healthy and free of hospitalization, antimicrobial therapy and invasive device in the six-month before sample collections. The swabs were collected from the anterior nares of the animals, using a transport system (Culture Swab Transport System; Copan Italia SpA.; Brescia Italy), and stored at 4°C for no more than 24 h. Written informed consent for participation was obtained from the owners of all animals included in the study. The study was approved (# IMPPG030) by the Animal Ethics Committee from Centro de Ciências da Saúde, Universidade Federal do Rio de Janeiro.
Bacterial identification
In the laboratory, the swab was introduced in an enrichment broth constituted of tryptic soy broth (TSB, BD; Becton, Dickinson and Company, BD; Becton DriveFranklin Lakes, NJ, USA) supplemented with 10 μg/mL methicillin and 7.5% of NaCl [40]. After incubation at 37°C for 18 h-48 h, aliquots of the turbid cultures were streaked on mannitol salt agar (BD; Becton, Dickinson and Company, BD; Becton Drive Franklin Lakes, NJ, USA) and incubated at 37°C for 18 h-24 h. Tube coagulase test was carried out for Gram-positive and catalase-positive colonies. Both coagulase-positive and -negative staphylococci were identified using Microscan WalkAway 96 SI automated system (Siemens Healthcare Diagnostic, West Sacramento, CA, USA) using Pos Combo Panel Type 33 (PC33) for bacterial identification and susceptibility tests. PCR amplification using the set of primers pse-F2 TRGGCAGTAGGATTCGTTAA and pse-R5 CTTTTGTGCTYCMTTTTGG was carried out for molecular identification of S. pseudintermedius, as previously described [41].
Antimicrobial susceptibility testing
Antibiograms were also carried out following the Clinical Laboratory Standard Institute (CLSI) [42] recommendations for the following antimicrobial disks: cefoxitin (30 μg; CEF) ciprofloxacin (5 μg; CIP), clindamycin (2 μg; CLI), chloramphenicol (30 μg; CHL), erythromycin (15 μg; ERY), gentamicin (10 μg; GEN), penicillin G (10 UI; PEN), rifampicin (5 μg; RIF), sulfamethoxazole-trimethoprim (25 μg; TMP), teicoplanin (30 μg; TEC) and tetracycline (30 μg; TET). The minimum inhibitory concentration (MIC) was determined for vancomycin (Sigma-Aldrich, St. Louis, MO, USA) according to CLSI guidelines [42]. The mecA was tested by polymerase chain reaction (PCR)-amplification of an internal fragment of the gene [43]. Staphylococcus aureus ATCC 25923 and S. aureus ATCC 29213 were used for quality control purposes.
Bacterial genotyping
SCCmec typing was carried out for all isolates using different typing systems [32, 43–46]. Pulsed-field gel electrophoresis (PFGE) of the Sma I-fragmented DNA was carried out for all isolates recovered, according to Teixeira et al. [47]. Multilocus sequence typing (MLST) was carried out as recommended for MRSA [48] and S. pseudintermedius[49] isolates. To assign the sequence types the allele sequences were correctly trimmed and submitted to the S. aureus MLST database (http://www.mlst.net), for the CA-MRSA BMBSA87, or to the MLST website (http://pubmlst.org/spseudintermedius/) sited at the University of Oxford for S. pseudintermedius[50].
Detection of virulence genes
The gene lukF-PV and those encoding for enterotoxins G, I, N and O of the egc cluster (seg, sei, sen and seo) were assessed by PCR, as described previously [20]. Additionally, another set of primers was used to test the genes lukSF-PV[21]. The pmsα 3 was tested with the primers described by Li et al. [25].
Abbreviations
- CA-MRSA:
-
Community-acquired methicillin-resistant Staphylococcus aureus
- CEF:
-
Cefoxitin
- CIP:
-
Ciprofloxacin
- CHL:
-
Chloramphenicol
- CLI:
-
Clindamycin
- CLSI:
-
Clinical Laboratory Standard Institute
- CoNS:
-
Coagulase-negative staphylococci
- egc:
-
Enterotoxin gene cluster
- ERY:
-
Erythromycin
- GEN:
-
Gentamicin
- MLST:
-
Multilocus locus sequence typing
- MRS:
-
Methicillin-resistant staphylococci (MRS)
- MRSA:
-
Methicillin-resistant Staphylococcus aureus
- MRCoNS:
-
Methicillin-resistant coagulase negative staphylococci
- MRSC:
-
Methicillin-resistant Staphylococcus conhii ssp. urealyticus
- MRSP:
-
Methicillin-resistant Staphylococcus pseudintermedius
- MRSS:
-
Methicillin-resistant Staphylococcus saprophyticus
- PCR:
-
Polymerase chain reaction
- PEN:
-
Penicillin G
- PFGE:
-
Pulsed-field gel electrophoresis
- PVL:
-
Panton-Valentine leukocidin
- RIF:
-
Rifampicin
- SCCmec:
-
Staphylococcal cassette chromosome mec
- ST:
-
Sequence Type
- SSTI:
-
Skin/soft tissue infections
- TEC:
-
Teicoplanin
- TET:
-
Tetracycline 30 μg
- TMP:
-
Sulfamethoxazole-trimethoprim
- TSB:
-
tryptic soy broth
- UTI:
-
Urinary tract infections
- WA-1:
-
Western-Austrália 1.
References
Pearman JW, Christiansen KJ, Annear DI, Goodwin CS, Metcalf C, Donovan FP, Macey KL, Bassett LD, Powell IM, Green JM, Harper WE, Mckelvie MS: Control of methicillin-resistant Staphylococcus aureus (MRSA) in an Australian metropolitan teaching hospital complex. Med J Aust. 1985, 142: 103-108.
Udo EE, Pearman JW, Grubb WB: Genetic analysis of community isolates of methicillin-resistant S. aureus in Western Australia. J Hosp Infect. 1993, 25: 97-108. 10.1016/0195-6701(93)90100-E.
DeLeo FR, Otto M, Kreiswirth BN, Chambers HF: Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010, 375: 1557-1568. 10.1016/S0140-6736(09)61999-1.
Tenover FC, Tickler IA, Goering RV, Kreiswirth BN, Mediavilla JR, Persing DH, MRSA Consortium: Characterization of nasal and blood culture isolates of methicillin-resistant Staphylococcus aureus from patients in United States Hospitals. Antimicrob Agents Chemother. 2012, 56: 1324-1330. 10.1128/AAC.05804-11.
Mishaan AM, Mason EO, Martinez-Aguilar G, Hammerman W, Propst JJ, Lupski JR, Stankiewicz P, Kaplan SL, Hulten K: Emergence of a predominant clone of community-acquired Staphylococcus aureus among children in Houston, Texas. Pediatr Infect Dis J. 2005, 24: 201-296. 10.1097/01.inf.0000151107.29132.70.
David MZ, Rudolph KM, Hennessy TW, Boyle-Vavra S, Daum RS: Molecular epidemiology of methicillin-resistant Staphylococcus aureus, rural southwestern Alaska. Emerg Infect Dis. 2008, 14: 1693-1699. 10.3201/eid1411.080381.
Rozenbaum R, Sampaio MG, Batista GS, Garibald AM, Terra GM, Souza MJ, Vieira EN, Silva-Carvalho MC, Teixeira LA, Figueiredo AM: The first report in Brazil of severe infection caused by community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA). Braz J Med Biol Res. 2009, 42: 756-760. 10.1590/S0100-879X2009005000007.
Wan MT, Fu SY, Lo YP, Huang TM, Cheng MM, Chou CC: Heterogeneity and phylogenetic relationships of community-associated methicillin sensitive/resistant Staphylococcus aureus isolates in healthy dogs, cats and their owners. J Appl Microbiol. 2012, 112: 205-213. 10.1111/j.1365-2672.2011.05179.x.
Lin Y, Barker E, Kislow J, Kaldhone P, Stemper ME, Pantrangi M, Moore FM, Hall M, Fritsche TR, Novicki T, Foley SL, Shukla SK: Evidence of multiple virulence subtypes in nosocomial and community-associated MRSA genotypes in companion animals from the upper midwestern and northeastern United States. Clin Med Res. 2011, 9: 7-16. 10.3121/cmr.2010.944.
Broens EM, Espinosa-Gongora C, Graat EA, Vendrig N, Van Der Wolf PJ, Guardabassi L, Butaye P, Nielsen JP, De Jong MC, Van De Giessen AW: Longitudinal study on transmission of MRSA CC398 within pig herds. BMC Vet Res. 2012, 8: 58-10.1186/1746-6148-8-58.
Perreten V, Kadlec K, Schwarz S, Andersson UG, Finn M, Greko C, Moodley A, Kania SA, Frank LA, Bemis DA, Franco A, Iurescia M, Battisti A, Duim B, Wagenaar JA, van Duijkeren E, Weese JS, Fitzgerald JR, Rossano A, Guardabassi L: Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J Antimicrob Chemother. 2010, 65: 1145-1154. 10.1093/jac/dkq078.
Ruscher C, Lübke-Becker A, Semmler T, Wleklinski-Claus G, Paasch A, Soba A, Stamm I, Kopp P, Wieler LH, Walther B: Multidrug-resistant Staphylococcus pseudintermedius (MRSP) genetic lineage in Europe. Vet Microbiol. 2010, 144: 340-346. 10.1016/j.vetmic.2010.01.008.
Penna B, Varges R, Martins R, Martins G, Lilenbaum W: In vitro antimicrobial resistance of staphylococci isolated from canine urinary tract infection. Can Vet J. 2010, 51: 738-742.
Bagcigil FA, Moodley A, Baptiste KE, Jensen VF, Guardabassi L: Occurrence, species distribution, antimicrobial resistance and clonality of methicillin- and erythromycin-resistant staphylococci in the nasal cavity of domestic animals. Vet Microbiol. 2007, 121: 307-15. 10.1016/j.vetmic.2006.12.007.
Palou J, Pigrau C, Molina I, Ledesma JM, Angulo J, Grupo Colaborador Español del Estudio ARESC: Etiology and sensitivity of uropathogens identified in uncomplicated lower urinary tract infections in women (ARESC Study): implications on empiric therapy. Med Clin. 2011, 136: 1-7.
Higashide M, Kuroda M, Omura CT, Kumano M, Ohkawa S, Ichimura S, Ohta T: Methicillin-resistant Staphylococcus saprophyticus isolates carrying staphylococcal cassette chromosome mec have emerged in urogenital tract infections. Antimicrob Agents Chemother. 2008, 52: 2061-2068. 10.1128/AAC.01150-07.
Söderquist B, Berglund C: Methicillin-resistant Staphylococcus saprophyticus in Sweden carries various types of staphylococcal cassette chromosome mec (SCCmec). Clin Microbiol Infect. 2009, 15: 1176-1178. 10.1111/j.1469-0691.2009.02771.x.
Svensson K, Hellmark B, Söderquist B: Characterization of SCCmec elements in methicillin-resistant Staphylococcus epidermidis isolated from blood cultures from neonates during three decades. APMIS. 2011, 119: 885-93. 10.1111/j.1600-0463.2011.02801.x.
Sasaki T, Kikuchi K, Tanaka Y, Takahashi N, Kamata S, Hiramatsu K: Reclassification of phenotypically identified Staphylococcus intermedius strains. J Clin Microbiol. 2007, 45: 2770-2778. 10.1128/JCM.00360-07.
Ribeiro A, Dias C, Silva-Carvalho MC, Berquó L, Ferreira FA, Santos RN, Ferreira-Carvalho BT, Figueiredo AM: First report of infection with community-acquired methicillin-resistant Staphylococcus aureus in South America. J Clin Microbiol. 2005, 43: 1985-1988. 10.1128/JCM.43.4.1985-1988.2005.
Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, Nesme X, Etienne J, Vandenesch F: Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002, 70: 631-641. 10.1128/IAI.70.2.631-641.2002.
Ribeiro A, Coronado AZ, Silva-Carvalho MC, Ferreira-Carvalho BT, Dias C, Rozenbaum R, Del Peloso PF, da Costa Ferreira Leite C, Teixeira LA, Figueiredo AM: Detection and characterization of international community-acquired infections by methicillin-resistant Staphylococcus aureus clones in Rio de Janeiro and Porto Alegre cities causing both community- and hospital-associated diseases. Diagn Microbiol Infect Dis. 2007, 59: 339-345. 10.1016/j.diagmicrobio.2007.05.007.
Srinivasan A, Seifried S, Zhu L, Bitar W, Srivastava DK, Shenep JL, Bankowski MJ, Flynn PM, Hayden RT: Short communication: methicillin-resistant Staphylococcus aureus infections in children and young adults infected with HIV. AIDS Res Hum Retroviruses. 2009, 25: 1219-1224. 10.1089/aid.2009.0040.
Li M, Cheung GY, Hu J, Wang D, Joo H-S, DeLeo FR, Otto M: Comparative analysis of virulence and toxin expression of global community-associated MRSA strains. J Infect Dis. 2010, 202: 1866-1876. 10.1086/657419.
Jarraud S, Peyrat MA, Lim A, Tristan A, Bes M, Mougel C, Etienne J, Vandenesch F, Bonneville M, Lina G: egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J Immunol. 2001, 166: 669-677.
Boost MV, Donoghue MMO, James A: Prevalence of Staphylococcus aureus carriage among dogs and their owners. Epidemiol Infect. 2008, 136: 953-964.
Loeffler A, Pfeiffer DU, Lindsay JA, Magalhães RJ, Lloyd DH: Prevalence of and risk factors for MRSA carriage in companion animals: a survey of dogs, cats and horses. Epidemiol Infect. 2010, 14: 1-10.
Lamaro-Cardoso J, de Lencastre H, Kipnis A, Pimenta FC, Oliveira LS, Oliveira RM, Nouer SS, Aires-de-Sousa M, Milheiriço C, Andrade AL: Molecular epidemiology and risk factors for nasal carriage of staphylococcus aureus and methicillin-resistant S. aureus in infants attending day care centers in Brazil. J Clin Microbiol. 2009, 47: 3991-3997. 10.1128/JCM.01322-09.
Soedarmanto I, Kanbar T, Ülbegi-Mohyla H, Hijazin M, Alber J, Lämmler C, Akineden Ö, Weiss R, Moritz A, Zschöck M: Genetic relatedness of methicillin-resistant Staphylococcus pseudintermedius (MRSP) isolated from a dog and the dog owner. Res Vet Sci. 2011, 91: 25-27. 10.1016/j.rvsc.2010.07.025.
Penna B, Varges R, Medeiros L, Martins GM, Martins RR, Lilenbaum W: Species distribution and antimicrobial susceptibility of staphylococci isolated from canine otitis externa. Vet Dermatol. 2010, 2: 292-296.
Feng Y, Tian W, Lin D, Luo Q, Zhou Y, Yang T, Deng Y, Liu YH, Liu JH: Prevalence and characterization of methicillin-resistant Staphylococcus pseudintermedius in pets from South China. Vet Microbiol. 2012, 160: 517-524. 10.1016/j.vetmic.2012.06.015.
Milheiriço C, Oliveira DC, de Lencastre H: Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother. 2007, 51: 3374-3377. 10.1128/AAC.00275-07.
Hauschild T, Wójcik A: Species distribution and properties of staphylococci from canine dermatitis. Res Vet Sci. 2007, 82: 1-6. 10.1016/j.rvsc.2006.04.004.
Eriksson A, Giske CG, Ternhag A: The relative importance of Staphylococcus saprophyticus as a urinary tract pathogen: distribution of bacteria among urinary samples analysed during 1 year at a major Swedish laboratory. APMIS. 2013, 121: 72-78. 10.1111/j.1600-0463.2012.02937.x.
Hussain Z, Stoakes L, Massey V, Diagre D, Fitzgerald V, El Sayed S, Lannigan R: Correlation of oxacillin MIC with mecA gene carriage in coagulase-negative staphylococci. J Clin Microbiol. 2000, 38: 752-754.
Swenson JM, Tenover FC: Cefoxitin disk study group. Results of disk diffusion testing with cefoxitin correlate with presence of mecA in Staphylococcus spp. J Clin Microbiol. 2005, 43: 3818-3823. 10.1128/JCM.43.8.3818-3823.2005.
Souza Antunes AL, Secchi C, Reiter KC, Rodrigues Perez LR, Peixoto de Freitas AL, Alves d’Azevedo P: Evaluation of oxacillin and cefoxitin disks for detection of resistance in coagulase negative staphylococci. Mem Inst Oswaldo Cruz. 2007, 102: 719-723. 10.1590/S0074-02762007005000078.
Zong Z, Lü X: Characterization of a new SCCmec element in Staphylococcus cohnii. PLoS One. 2010, 5: e14016-10.1371/journal.pone.0014016.
de Araújo BE, Borchert JM, Manhães PG, Ferreira FA, Ramundo MS, Silva-Carvalho MC, Seabra AC, Victal SH, Sá Figueiredo AM: A rare case of pyomyositis complicated by compartment syndrome caused by ST30-staphylococcal cassette chromosome mec type IV methicillin-resistant Staphylococcus aureus. Am J Emerg Med. 2010, 28: 537.e3-6-
Silva FR, Mattos EM, Coimbra MV, Ferreira-Carvalho BT, Figueiredo AM: Isolation and molecular characterization of methicillin-resistant coagulase-negative staphylococci from nasal flora of healthy humans at three community institutions in Rio de Janeiro City. Epidemiol Infect. 2001, 127: 57-62.
Sasaki T, Tsubakishita S, Tanaka Y, Sakusabe A, Ohtsuka M, Hirotaki S, Kawakami T, Fukata T, Hiramatsu K: Multiplex-PCR method for species identification of coagulase-positive staphylococci. J Clin Microbiol. 2010, 48: 765-769. 10.1128/JCM.01232-09.
CLSI: Performance standards for antimicrobial disk and dilution susceptibility testing; twenty-first informational supplement. M100-S21, Vol. 31 No. 1. 2011, PA, USA: Clinical and Laboratory Standards Institute, Wayne
Oliveira DC, de Lencastre H: Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002, 46: 2155-2161. 10.1128/AAC.46.7.2155-2161.2002.
Boye K, Bartels MD, Andersen IS, Moller JA Westh H: A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I-V. Clin Microbiol Infect. 2007, 13: 725-727. 10.1111/j.1469-0691.2007.01720.x.
Berglund C, Ito T, Ikeda M, Ma XX, Söderquist B, Hiramatsu K: Novel type of staphylococcal cassette chromosome mec in a methicillin-resistant Staphylococcus aureus strain isolated in Sweden. Antimicrob Agents Chemother. 2008, 52: 3512-3516. 10.1128/AAC.00087-08.
Zhang K, McClure JA, Elsayed S, Conly JM: Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009, 53: 531-540. 10.1128/AAC.01118-08.
Teixeira LA, Resende CA, Ormonde LR, Rosenbaum R, Figueiredo AM, de Lencastre H, Tomasz A: Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J Clin Microbiol. 1995, 33: 2400-2404.
Enright MC, Day NPJ, Davies CE, Peacock SJ, Spratt BG: Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000, 38: 1008-1015.
Solyman SM, Black CC, Duim B, Perreten V, van Duijkeren E, Wagenaar JA, Eberlein LC, Sadeghi LN, Videla R, Bemis DA, Kania SA: Multilocus sequence typing for characterization of Staphylococcus pseudintermedius. J Clin Microbiol. 2013, 51: 306-310. 10.1128/JCM.02421-12.
Jolley KA, Maiden MC, BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010, 11: 595-10.1186/1471-2105-11-595.
Acknowledgments
This work was supported in part by grants of Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação Carlos Chagas de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and European Commission’s Seventh Framework Programme (FP7), through the Marie Curie International Research Staff Exchange Scheme NANO_GUARD (PIRSES-GA-2010-269138).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
All authors declare that they have no competing interest.
Authors’ contributions
IMZQ, MSR, MCSC and LCR carried out the bacterial isolation, identification and participate in the majority of the experiments of this paper. COB and RRS carried out PFGE analysis. TFO has been involved in the study design and in the draft of the manuscript. RO was responsible for examining and selecting healthy animals, for collecting bacterial specimens and also for revising critically the manuscript with important intellectual contributions. PDP carried out experiments for identification and speciation of the coagulase-negative isolates. AMSF has been involved in the study design and in the revision of the manuscript critically for final approval of this version. All authors read and approved the final manuscript.
Isidório Mebinda Zuco Quitoco, Mariana Severo Ramundo contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Quitoco, I.M.Z., Ramundo, M.S., Silva-Carvalho, M.C. et al. First report in South America of companion animal colonization by the USA1100 clone of community-acquired meticillin-resistant Staphylococcus aureus (ST30) and by the European clone of methicillin-resistant Staphylococcus pseudintermedius (ST71). BMC Res Notes 6, 336 (2013). https://doi.org/10.1186/1756-0500-6-336
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-0500-6-336