Abstract
Background
An accurate gene expression quantification using TaqMan Arrays (TA) could be limited by the low RNA quantity obtained from some clinical samples. The novel cDNA preamplification system, the TaqMan PreAmp Master Mix kit (TPAMMK), enables a multiplex preamplification of cDNA targets and therefore, could provide a sufficient amount of specific amplicons for their posterior analysis on TA.
Findings
A multiplex preamplification of 47 genes was performed in 22 samples prior to their analysis by TA, and relative gene expression levels of non-preamplified (NPA) and preamplified (PA) samples were compared. Overall, the mean cycle threshold (CT) decrement in the PA genes was 3.85 (ranging from 2.07 to 5.01). A high correlation (r) between the gene expression measurements of NPA and PA samples was found (mean r = 0.970, ranging from 0.937 to 0.994; p < 0.001 in all selected cases). High correlation coefficients between NPA and PA samples were also obtained in the analysis of genes from degraded RNA samples and/or low abundance expressed genes.
Conclusion
We demonstrate that cDNA preamplification using the TPAMMK before TA analysis is a reliable approach to simultaneously measure gene expression of multiple targets in a single sample. Moreover, this procedure was validated in genes from degraded RNA samples and low abundance expressed genes. This combined methodology could have wide applications in clinical research, where scarce amounts of degraded RNA are usually obtained and several genes need to be quantified in each sample.
Similar content being viewed by others
Findings
Background
TaqMan Arrays (TAs) have recently been introduced as a novel approach to measure gene expression. They combine the high sensitivity provided by the real time quantitative reverse-transcription polymerase chain reaction (qRT-PCR) with the ability to simultaneously assay RNA expression levels of up to 384 target genes in a single sample [1, 2]. However, an accurate quantification with this approach could be limited by the low sample amount commonly encountered in clinical research. Therefore, it is necessary to preamplify samples to generate enough cDNA copies to enable an accurate quantification of transcripts and maintain high sensitivity.
Most of the current protocols for increasing small amounts of mRNA are designed to globally amplify all the transcriptome instead of the specific targets of interest and are usually validated by DNA array, which is a more imprecise methodology than qRT-PCR to quantify individual genes [3, 4].
Recently, a novel system, the TPAMMK, to increase cDNA quantity prior to gene expression analysis by conventional quantitative PCR has been described [5–8]. However, there is no study validating this approach prior to gene expression quantification by means of TA. The possibility of simultaneously measuring gene expression of dozens to hundreds of genes in those samples that yield scarce quantities of RNA would be of great interest.
Here, we evaluate this cDNA preamplification system (TPAMMK) prior to gene expression quantification by TA. We test the approach in the context of bladder cancer detection, by analyzing RNA samples obtained from bladder fluids. Furthermore, this type of sample provides us the possibility to analyze the reliability of this combined methodology for preamplifying cDNA from degraded RNA samples and/or low abundance expressed genes.
Results and discussion
One of the major drawbacks for the clinical use of RNA based techniques is the difficulty of obtaining sufficient quantities of high quality RNA from some human samples. The novel multiplexed miniaturized format provided by TAs could be limited by the scarce quantities of isolated RNA, since the sample is divided into a large number of aliquots [9]. Several strategies for amplifying RNA [10–12] or cDNA have been described [5–8, 13, 14], but to date, there is no published study validating that cDNA preamplification prior to TA analysis provides a reliable representation of gene expression profiling.
In this study, we have used the TPAMMK (AB) to preamplify 47 genes in 22 bladder fluids samples prior to their analysis by TA. This preamplification process allowed us to load a 30 fold greater quantity of cDNA in each TA port (500 ng for NPA vs ~15000 ng for PA samples) which should theoretically result in a decrement of 4.9 CTs. However, we found an overall mean CT decrement of 3.85 (ranging from 2.07 to 5.01) in PA samples (Table 1). Although variations in amplification efficiencies (AE) of the primers/probe sets used in TA methodology have been found in this study and others [1], they do not help to explain differences between the theoretical and the experimental CT decrement described, since there is no significant correlation between the experimental CT decrement and the primers/probe set AE, neither before (r = 0.062; p = 0.678) nor after (r = 0.060; p = 0.097) the cDNA preamplification process (Table 1).
On the other hand, linear regression analysis showed a high correlation (r) between gene expression measurements of NPA and PA samples for the validated genes (mean r values = 0.970, ranging from 0.937 to 0.994, p < 0.001 in all cases, except for sample number 15 where only 3 genes could be analyzed) (Table 2). Thus, albeit CT decrement for each individual gene is not uniform, the overall relative gene expression levels in PA samples remained proportional to the original gene expression levels in NPA samples. However, when checking preamplification uniformity individually for each gene (ΔΔCT values within ± 1.5) we found that there were three genes (IGF2, FGFR3 and CRH) that were consistently non-uniformly preamplified in a significant number of samples and therefore were not suitable for preamplification (Table 2). We do not have a clear explanation for this fact, although transcript abundance, primers and probe localization or amplicon length/sequence could influence the preamplification efficiency. From these results, it becomes clear that cDNA preamplification before gene expression quantification by TA can facilitate the analysis of multiple target genes from very low quantities of RNA in a single experiment. However, checking the preamplification uniformity in each target gene with control material before its evaluation in testing samples is mandatory.
As the described CT decrement would not be sufficient to accurately quantify gene expression in some cases, it must be mentioned that, according to kit manufacturers, it is possible to perform up to 14 preamplification cycles, as well as to increase the quantity of cDNA in the preamplification reaction up to 250 ng or to load more preamplified volume of cDNA into each TA port to achieve the desired decrement.
Since our goal was to test the suitability of the cDNA preamplification of specific target genes before their quantification by TA, we have only analyzed those genes with a reliable CT value in NPA samples (CT ≥ 31). However, to determine the linearity of the cDNA preamplification in low abundant expressed genes (those target genes with a CT value > 31 in NPA samples), we performed two serial dilutions (1/20 and 1/400) of 3 cDNAs from NPA samples with different RNA degradation levels (sample number 2, 11 and 22) and preamplified them with the same protocol used for the non diluted NPA samples. Subsequently, we compared the ΔCT values from diluted PA samples with the corresponding ΔCT of the initial cDNA (non diluted NPA) samples. This comparison yielded correlation coefficients close to 1, indicating that the vast majority of low abundant expressed genes are correctly preamplified and that the preamplification process maintains relative gene expression levels of the initial RNA over a broad range of CT values (Table 3).
For some time, it has been believed that degraded RNA samples were not suitable for gene expression studies. Nevertheless, many authors have recently reported the use of this material for gene profiling using DNA microarrays as well as qRT-PCR approaches [12, 15]. In order to investigate whether RNA degradation influences the efficiency of preamplification, gene expression measurements from those samples with a high RNA quality (RIN > 8; n= 6), those with good RNA quality (5 < RIN < 8; n= 8) and those with low RNA quality (RIN < 5; n= 6) [16] were compared (Table 2). The two samples with non available RIN were excluded from this part of the study. The mean CT decrement after preamplification was very similar in the three groups of samples; 3.90, 3.68 and 4.00, respectively. As expected, we initially found that the average number of validated genes corresponds to the RNA integrity (35.8, 34.6 and 16.5 in high, good and low RNA quality samples, respectively). The average number of validated genes in the three groups of samples after the cDNA preamplification was 45.2, 43.9 and 32.8 in high, good and low RNA quality samples, respectively, resulting from the increment in the number of validated genes being much higher in degraded RNA than from high/good quality RNA samples.
Thus, we have been able to demonstrate that preamplifed cDNA from samples with different RNA degradation states is a suitable material for TA analysis, facilitating the simultaneous analysis of multiple targets in a single experiment from archived pathology specimens in retrospective studies.
Conclusion
To our knowledge, this is the first quantitative gene expression report validating that cDNA preamplification using the TPAMMK prior to TA analysis preserves relative transcript expression levels of individual genes. The possibility to increase cDNA quantity before its analysis by TA opens up the possibility of analysing multiple target genes in a single experiment in those samples that yield scarce quantities of RNA. Furthermore, this approach is suitable for preamplifying genes from degraded RNA and low abundance expressed genes. This combined methodology could have wide applications in clinical research, where scarce amounts of degraded RNA are usually obtained and several genes needs to be quantified in each sample.
Methods
Patients and samples
Ten bladder washings (BW) and 7 voided urine specimens from patients with pathologically diagnosed bladder cancer (BC) [17, 18] and 4 BW and 1 urine sample from patients without history of BC (controls) were collected between April 2004 and September 2005 after informed consent (Table 2).
Ice cooled BW or urine samples were mixed with 1/25 volumes of 0.5 M EDTA, pH 8.0 and were centrifuged at 1000 × g for 10 minutes. The cell pellets were re-suspended in 1 ml of TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and frozen at -80°C until RNA extraction.
RNA extraction and cDNA synthesis
RNAs were extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer's instructions. Total RNA was quantified by spectrophotometric analysis at 260 nm and RNA degradation was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany) [19] (Table 2).
cDNA was synthesized from 1 μg of total RNA using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, USA, hereafter referred as AB) following manufacturer instructions, except that the final volume of the reaction was 50 μl.
Multiplex preamplification of cDNA targets
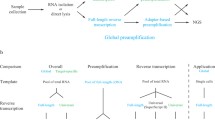
A multiplex PCR preamplification of the 46 specific cDNA targets and the endogenous control GUSB (Table 1) was performed using TPAMMK following manufacturer's instructions (AB). The 47 TaqMan Gene Expression Assays (AB) were pooled together at 0.2× final concentration. Subsequently, 12.5 μl of the pooled assay mix (0.2X) were combined with 25 ng of each cDNA sample and 25 μl of the TaqMan PreAmp Master Mix (2X) in a final volume of 50 μl (Figure 1). Thermal cycling conditions were as follows: initial hold at 95°C during 10 min and ten preamplification cycles of 15 sec at 95°C and 4 min at 60°C.
Workflow for the entire preamplification process. Detailed steps are described in the Material and Methods section. Briefly, to increase the quantity of the specific cDNA targets for gene expression analysis using TaqMan methodology, cDNA from the reverse transcription is mixed with pooled TaqMan Gene Expression Assays and with TaqMan PreAmp MasterMix. After the multiplex preamplification of desired cDNA targets, quantitative real time PCR amplification of preamplified target cDNAs, using sequence-specific primers and TaqMan probes from the TaqMan Gene Expression Assays and TaqMan Gene Expression Master Mix, is performed.
For samples n° 2, 11 and 22, two dilutions (1/20 and 1/400) of the NPA cDNA were prepared and 1.25 μl of each cDNA dilution (1.25 ng and 0.0625 ng, respectively) were subsequently preamplified with the same protocol described above.
TaqMan Arrays (TA)
The NPA and PA target cDNAs were then amplified in singleplex reactions using TA following manufacturer's recommendations (AB). Commercially available TaqMan Gene Expression Assays (AB) for all the 46 genes differentially expressed in bladder cancer specimens (data not shown) and the endogenous control GUSB were used (Table 1). Twenty-five μl of NPA cDNAs and 30 μl of PA cDNAs were mixed with 50 μl of 2× TaqMan Universal PCR Master Mix (AB) in a final volume of 100 μl. After loading mixes into the TA ports, cards were centrifuged twice for 1 min at 1200 rpm, sealed and run in an ABI PRISM 7900HT SDS with the following thermal conditions: 2 min at 50°C, 10 min at 94.5°C, 40 cycles of denaturation at 97°C for 30 sec and annealing and extension at 59.7°C for 1 min.
Data analysis
Quantitative real time PCR data were processed with SDS 2.1 software package (AB). A defined baseline of 3 to 12 cycles and a defined threshold of 0.35 were used for all the genes to record the cycle thresholds (CTs). Since precision on TA starts dropping off at around 30–32 CTs, assays that yielded a CT > 31 cycles were excluded from the analysis and comparisons between PA and NPA genes were performed only taking into account those genes with a CT value ≤ 31 in NPA samples (named validated genes). Data normalization was carried out with reference gene GUSB.
Linear regression analysis was performed to compare gene expression data (ΔCT) from NPA targets (ΔCTNPA = CTNPA target -CTNPA GUSB) vs PA targets (ΔCTPA = CTPA target -CTNPA GUSB). Those regressions with less than 4 points were excluded from the analysis. Specific gene preamplification uniformity was checked calculating the ΔCTNPA and ΔCTPA, and determining the ΔΔCT between NPA and PA targets (ΔΔCT = ΔCTPA -ΔCTNPA). ΔΔCT values close to zero indicated preamplification uniformity. Targets that produce ΔΔCT values within ± 1.5 were considered uniformly preamplified (TPAMMK Protocol, AB). AE of each assay was calculated from fluorescent data using the DART-PCR software version 1.0 [20, 21]. Pfaffl (2001) definition for AE has been used [22].
References
Goulter AB, Harmer DW, Clark KL: Evaluation of low density array technology for quantitative parallel measurement of multiple genes in human tissue. BMC Genomics. 2006, 7: 34-10.1186/1471-2164-7-34.
Tran DC, Brazeau DA, Fung HL: Determination of nitric oxide-donor effects on tissue gene expression in vivo using low-density gene arrays. Methods Enzymol. 2005, 396: 387-395. 10.1016/S0076-6879(05)96033-3.
Seth D, Gorrell MD, McGuinness PH, Leo MA, Lieber CS, McCaughan GW, Haber PS: SMART amplification maintains representation of relative gene expression: quantitative validation by real time PCR and application to studies of alcoholic liver disease in primates. J Biochem Biophys Methods. 2003, 55: 53-66. 10.1016/S0165-022X(02)00177-X.
Provenzano M, Mocellin S: Complementary techniques: validation of gene expression data by quantitative real time PCR. Adv Exp Med Biol. 2007, 593: 66-73.
Wang Y, Barbacioru CC, Shiffman D, Balasubramanian S, Iakoubova O, Tranquilli M, Albornoz G, Blake J, Mehmet NN, Ngadimo D, Poulter K, Chan F, Samaha RR, Elefteriades JA: Gene expression signature in peripheral blood detects thoracic aortic aneurysm. PLoS ONE. 2007, 2: e1050-10.1371/journal.pone.0001050.
Barrachina M, Moreno J, Juves S, Moreno D, Olive M, Ferrer I: Target genes of neuron-restrictive silencer factor are abnormally up-regulated in human myotilinopathy. Am J Pathol. 2007, 171: 1312-1323. 10.2353/ajpath.2007.070520.
Okamoto N, Chihara R, Shimizu C, Nishimoto S, Watanabe T: Artificial lymph nodes induce potent secondary immune responses in naive and immunodeficient mice. J Clin Invest. 2007, 117: 997-1007. 10.1172/JCI30379.
Denning KM, Smyth PC, Cahill SF, Finn SP, Conlon E, Li J, Flavin RJ, Aherne ST, Guenther SM, Ferlinz A, O'Leary JJ, Sheils OM: A molecular expression signature distinguishing follicular lesions in thyroid carcinoma using preamplification RT-PCR in archival samples. Mod Pathol. 2007, 20: 1095-1102. 10.1038/modpathol.3800943.
Dafforn A, Chen P, Deng G, Herrler M, Iglehart D, Koritala S, Lato S, Pillarisetty S, Purohit R, Wang M, Wang S, Kurn N: Linear mRNA amplification from as little as 5 ng total RNA for global gene expression analysis. Biotechniques. 2004, 37: 854-857.
Xiang CC, Chen M, Kozhich OA, Phan QN, Inman JM, Chen Y, Brownstein MJ: Probe generation directly from small numbers of cells for DNA microarray studies. Biotechniques. 2003, 34: 386-3.
Marko NF, Frank B, Quackenbush J, Lee NH: A robust method for the amplification of RNA in the sense orientation. BMC Genomics. 2005, 6: 27-10.1186/1471-2164-6-27.
Mengual L, Burset M, Ars E, Ribal MJ, Lozano JJ, Minana B, Sumoy L, Alcaraz A: Partially Degraded RNA from Bladder Washing is a Suitable Sample for Studying Gene Expression Profiles in Bladder Cancer. Eur Urol. 2006, 50: 1347-1356. 10.1016/j.eururo.2006.05.039.
Max N, Wolf K, Spike B, Thiel E, Keilholz U: Nested quantitative real time PCR for detection of occult tumor cells. Recent Results Cancer Res. 2001, 158: 25-31.
Bittman B, Berk L, Shannon M, Sharaf M, Westengard J, Guegler KJ, Ruff DW: Recreational music-making modulates the human stress response: a preliminary individualized gene expression strategy. Med Sci Monit. 2005, 11: BR31-BR40.
Cronin M, Pho M, Dutta D, Stephans JC, Shak S, Kiefer MC, Esteban JM, Baker JB: Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol. 2004, 164: 35-42.
Fleige S, Pfaffl MW: RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006, 27: 126-139. 10.1016/j.mam.2005.12.003.
Sobin LH, Wittekind CH: TNM Classification of Malignant Tumours. International Union Against Cancer. 2002, New York, Jonh Wiley & Sons, 6th
Lopez-Beltran A, Sauter G, Gasser T, Hartmann A, Schmitz-Dräger BJ, Helpap B, Ayala AG, Tamboni P, Knowles MA, Sidransky D, Cordon-Cardo C, Jones PA, Cairns P, Simon R, Amin MB, Tyczynsky JE: Tumours of the Urinary System. Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. World Health Organization Classification of Tumours. Edited by: Eble JN, Sauter G, Epstein JI and Sesterhenn IA. 2004, Lyon, IARC Press, 89-157.
Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, Mueller O, Schroeder A, Auffray C: Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005, 33: e56-10.1093/nar/gni054.
Peirson SN, Butler JN, Foster RG: Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003, 31: e73-10.1093/nar/gng073.
Gene quantification. 2008, [http://www.gene-quantification.de/download.html]
Pfaffl MW: A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29: e45-10.1093/nar/29.9.e45.
Acknowledgements
This research was partly supported by grants from the Spanish Urological Association (FIU 2007) and from Fondo de Investigaciones Sanitarias (PI070040) and by private funding (Laboratorios FINA BIOTECH). Advice from Dr María Luisa Checa is gratefully acknowledged. We thank Jon Sherlock for the critical revision of the manuscript and Helena Kruyer for the English correction of the manuscript. We also thank Dr. Dolors Colomer for her assistance in the Real Time PCR management.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
All authors participated in study concept, design and interpretation of data. LM carried out the experiments, participated in acquisition and analysis of data and drafting of the manuscript. MB participated in analysis of data, provided statistical expertise and critical revision of the manuscript. MM-A participated in collection of samples, assisted some experiments and critical revision of the manuscript. MJR participated in collection of samples, provided clinical expertise and critical revision of the manuscript. AA provided clinical expertise, critical revision of the manuscript, obtained funding and supervised conduct of the study. All the authors have read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Mengual, L., Burset, M., Marín-Aguilera, M. et al. Multiplex preamplification of specific cDNA targets prior to gene expression analysis by TaqMan Arrays. BMC Res Notes 1, 21 (2008). https://doi.org/10.1186/1756-0500-1-21
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-0500-1-21