Abstract
Background
The Ay allele at the agouti locus causes obesity and promotes linear growth in mice. The effect of the Ay allele on obesity has been extensively investigated, whereas its effect on body length is only poorly analyzed. To gain insight into the genetic control of body length, quantitative trait locus (QTL) analysis was performed in F2 female mice produced by crossing C57BL/6 J females and DDD.Cg-Ay males. A congenic DDD.Cg-Ay strain was established by introgressing the Ay allele from the B6.Cg-Ay strain by backcrossing for 12 generations. DDD.Cg-Ay females were longer than B6.Cg-Ay females; therefore, QTLs that interact with the Ay allele may be identified for body length. In addition, QTL analysis was also performed for plasma insulin-like growth factor 1 (IGF1) levels because IGF1 is known to play essential roles in growth and development. If QTLs for IGF1 levels coincide with those for body length, we can gain endocrinological insight into the QTLs for body length.
Results
Correlations between body length and IGF1 levels were statistically significant in F2 populations. For body length, two significant QTLs were identified on chromosomes 15 and 17. For IGF1 levels, three significant QTLs were identified on chromosomes 10, 12, and 19. QTLs on chromosomes 12 and 19 appeared to be novel, and the latter interacted with the Ay allele.
Conclusion
QTLs for body length and IGF1 levels contained candidate genes that were components of the growth hormone/insulin-like growth factor axis. However, there was no overlap between QTLs for these two traits. Contrary to our expectations, QTLs that interacted with the Ay allele were identified not for body length but for IGF1 levels. Body length and IGF1 levels were, thus, controlled by different sets of genes.
Similar content being viewed by others
Background
Traditionally, five single gene obesity mutations, Cpefat, Tubtub, Lepob, Leprdb, and Ay, have been identified in mice[1]. Among the five mutations, only the Ay allele is dominant and homozygous lethal; therefore, living Ay mice are invariably heterozygotes. Obesity in Ay mice is moderate and occurs late compared with that in the other four mutants. The Ay allele is known not only to cause obesity but also to promote linear growth[2].
In normal mice, the agouti gene is expressed only in the skin[3, 4], and it regulates pigmentation by serving as an inverse agonist of the melanocortin 1 receptor[5, 6]. However, in Ay mice, the Ay allele is associated with a large deletion, causing agouti gene expression to be aberrantly controlled by the unrelated Raly gene promoter and leading to its ectopic overexpression[4, 7–9]. As a result, Ay mice have a yellow coat color and develop maturity onset obesity. Obesity in Ay mice is believed to be a consequence of the agouti protein serving as a constitutive antagonist of the melanocortin 3 receptor (MC3R) and melanocortin 4 receptor (MC4R) by mimicking the action of the agouti-related protein[10–12].
Two mouse strains congenic for the Ay allele are available to date: B6.Cg-Ay (C57BL/6 J background, hereafter B6-Ay) and KK.Cg-Ay (KK/Ta background, hereafter KK-Ay) strains. We developed a novel strain congenic for the Ay allele in an inbred DDD/Sgn (hereafter DDD) strain background, i.e., DDD.Cg-Ay (hereafter DDD-Ay) strain[13]. DDD-Ay females are characterized by their massive obesity compared with KK-Ay and B6-Ay females[14], i.e., the average body weight at 16 weeks was 54.2 g in DDD-Ay, 52.2 g in KK-Ay, and 38.5 g in B6-Ay. Although KK-Ay and B6-Ay females did not weigh more than 60 g (body weights were measured by 29 weeks), DDD-Ay females weighed more than 60 g at 19 weeks and older and some weighed more than 70 g by 22 weeks. The magnitude of phenotypic effect of the Ay allele was thus strongly influenced by the genetic background.
To determine the genetic basis of obesity in DDD-Ay mice and to determine whether or not their high body weight was because of the presence of DDD background-specific modifiers, quantitative trait locus (QTL) analyses for body weight and obesity (defined by body mass index, BMI) were previously performed in two types of F2 female mice [F2Ay (F2 mice with the Ay allele) and F2 non- Ay mice (F2 mice without the Ay allele)] produced by crossing C57BL/6 J females and DDD-Ay males[14]. The presence of DDD background-specific modifiers was not confirmed, and a multifactorial basis for obesity in DDD-Ay females was revealed.
In this study, the genetic basis of body length was analyzed in the same F2 population. In addition to the results of the analysis of body weight, we will gain insight into the genetic control of body size because body length also serves as a representative body size parameter. Furthermore, the Ay allele is known not only to cause obesity but also to promote linear growth[2]. The effect of the Ay allele on body weight has been extensively investigated, whereas its effect on body length is only poorly analyzed[15, 16]. As with obesity, the effect of the Ay allele on body length was considered to be mediated by the melanocortin 4 receptor[11]. DDD.Cg-Ay strain was longer than B6.Cg-Ay strain; therefore, DDD background-specific modifiers may be identified for body length.
In addition, QTL analysis was also performed for plasma insulin-like growth factor 1 (IGF1) levels because QTLs identified for body length contained candidate genes that were components of the growth hormone/insulin-like growth factor axis and because IGF1 is known to play essential roles in growth and development[17–21]. Thus, we hypothesized that some QTLs for IGF1 levels will be involved in the control of body length. If QTLs for IGF1 levels coincide with those for body length, we can gain endocrinological insight into the QTLs for body length.
Results
Body length and plasma IGF1 levels in parental, F1, and F2 female mice are summarized in Table1. In parental female mice, Ay mice were significantly longer and had significantly higher IGF1 levels than non-Ay mice. Furthermore, DDD-Ay females were significantly longer and had significantly higher IGF1 levels than B6-Ay females and DDD females were significantly longer and had significantly higher IGF1 levels than B6 females. In F2 females, Ay mice were significantly longer than non-Ay mice, but IGF1 levels did not significantly differ between Ay and non-Ay mice.
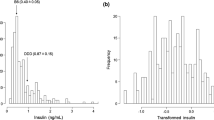
Histograms showing the distribution of body length in F2 females are shown in Figure1 [(A) F2 non-Ay females, n = 137, (B) F2Ay females, n = 139]. Mean ± S.E. body length was significantly larger in F2Ay females (99.73 ± 0.22 mm) than F2 non-Ay females (95.42 ± 0.27 mm) (P < 5.2 × 10–28). In combined F2 females (F2 non-Ay plus F2Ay), body length was normally distributed. Histograms showing the distribution of IGF1 levels in F2 females are shown in Figure2 [(A) F2 non-Ay, n = 148, (B) F2Ay females, n = 150]. Mean ± S.E. IGF1 levels did not significantly differ between F2 non-Ay (459.5 ± 5.8 ng/dl) and F2Ay (445.6 ± 5.8 ng/dl) (P > 0.09) females. In combined F2 females, IGF1 levels were not normally distributed; therefore, IGF1 levels were normalized by Box-Cox transformation. Compared to a relatively high correlation between body length and body weight, correlations between body weight and IGF1 levels and between body length and IGF1 levels were modest albeit all correlations were statistically significant in both the F2 non-Ay and F2Ay females (Table2).
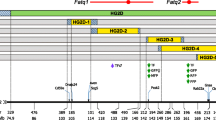
Because the Ay allele had a large phenotypic effect on IGF1 levels, the agouti locus genotype (non-Ay or Ay) was included as an additive covariate in the following QTL mapping analyses. For body length, two significant QTLs were identified on chromosomes 15 and 17, and two suggestive QTLs were identified on chromosomes 6 and 11 (Table3 and Figure3). We assigned the gene symbols Blndq1 (body length in DDD QTL no. 1) and Blndq2 to the significant QTLs. The DDD allele was associated with increased body length at the Blndq1(Figure4A), whereas the DDD allele was associated with decreased body length at the blndq2 (Figure4B). There were no significant pair-wise interactions.
LOD score plots for body length and IGF1 levels by single QTL scan with the agouti locus genotype as an additive covariate. The x-axis shows the chromosome numbers and the y-axis shows the LOD scores at these locations. Blue lines indicate the LOD scores for body length, and red lines indicate the LOD scores for IGF1 levels. Horizontal dashed lines (color-coded by each trait) indicate significant threshold LOD scores determined by 1,000 permutations.
Allelic contributions to QTLs for body length on (A) chromosomes 15 and (B) 17, and those to QTLs for IGF1 levels on (C) chromosomes 10, (D) 12, and (E) 19. Homozygous DDD alleles are represented by DDD/DDD, homozygous B6 alleles by B6/B6, and heterozygous alleles by DDD/B6. The y-axes show the mean trait values and the error bars show SE.
Because the difference between the LOD score with the agouti locus genotype as an interactive covariate and the LOD score with the agouti locus genotype as an additive covariate concerns the test of the QTL × agouti locus genotype interaction, this was performed. However, there were no significant QTLs that interacted with the Ay allele.
For IGF1 levels, two significant QTLs were identified on chromosomes 10 and 12, and four suggestive QTLs were identified on chromosomes 8, 13, 14, and 19 (Table3 and Figure3). We assigned the gene symbols Igfdq1 (IGF1 levels in DDD QTL no. 1) and Igfdq2 to the significant QTLs. The DDD allele was associated with increased IGF1 levels at the Igfdq1 (Figure4C), whereas the heterozygous genotype was associated with increased IGF1 levels at the Igfdq2 (Figure4D). There were no significant pair-wise interactions.
When the QTL × agouti locus genotype interaction was tested, one significant interacting QTL was identified on chromosome 19 (Table4 and Figure5). We assigned the gene symbol Igfdq3 to this locus. Indeed, a direction of allele effect of this QTL differed between F2 non-Ay and F2Ay females (Figure4E). Thus, Igfdq3 interacted with the Ay allele.
Interaction LOD score plots for the IGF1 levels. A horizontal dashed line indicates significant threshold LOD scores determined by 1,000 permutations. Solid lines denote the LOD scores when the agouti genotypes (non-Ay or Ay) were included as covariates. Horizontal dashed lines indicate significant threshold LOD scores determined by 1,000 permutations.
Discussion
Blndq1 (chromosome 15) and Blndq2 (chromosome 17) are major determinants of body length. These QTLs did not interact with the Ay allele, and the allele effect of these QTLs was in the same direction in F2 non-Ay and F2Ay mice. When the agouti locus genotype was included as a covariate in the analysis, no significant QTL × covariate interactions were identified. When the same analysis was applied to the body weight data, which was previously analyzed, no significant QTL × covariate interactions were identified. Thus, although DDD-Ay females were heavier and longer than B6-Ay mice, we could not identify any QTLs that interacted with the Ay allele. The effect of the Ay allele on size is probably rather complex in its physiologic mechanism of action. Indeed, we previously observed that the Ay allele decreased the size of the mandibular bone and the testis weight significantly[13, 22].
In agreement with the results by Reed et al.[23], in which the correlation coefficient between body weight and body length in a mouse F2 intercross was 0.67, the correlation coefficient between weight and body length was similarly high in F2 non-Ay and F2Ay mice in this study. Blndq2 colocalized with Bwdq3, which was identified for body weight in a previous study using the same F2 intercross[14]. At both the QTLs, the DDD allele was associated with decreased trait values, which suggests that these QTLs may be allelic and play a role in determining overall body size. Also, Blndq1 colocalized with a suggestive QTL for body weight. A suggestive QTL for body length identified on chromosome 6 overlapped with a significant QTL for body weight, Bwdq2. Thus, there were some overlaps between QTLs for body length and body weight.
Although there are only a limited number of studies on body length as compared with studies on body weight, body length QTLs have been reported by others. Among the results of such studies, QTLs for body length have been mapped to chromosome 2 several times. Reed et al.[23] identified significant QTLs on chromosome 2 (Bdln3), near the agouti locus, in an F2 intercross between 129P3/J and C57BL/6ByJ. Farber and Medrano[24] identified a significant QTL (Bdlnq7) on distal chromosome 2 in an F2 intercross between the CAST/EiJ and B6-hg/hg strains. Chiu et al.[25] presented evidence that a significant QTL was present on distal chromosome 2 using subcongenic strains for obesity. Masinde et al.[19] identified Lgth1 and Lgth2 on chromosome 2 in an F2 intercross between the MRL/MPJ and SJL/J strains. Apparently, the agouti locus is one of the plausible candidate genes for these QTLs.
Body length QTLs were also reported for other chromosomes. Reed et al.[23] identified Bdln6, which was significant only for males, on chromosome 4 in the same F2 intercross described previously. Masinde et al.[19, 20] identified Lgth3 on chromosome 4, Lgth4 and Lgth5 on chromosome 9, Lgth6 on chromosome 11, Lgth7 and Lgth8 on chromosome 13, and Lgth9 on chromosome 17 in the same F2 intercross described previously. The 95% CI for Lgth6 overlapped with that for suggestive QTL on chromosome 11. Similarly, Lgth9 overlaps with Blndq2, which suggests that these loci are allelic.
It is important to note that more than a few of these QTLs for body length have candidate genes that were contained in growth hormone/insulin-like growth factor axis. Indeed, the 95% CI for Blndq1 on the proximal part of chromosome 15 contains growth hormone receptor (Ghr) locus and the 95% CI for Blndq2 on proximal chromosome 17 contains insulin-like growth factor 2 receptor (Igf2r) as candidate genes. Therefore, we next performed QTL analysis on IGF1 levels.
Surprisingly, there was no overlap between QTLs for body length and IGF1 levels. This suggests that genetic variations affecting IGF1 levels did not have substantial effects on body length. Igfdq1 (chromosome 10) and Igfdq2 (chromosome 12) are primary determinants of IGF1 levels. Igfdq1 and Igfdq2 did not interact with the Ay allele, and the allele effect of these QTLs was in the same direction in F2 non-Ay and F2Ay mice. In contrast, Igfdq3 interacted with the Ay allele. The 95% CI for Igfdq3 contains Kazal-type serine peptidase inhibitor domain 1 (Kazald1), which is also known as Igfbp-rp10, as a candidate gene. Kazald1 is shown to promote proliferation of osteoblasts during bone formation and bone regeneration[26].
To date, four other studies have addressed blood (plasma or serum) IGF1 levels in mouse intercrosses[27–30]. Brockmann et al.[27] identified two significant QTLs on chromosomes 10 and 18 in an F2 intercross between Du6i and DBA/2 mice. They reported that chromosomal regions harboring these QTLs did not show any linkage to body, muscle, or fat weight. Rosen et al.[28] identified three significant QTLs on chromosomes 6, 10, and 15 in an F2 intercross between C3H/HeJ and B6. Harper et al.[29] identified five significant QTLs on chromosomes 1, 3, 8, 10, and 17 in genetically heterogeneous mice. Leduc et al.[30] identified four significant QTLs on chromosomes 9, 10, 15, and 17 in F2 mice between MRL/MpJ and SM/J. Thus, Igfdq2 (chromosome 12) and Igfdq3 (chromosome 19), identified in this study, are considered to be novel QTLs for blood IGF1 levels. In particular, Igfdq3 was shown to interact with the Ay allele. Most importantly, all five studies, including the present study, identified a significant QTL on chromosome 10 at a position containing the Igf1 locus, and it was considered that the Igf1 itself was a plausible candidate gene for the QTL. To determine whether or not Igf1 is responsible for Igfdq2, further studies, including sequence and expression analyses, will be required[28, 30, 31].
Finally, we could not analyze the distal portion of chromosome 2 (surrounding the agouti locus) and the mid-part of chromosome 7 (surrounding tyrosinase locus) because these chromosomal regions in DDD-Ay strain were derived from B6-Ay strain. In particular, the allele at the distal chromosome 2 was biased toward the B6 allele in F2 non-Ay mice, whereas the allele was biased toward the DDD allele in F2Ay mice. Also, we could not sufficiently analyze the entire part of X chromosome due to the cross direction of parental strains. Therefore, we cannot deny a possibility that there are additional QTLs in these chromosomal regions. Most importantly, because the present study was conducted in females, the QTL effect may not necessarily be confirmed in males. We are currently establishing QTL congenic strains to confirm the presence and effect of the QTLs in both sexes.
Conclusion
In summary, QTLs for body length and IGF1 levels contained candidate genes that were components of the growth hormone/insulin-like growth factor axis. However, there was no overlap between QTLs for each trait. Contrary to our expectations, a QTL that interacted with the Ay allele was identified not for body length but for IGF1 levels. Body length and IGF1 levels were thus controlled by different sets of genes.
Methods
Mice
The inbred mouse DDD strain (agouti locus genotype, A/A) and the congenic mouse DDD-Ay strain (Ay/A) were maintained at the National Institute of Agrobiological Sciences (NIAS). The inbred mouse C57BL/6 J strain (hereafter designated B6, a/a) was purchased from Clea Japan (Clea Japan Inc., Tokyo). The congenic mouse B6-Ay strain (Ay/a) was purchased from The Jackson Laboratory (Bar Harbor, ME, USA). The DDD-Ay strain was established by introgressing the Ay allele from the B6-Ay strain in the DDD strain by backcrossing for 12 generations[13]. Hereafter, both DDD-Ay and B6-Ay are referred to as “Ay mice.” Similarly, their control littermates, DDD and B6, are referred to as “non-Ay mice.”
DDD-Ay males were crossed with B6 females to produce the F1 generation, and F1Ay (Ay/a) mice were intercrossed with F1 non-Ay (A/a) mice to produce the F2 generation. F2 females were weaned at 4 weeks. The mice were housed in groups of 4–5 for 16 weeks.
All mice were maintained in a specific pathogen-free facility with a regular light–dark cycle (12 h light and 12 h dark) and controlled temperature (23 ± 1°C) and humidity (50%). Food (CRF-1; Oriental Yeast Co Ltd., Tokyo, Japan) and water were freely available throughout the experimental period. All animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of NIAS.
Phenotyping
At the age of 16 weeks, the body weight of the mice, fasted for 4 h, was determined using an electric balance to the nearest 0.01 g. The mice were euthanized with an overdose of ether. Whole blood was drawn from the heart into a plastic tube using heparin as an anticoagulant. Sample tubes were centrifuged at 7,000 rpm for 5 min at −4°C to separate the plasma. The plasma samples were maintained at −80°C until use. The IGF1 concentration was determined by ELISA (R&;D Systems, Inc., Minneapolis, MN55413, USA). The anal–nasal length of each mouse was measured by a pair of digital calipers to the nearest 0.01 mm just after the blood collection to avoid rigor mortis. Body length is defined as the anal–nasal length in this study.
Normality of the distribution of trait data for combined F2 females (F2 non-Ay plus F2Ay, n = 298) was tested using the Shapiro–Wilk W test (JMP 8.0.2, SAS Institute Japan, Tokyo, Japan). If the trait values did not follow a normal distribution, they were appropriately normalized using the Box–Cox transformation.
Genotyping and QTL analysis
Genomic DNA isolation and genotyping of microsatellite markers were performed according to the procedure described in our previous study[14]. Microsatellite markers used in this study were listed in the Table5 with their map position (cM) calculated using combined F2 females.
It should be noted that chromosome 7 is divided into two parts. Due to the introgression of the Tyr locus from the B6 strain, a mid-part of the DDD genome on chromosome 7 is replaced by a B6 genome in DDD-Ay mice. In this study, a region proximal to the B6 region was defined as “chromosome 7.1 (D7Mit250).” whereas a region distal to the B6 region was defined as “chromosome 7.2 (D7Mit362).”
Of a total of 298 F2 females, 148 were F2 non-Ay and 150 were F2Ay mice. QTL analysis was performed using R/qtl[33, 34]. Data for F2 non-Ay and F2Ay mice were combined and analyzed using the agouti locus genotype (i.e., Ay or non-Ay) as a covariate. Threshold logarithm of odds (LOD) scores for suggestive (P < 0.63) and significant (P < 0.05) linkages were determined by performing 1,000 permutations for each trait[35]. For significant QTLs, a 95% confidence interval (CI) was defined by a decline of 1.5 LOD. After these single QTL scans, pair-wise evaluations for potential interactions between loci were made. At this stage, threshold LOD scores were based strictly on those recommended by Broman in “A Brief Tour of R/qtl” (http://www.rqtl.org).
Other statistics
Statistical analysis for the two groups was performed using Student’s or Welch’s t-test, while that for more than two groups was performed using Tukey-Kramer HSD test (JMP 8, SAS Institute Inc., Cary, NC 27513, USA). Strength of association between body weight and body length was evaluated by using the Pearson product moment correlation coefficient. P < 0.05 was considered to be statistically significant.
References
Leibel RL, Chung WK, Chua SC: The molecular genetics of rodent single gene obesities. J Biol Chem. 1997, 272: 31937-31940. 10.1074/jbc.272.51.31937.
Heston WE, Vlahakis G: Influence of the Ay gene on mammary-gland tumors, hepatomas, and normal growth in mice. J Natl Cancer Inst. 1961, 26: 969-983.
Bultman SJ, Michaud EJ, Woychik RP: Molecular characterization of the mouse agouti locus. Cell. 1992, 71: 1195-1204. 10.1016/S0092-8674(05)80067-4.
Miller MW, Duhl DM, Vrieling H, Cordes SP, Ollmann MM, Winkes BM, Barsh GS: Cloning of the mouse agouti gene predicts a secreted protein ubiquitously expressed in mice carrying the lethal yellow mutation. Genes Dev. 1993, 7: 454-467. 10.1101/gad.7.3.454.
Robbins LS, Nadeau JH, Johnson KR, Kelly MA, Rosell-Rehfuss L, Baack E, Mountjoy KG, Cone RD: Pigmentation phenotypes of variant extension locus alleles result from point mutations that alter MSH receptor function. Cell. 1993, 26: 827-834.
Lu D, Willard D, Patel IR, Kadwell S, Overton L, Kost T, Luther M, Chen W, Woychik RP, Wilkinson WO, Cone RD: Agouti protein is an antagonist of the melanocyte-stimulating-hormone receptor. Nature. 1994, 371: 799-802. 10.1038/371799a0.
Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS: Neomorphic agouti mutations in obese yellow mice. Nat Genet. 1994, 8: 59-65. 10.1038/ng0994-59.
Michaud EJ, Bultman SJ, Stubbs LJ, Woychik RP: The embryonic lethality of homozygous lethal yellow mice (Ay/Ay) is associated with the disruption of a novel RNA-binding protein. Genes Dev. 1993, 7: 1203-1213. 10.1101/gad.7.7a.1203.
Michaud EJ, Bultman SJ, Klebig ML, van Vugt MJ, Stubbs LJ, Russell LB, Woychik RP: A molecular model for the genetic and phenotypic characteristics of the mouse lethal yellow (Ay) mutation. Proc Natl Acad Sci USA. 1994, 91: 2562-2566. 10.1073/pnas.91.7.2562.
Chen AS, Marsh DJ, Trumbauer ME, Frazier EG, Guan XM, Yu H, Rosenblum CI, Vongs A, Feng Y, Cao L, Metzger JM, Strack AM, Camacho RE, Mellin TN, Nunes CN, Min W, Fisher J, Gopal-Truter S, Maclntyre DE, Chen HY, Van der Ploeg LH: Inactivation of the mouse melanocortin-3 receptor results in increased fat mass and reduced lean body mass. Nat Genet. 2000, 26: 97-102. 10.1038/79254.
Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, Gu W, Kesterson RA, Boston BA, Cone RD, Smith FJ, Campfield LA, Burn P, Lee F: Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997, 88: 131-141. 10.1016/S0092-8674(00)81865-6.
Ollmann MM, Wilson BD, Yang YK, Kerns JA, Chen Y, Gantz I, Barsh GS: Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science. 1997, 278: 135-138. 10.1126/science.278.5335.135.
Suto J: The Ay allele at the agouti locus reduces the size and alters the shape of the mandible in mice. Proc Jpn Acad Ser B. 2009, 85: 248-257. 10.2183/pjab.85.248.
Suto J: Quantitative trait loci that control body weight and obesity in an F2 intercross between C57BL/6J and DDD.Cg-Ay mice. J Vet Med Sci. 2011, 73: 907-915. 10.1292/jvms.10-0573.
Martin NM, Houston PA, Patterson M, Sajedi A, Carmignac DF, Ghatei MA, Bloom SR, Small CJ: Abnormalities of the somatotrophic axis in the obese agouti mouse. Int J Obes. 2006, 30: 430-438. 10.1038/sj.ijo.0803076.
Martinelli CE, Keogh JM, Greenfield JR, Henning E, van der Klaauw AA, Blackwood A, O’Rahilly S, Roelfsema F, Camacho-Hübner C, Pijl H, Farooqi IS: Obesity due to melanocortin 4 receptor (MC4R) deficiency is associated with increased linear growth and final weight, fasting hyperglycemia, and incompletely suppressed growth hormone secretion. J Clin Endocrinol Metab. 2011, 96: E181-E188. 10.1210/jc.2010-1369.
Baker J, Liu JP, Robertson EJ, Efstratiadis A: Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993, 75: 73-82.
Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A: Mice carrying null mutations of the genes encoding insulin-like growth factor 1 (Igf-1) and type 1 IGF receptor (Igf1r). Cell. 1993, 75: 59-72.
Masinde GL, Li X, Gu W, Davidson H, Hamilton-Ulland M, Wergedal J, Mohan S, Baylink DJ: Quantitative trait loci (QTL) for lean body mass and body length in MRL/MPJ and SJL/J F2 mice. Funct Integr Genomics. 2002, 2: 98-104. 10.1007/s10142-002-0053-7.
Masinde GL, Li X, Gu W, Hamilton-Ulland M, Mohan S, Baylink DJ: Quantitative trait loci that harbor genes regulating muscle size in (MRL/MPJ × SJL/J) F2 mice. Funct Integr Genomics. 2002, 2: 120-125. 10.1007/s10142-002-0067-1.
Powell-Braxton L, Hollingshead P, Warburton C, Dowd M, Pitts-Meek S, Dalton D, Gillett N, Stewart TA: IGF-1 is required for normal embryonic growth in mice. Genes Dev. 1993, 7: 2609-2617. 10.1101/gad.7.12b.2609.
Suto J: Identification of multiple quantitative trait loci affecting the size and shape of the mandible in mice. Mamm Genome. 2009, 20: 1-13. 10.1007/s00335-008-9154-5.
Reed DR, Li X, McDaniel AH, Lu K, Li S, Tordoff MG, Price RA, Bachmanov AA: Loci on chromosomes 2, 4, 9, and 16 for body weight, body length, and adiposity identified in a genome scan of an F2 intercross between the 129P3/J and C57BL/6ByJ mouse strains. Mamm Genome. 2003, 14: 302-313. 10.1007/s00335-002-2170-y.
Farber CR, Medrano JF: Fine mapping reveals sex bias in quantitative trait loci affecting growth, skeletal size and obesity-related traits on mouse chromosomes 2 and 11. Genetics. 2007, 175: 349-360.
Chiu S, Kim K, Haus KA, Espinal GM, Millon LV, Warden CH: Identification of positional candidate genes for body weight and adiposity in subcongenic mice. Physiol Genomics. 2007, 31: 75-85. 10.1152/physiolgenomics.00267.2006.
Shibata Y, Tsukazaki T, Hirata K, Xin C, Yamaguchi A: Role of a new member of IGFBP superfamily, IGFBP-rP10, in proliferation and differentiation of osteoblastic cells. Biochem Biophys Res Commun. 2004, 325: 1194-1200. 10.1016/j.bbrc.2004.10.157.
Brockmann GA, Kratzsch J, Haley CS, Renne U, Schwerin M, Karle S: Single QTL effects, epistasis, and pleiotropy account for two-thirds of the phenotypic F2 variance of growth and obesity in Du6i × DBA/2 mice. Genome Res. 2000, 10: 1941-1957. 10.1101/gr.GR1499R.
Rosen CJ, Churchill GA, Donahue LR, Shultz KL, Burgess JK, Powell DR, Beamer WG: Mapping quantitative trait loci for serum insulin-like growth factor-1 levels in mice. Bone. 2000, 27: 521-528. 10.1016/S8756-3282(00)00354-9.
Harper JM, Galecki AT, Burke DT, Pinkosky SL, Miller RA: Quantitative trait loci for insulin-like growth factor 1, leptin, throxine, and corticosterone in genetically heterogeneous mice. Physiol Genomics. 2003, 15: 44-51.
Leduc MS, Hageman RS, Meng Q, Verdugo RA, Tsaih S-W, Churchill GA, Paigen B, Yuan R: Identification of genetic determinants of IGF-1 levels and longevity among mouse inbred strains. Aging Cell. 2010, 9: 823-836. 10.1111/j.1474-9726.2010.00612.x.
Adamo ML, Ma X, Ackert-Bicknell CL, Donahue LR, Beamer WG, Rosen CJ: Genetic increase in serum insulin-like growth factor-1 (IGF-1) in C3H/HeJ compared with C57BL/6 J mice is associated with increased transcription from the IGF-1 exon 2 promoter. Endocrinology. 2006, 147: 2944-2955. 10.1210/en.2005-0742.
Suto J, Matsuura S, Yamanaka H, Sekikawa K: Quantitative trait loci that regulate plasma lipid concentration in hereditary obese KK and KK-Ay mice. Biochim Biophys Acta. 1999, 1453: 385-395. 10.1016/S0925-4439(99)00013-7.
Broman KW, Sen Ś: A guide to QTL mapping with R/qtl. 2009, New York: Springer
Broman KW, Wu H, Sen Ś, Churchill GA: R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003, 19: 889-890. 10.1093/bioinformatics/btg112.
Lander E, Kruglyak L: Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995, 11: 241-247. 10.1038/ng1195-241.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author declares that he has no competing interests.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Suto, Ji. Quantitative trait locus mapping of genes that control body length and plasma insulin-like growth factor 1 level in mice. BMC Res Notes 5, 547 (2012). https://doi.org/10.1186/1756-0500-5-547
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-0500-5-547