Abstract
Background
Late-onset Alzheimer's disease (LOAD) is an age related neurodegenerative disease with a high prevalence that places major demands on healthcare resources in societies with increasingly aged populations. The only extensively replicable genetic risk factor for LOAD is the apolipoprotein E gene. In order to identify additional genetic risk loci we have conducted a genome-wide association (GWA) study in a large LOAD case – control sample, reducing costs through the use of DNA pooling.
Methods
DNA samples were collected from 1,082 individuals with LOAD and 1,239 control subjects. Age at onset ranged from 60 to 95 and Controls were matched for age (mean = 76.53 years, SD = 33), gender and ethnicity. Equimolar amounts of each DNA sample were added to either a case or control pool. The pools were genotyped using Illumina HumanHap300 and Illumina Sentrix HumanHap240S arrays testing 561,494 SNPs. 114 of our best hit SNPs from the pooling data were identified and then individually genotyped in the case – control sample used to construct the pools.
Results
Highly significant association with LOAD was observed at the APOE locus confirming the validity of the pooled genotyping approach.
For 109 SNPs outside the APOE locus, we obtained uncorrected p-values ≤ 0.05 for 74 after individual genotyping. To further test these associations, we added control data from 1400 subjects from the 1958 Birth Cohort with the evidence for association increasing to 3.4 × 10-6 for our strongest finding, rs727153.
rs727153 lies 13 kb from the start of transcription of lecithin retinol acyltransferase (phosphatidylcholine – retinol O-acyltransferase, LRAT). Five of seven tag SNPs chosen to cover LRAT showed significant association with LOAD with a SNP in intron 2 of LRAT, showing greatest evidence of association (rs201825, p-value = 6.1 × 10-7).
Conclusion
We have validated the pooling method for GWA studies by both identifying the APOE locus and by observing a strong enrichment for significantly associated SNPs. We provide evidence for LRAT as a novel candidate gene for LOAD. LRAT plays a prominent role in the Vitamin A cascade, a system that has been previously implicated in LOAD.
Similar content being viewed by others
Background
Late-onset Alzheimer's disease (LOAD) is an age related neurodegenerative disease and the most common form of dementia in the over 65 age group. It affects 20% of people aged 75 – 84 years, rising to 50% in the over 85's, thus placing major demands on healthcare resources in societies with increasingly aged populations [1]. It has a high heritability with estimates ranging between 60 – 80% [2]. The only extensively replicable genetic risk factor for LOAD is the apolipoprotein E gene, in which the ε4 genotype is overrepresented in LOAD cases compared to controls. The ε2 genotype is underrepresented and believed to have a protective effect on disease development. However the presence of the APOE-ε4 genotype is neither necessary nor sufficient for the development of the disease, indeed 40 – 70% of European LOAD patients do not carry an ε4 variant, and additional genetic loci remain to be identified [3].
Recent advances in genotyping technology make it possible to conduct genome-wide association (GWA) studies, testing the whole genome with hundreds of thousands of single nucleotide polymorphisms (SNPs). For a complex disease such as LOAD, in which multiple genetic and environmental factors are thought to contribute to risk [2, 4] GWA studies offer the potential to detect susceptibility genes with greater confidence than with linkage analysis. It has been estimated that over 80% of genetic variation of common SNPs of the human genome in European populations can be captured at an r 2 > 0.8 by using current SNP genotyping arrays [5].
In order to detect variants of small effect, particularly if the association is indirect, and to overcome the issue of multiple testing, large sample sizes are required [6, 7]. Currently, GWA studies are expensive, generally restricting this type of work to groups or consortia with substantial funding for that purpose. Genome-wide association analysis of pools of case and control DNA offers an economic approach with the potential to identify disease loci [8–10]. In DNA pooling, equal amounts of DNA from each sample are combined to form pools from cases and controls, which are genotyped to get an estimate of the true allele frequency difference for each variant. This estimate is then used to test a limited number of SNPs for genetic association at a fraction of the cost of individual genotyping [9, 11, 12].
In this study, 561,494 SNPs were genotyped in DNA pools constructed from the Medical Research Council (MRC) Genetic Resource for LOAD case – control samples. In order to select SNPs for testing by individual genotyping, we applied three complimentary approaches to select the highest-ranked SNPs. We successfully genotyped individually 114 SNPs. We found association with LOAD of several SNPs close to the APOE locus (2.08 × 10-9 – 8.24 × 10-11) thus confirming the validity of the pooled genotyping approach. In addition, we obtained evidence for several novel genetic associations to LOAD, our most significant findings being association of SNPs in the lecithin retinol acyltransferase (phosphatidylcholine – retinol O-acyltransferase, LRAT) gene.
Results
Before we started work with Illumina arrays, we validated the pool construction with the SNaPshot method. The pools gave an accurate estimate of the real difference in allele frequencies for 3 SNPs previously individually genotyped in this sample. rs11084424, rs157580 and rs157581 showed allele frequency differences between cases and controls in pools of 4%, 9% and 17% respectively which compared well with the real differences of 3%, 10% and 19%. Published estimates of the accuracy of DNA pooling report similar high accuracy with errors of < 2% between predicted and true allele frequency differences using different methods [11].
Genome-wide pooled genotyping was carried out on the Illumina HumanHap300 and Illumina Sentrix HumanHap240S arrays assaying 561,494 SNPs. Frequencies for each SNP were averaged over four replicate case and three replicate control arrays for the Illumina HumanHap300 and eight each for the Sentrix HumanHap240S arrays. The predicted averaged patient and control allele frequencies showed as expected a very high Pearson correlation with each other of r = 0.998, indicating a low technical variability of the method. Figure 1 shows predicted allele frequencies in case and control pools determined using the Illumina HumanHap300 platform. Data from the Illumina Sentrix HumanHap240S arrays showed similarly high correlations (r = 0.997). Predicted allele frequencies were compared with actual population allele frequencies (1958 Birth Cohort controls genotyped with the same HumanHap300) and gave a very good correlation of 0.969 (Figure 2). This indicates that even uncorrected data from pooling on this platform predict fairly well the true absolute allele frequencies of SNPs.
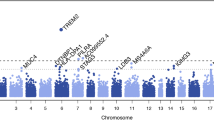
Figure 3 shows the combined Z-test p-value results for the whole genome. On the X-axis we have plotted the position in the genome by chromosome, and on the Y-axis the negative logarithm of the p-value. The strongest evidence for association with LOAD was observed with SNPs on chromosome 19 surrounding the APOE gene. In all, 7 SNPs within 71 kb were predicted by the pools to have allele frequency differences between 6% – 14% and "combined" p-values ranging from 9.0 × 10-5 to 3.6 × 10-22. No other region of the genome showed such a large number of significant markers over a relatively small region. Five of the seven SNPs were individually genotyped and were confirmed to be highly significant (p-value range 2.08 × 10-9 – 8.24 × 10-11, Table 1). All are in high linkage disequilibrium (LD) with the SNPs that define the APOE genotypes which are not themselves typed on the Illumina platform (Figure 4). The distributions of APOE alleles in this population are as follows:
Plot of combined Z-test p-values against chromosomal location for pooled data. Out of 561,494 SNPs that were genotyped in our case – control pools, only 3 would have remained significant after Bonferroni correction for multiple testing, two of these SNPs are near the APOE gene on Chromosome 19. For a genome-wide association study on 500,000 markers (assuming that markers are independent) the significance level is 10-7. The line of genome-wide significance is plotted therefore at this level, although the combined Z-statistic provides only an approximation of the p-values produced from individual genotyping.
APOE-ε2, APOE-ε3, APOE-ε4 = 5.1, 57.9, 37.0 and 9.5, 77.3, 13.2% in cases and controls respectively, giving an allelic p-value of p = 1.9 × 10-73 and an odds ratio (95%CI) = 3.85 (3.55 – 4.15) for the APOE-ε4 allele.
Table 1 shows results for our most significant SNPs, in addition to those at the APOE locus, following individual genotyping. The most significant SNP, rs727153, reached a p-value of 2.4 × 10-5.
To further test the associations, we added control data from a set of controls comprising approximately 1400 subjects from the 1958 Birth Cohort for the 79 SNPs that showed individual genotype p-value ≤ 0.05. The association for five SNPs becomes more significant with the inclusion of the additional controls. The majority of SNPs however became less significant indicating that for these SNPs we had probably identified false positives, although this could also happen to true-positive findings, as the initial discovery study tends to over-estimate the effect size. Our strongest finding, rs727153 remained the most significant individually genotyped SNP, with the evidence for association increasing from 2.4 × 10-5 to 3.4 × 10-6.
rs727153 is an intergenic SNP approximately 13 kb from the start of transcription of lecithin retinol acyltransferase (phosphatidylcholine – retinol O-acyltransferase, LRAT). We genotyped additional SNPs in this region to test if our significant association extended into the LRAT gene. rs727153 is within an LD block flanked by SNPs rs11935519 and rs149225 (Chr4:156,005,695..156,040,821 – HapMap data Rel 21a/phaseII Jan07), which includes LRAT (Figure 5). Using a pair-wise approach in Haploview we identified 7 tagging SNPs required to cover the LD block, capturing all common SNPs with an r 2 > 0.8 and individually genotyping them in our LOAD case – control sample. The results are presented in Table 2. Five SNPs are significantly associated with LOAD in this region, with a SNP in intron 2 of LRAT, rs201825 showing the strongest evidence (p-value = 1.7 × 10-6). After the addition of extra controls data from the 1958 birth cohort for the 4 SNPs in LRAT which have been genotyped in that sample, 3 became more significant (rs201825, p-value = 6.1 × 10-7) and the fourth remained unchanged.
Analysis of the tagSNPs in this LD block using all possible 2- and 3-marker haplotypes resulted in no evidence for association greater than that reached by rs201825 (data not shown).
Table 3 provides the distribution of the individual genotyping p-values for 109 SNPs outside the APOE locus, in addition to the p-value distributions for our three methods used to choose the SNPs for individual genotyping. After individual genotyping, we obtained uncorrected p-values ≤ 0.05 for 74 of the 109 SNPs compared with expected 5.5 for a random selection of SNPs, an enrichment of ~13 fold across all selection methods. The cluster method appeared to generate the highest percentage of significantly associated SNPs (84.8%), at greater levels of significance, identifying the 3 most significant SNPs after individual genotyping. One of the three most significant SNPs (rs3754675) was also in the top 115 SNPs ranked by the "combined" Z-test. The most significant SNP by individual genotyping chosen by the allele frequency method (rs12510838) had a p-value of 0.00024, an order of magnitude less than our most significant SNP chosen by the other methods (Table 1). Thus it appears that the cluster method provided the best way of identifying true associations and that the combined Z-test improves on the identification of single significant SNPs over a simple method of following-up only the highest differences in allele frequencies.
Discussion
In this study we used DNA pooling to offset the high costs of conducting a GWA study for LOAD. Using Illumina HumanHap300 and Illumina Sentrix HumanHap240S arrays we estimated the allele frequencies of 561,494 SNPs across the genome in pools constructed from 1,082 LOAD cases and 1,239 age-matched controls. We unequivocally identified the APOE locus as the major genetic risk factor for the disease. As noted by others this can be seen as a positive control that the pooling method is a viable alternative to individual genotyping in GWA studies for complex disorders [13]. The association of APOE with LOAD is well replicated [14, 15] and has been observed in a GWA study that used an alternative genotyping platform [16]. Neither the Illumina platform used here nor the Affymetrix system used in that study directly test the SNPs that define the APOE-ε4 genotype, highlighting the strength of high density GWA studies to detect indirect association. Our findings are in agreement with others that APOE is the major pathogenic locus for LOAD, and that further loci of smaller effect remain to be identified [16, 17].
Following pooled genotyping, it is necessary to follow up positive results with individual genotyping to confirm the observed associations. We used three methods to identify SNPs to follow up. The cluster method appeared to generate more true associations than the other methods we used. This is perhaps not surprising, as this method should minimise false positives due to technical artefacts caused by pooled genotyping, as each SNP showing evidence of association was required to be supported by one or more highly significant neighbouring SNP. In contrast, a singleton highly-ranked SNP found by any other method could be due to a technical artefact, no matter how carefully it is filtered. However, the cluster method can miss true positive signals from functional variants that are not in high LD with other tested SNPs. Therefore, we believe that unsupported SNPs have to be followed up as well, unless they are in high LD with other SNPs on the array, which do not support the association. As has been proposed by others, the data presented here suggests that the best way of choosing unsupported SNPs would be to use a statistical test that takes into account technical variation, such as the combined Z-test [11]. We acknowledge that this study has not exhaustively followed up all positive signals – to do this would negate the cost efficiency of the pooling method but it is possible that more significantly associated SNPs could be confirmed by individual genotyping. In fact, we only genotyped individually half of the SNPs that we considered worth following up. However it appears clear that a signal with the strength of association, in terms of a number of highly significant SNPs in a narrow region, as observed at the APOE locus, has not been overlooked.
In order to confirm our individual genotyping results, data from the 1958 Birth Cohort were used to form an additional set of controls genotyped with the Illumina platform. These controls have previously been used by the Wellcome Trust Case Consortium for a GWA study for 7 common diseases [18]. There are a number of explanations for our observation that for the majority of SNPs significant in this study the strength of association decreased with additional control data. Firstly, and we believe most likely, the initial findings may have been false positives due to an inflation of the effect size in the original analysis due to sampling variance in the control allele frequencies (i.e. some of the most significant SNPs have the highest sampling variance in either cases or controls). Secondly, the use of unscreened controls in an association study of a disorder of old age is expected to reduce power [19], this is particularly clear in the APOE locus. Some degree of genotyping discrepancies between the two platforms used is also possible (Sequenom in our lab and Illumina in the 1958 Birth Cohort), however very unlikely to account for such large differences.
We identified multiple SNP associations (best p-value = 6.12 × 10-7) in the gene encoding lecithin retinol acyltransferase (LRAT, 4q32.1, MIM: 604863). This is a highly plausible functional candidate gene for LOAD. LRAT plays a prominent role in the Vitamin A (retinoid) cascade by producing retinyl esters, storage forms of retinoid. The retinoid system has been previously implicated in LOAD. Retinoid levels in plasma, serum and brain are lowered in LOAD patients, and the restriction of dietary retinoid in mice results in memory impairment [20, 21]. Furthermore disruption of the retinoid signalling pathway in adult rats by a dietary deficiency of vitamin A leads to deposition of amyloid beta in the cerebral blood vessels. There is a down regulation of retinoic acid receptor alpha in the forebrain neurons of the retinoid-deficient rats and a loss of choline acetyl transferase expression, which precedes amyloid beta deposition. In neocortex of pathology samples of patients with Alzheimer's disease, the same retinoic acid receptor alpha deficit in the surviving neurons is observed suggesting that retinoids are important for the maintenance of the adult nervous system and their loss may in part play a role in Alzheimer's disease [22]. Nevertheless, despite the functional plausibility, our genetic findings in LRAT fall short of the degree of statistical significance required to provide unequivocal evidence for association, given the large number of comparisons made in a GWA study. Thus our findings, whilst highly suggestive, will require confirmation in independent samples.
Conclusion
In summary, we have validated the pooling method for GWA studies by both identifying the APOE locus, a known risk gene for LOAD, and by observing a strong enrichment for significantly associated SNPs. We have also compared methods for prioritising SNPs for individual genotyping. Finally, we provide evidence for LRAT as a novel candidate gene for LOAD. GWA studies with pooled DNA provide a viable, quick and inexpensive approach to identifying susceptibility genes. Inaccuracies, however, mean that some loci that might be detected by individual genotyping will remain undetected.
Methods
Subjects
Written informed consent was obtained from all subjects for publication of this case report. A copy of the written consent is available for review by the Editor-in-Chief of this journal. The sample consisted of individuals ascertained from both community and hospital settings in the UK collected as part of the MRC genetic resource for LOAD. Clinical data and DNA samples were collected from 1,082 individuals (71% females) with late-onset AD (LOAD) and 1,239 control subjects (referred to in the text as "MRC controls", 62% females). Age at onset ranged from 60 to 95 years (mean = 75.84 years, SD = 6.79). Controls were matched for age (mean = 76.53 years, SD = 6.33), gender and ethnicity. AD cases and controls described here were ascertained by three collaborating centres: Department of Psychological Medicine, Cardiff University, Cardiff (coordinating centre); Institute of Psychiatry, London; and Cambridge University, Cambridge, as previously described [23]. Ethical approval was obtained from the Multi-centre Research Ethics Committee (MREC), relevant local ethics committees and NHS trusts, in the regions where subjects were recruited.
All cases were Caucasian, of UK origin (parents born in the UK) and diagnosed with probable AD in accordance with the National Institute of Neurological and Communication Disorders and Stroke and the Alzheimer's disease and Related Disorders Associations (NINCDS-ADRDA) clinical diagnostic criteria for AD [24]. All diagnoses were made based on a semi-structured interview with known validity for AD pathology (i.e. positive predictive value of 92–95% [25, 26] which included: The Mini Mental State Examination (MMSE) [27]; The Cambridge Mental Disorders of the Elderly Examination (CAMDEX; informant interview) [28]; The Blessed Dementia Scale [29]; The Bristol Activities of Daily Living Scale [30]; Webster Rating Scale [31]; Global Deterioration Scale (GDS) [32]; Cornell Scale for Depression in Dementia [33]; Neuropsychiatric Inventory (NPI) (12-item version) [34]. Interviews were primarily conducted with the AD sufferer's next of kin. Ethical permission was obtained from the Multi-centre Research Ethics Committee, relevant local ethics committees, and National Health Service trusts.
Control subjects were either spouses of AD patients or selected from primary-care practices situated in the same geographical areas as AD patients. All controls were 60 years or above and of UK origin. Control individuals were screened for cognitive decline using the MMSE, and a cut-off score of ≥ 28 was adopted. Assessment of controls also included a section of the Cambridge Mental Disorders of the Elderly Examination and the Geriatric Depression Scale [35]. Exclusion criteria were the presence of dementia, depression, delirium or other illnesses likely to significantly reduce cognitive function.
DNA Pool Construction
DNA was obtained from blood samples by phenol/choloroform extraction, followed by precipitation in ethanol and storage in TE buffer. DNA quality was assessed by PCR amplification of microsatellite markers under standard conditions, with those samples showing robust amplification being included in pools (1,082 LOAD cases and 1,239 controls). Initial DNA concentrations were determined by UV spectrophotometry using absorbance at 260 nm readings. Dilutions were made using water to bring the concentration of each sample to a target of 20 ng/ul. The concentration of each sample was then determined using the PicoGreen dsDNA Quantitation Reagent (Molecular Probes, Eugene, Ore.) in a Labsystems Ascent Fluoroskan (LifeSciences Int., Basingstoke, UK). Each sample was then diluted to 4 ng/ul (± 0.5 ng/ul), allowed to equilibrate at 4°C for 48 h before another quantification using the PicoGreen method. Samples out of the 4 ng/ul (± 0.5 ng/ul) range were diluted/concentrated, incubated and re-quantified until they were within the required range. Equal volumes of each sample were then added to either a case or control pool using a Biomek® FX Laboratory Automation Workstation (Beckman Coulter, Inc., Fullerton, CA). We chose to make single pools of cases and controls and hybridise them multiple times, rather than construct many small pools and hybridise them on single arrays, as this method has been shown to be more powerful [36].
Pool validation with the SNaPshot method
In order to test the accuracy of our pool construction, we genotyped 3 significantly associated SNPs in the pools for which we had individual genotype data on the samples used to create the pools (rs11084424, rs157580 and rs157581). SNaPshot genotyping was carried out as previously described [12]. Briefly, forward and reverse primers were designed using primer 3 software http://primer3.sourceforge.net/. Extension primers for the SNaPshot assay were designed using FP PRIMER 1.0.1 b software http://m034.pc.uwcm.ac.uk/FP_Primer.html. PCR was performed under standard conditions, using 15 ng pooled genomic DNA and HotStar Taq DNA polymerase (Qiagen). Primer extension products were run on a 3100 DNA sequencer (Applied Biosystems) and the data were processed by the GeneScan Analysis 3.7 (Applied Biosystems). SNP allele frequencies in DNA pools were estimated from peak heights obtained by using Genotyper 2.5 (PE Biosystems, Cheshire, UK). Estimated allele frequencies from pools were corrected for unequal representation of alleles using the mean of the ratios obtained from four analyses of a heterozygote [37].
Pooled DNA Genotyping using Illumina Platform
Genome-wide genotyping was performed using Illumina HumanHap300 and Illumina Sentrix HumanHap240S arrays (Illumina Inc., San Diego, CA, USA) according to the manufacturer's protocols. Chips were scanned in standard mode on a BeadStation 500 GX (Illumina) at the University of Bonn and raw data were extracted for statistical analysis with BeadStudio v3.1 software. Because of the expected inter-experiment variation, replicate arrays were genotyped for each pool: four arrays on the Illumina HumanHap300, and 8 arrays on the Illumina Sentrix HumanHap240S array. The reason for using more HumanHap240S arrays is that we first performed the work on HumanHap300, and noticed the need for more replications. Replicate arrays were excluded if the estimated allele frequencies produced a Pearson correlation of r ≤ 0.991 with two or more other replicate arrays of the same pool (i.e. only arrays which correlated at r ≥ 0.992 with each other were retained for analysis). This cut-off was adopted during other pooling work in our department, but in this experiment resulted in the exclusion of data from only a single array (from a control pool) genotyped on the HumanHap300 array.
Analysis of pooled DNA
Approximation of allele A frequencies for each replicate was produced on the basis of the raw data as follows: f_alleleA = Xraw/(Xraw+Yraw), averaged over the number of replicates in each pool (Xraw and Yraw are the intensities of the two dyes Cy5 and Cy3, used to genotype SNPs on the Illumina platform).
Selection of SNPs for individual genotyping
SNPs from the pooling data were prioritised for individual genotyping based on three different methods, outlined below, since, at the time of this experiment we did not know which method would perform best. For all methods, we excluded rare SNPs (true allele frequency less than 5% in the CEU population of the HapMap, http://www.hapmap.org/) and the 5% of SNPs showing the highest technical variability as indicated by the size of the standard deviation amongst measures from the replicate arrays.
1) Cluster Method
We plotted the differences in allele frequencies between cases and controls for each SNP against their position in the genome. We identified 58 clusters where at least two SNPs within 100 kb had a predicted allele frequency difference greater than 5%. One SNP from each cluster was selected for individual genotyping.
2) Allele Frequency Difference Method
Separately for the Illumina HumanHap300 and Illumina Sentrix HumanHap240S arrays (to account for any differences caused by technical artefacts between the two arrays) the pooled data were sorted by predicted allele frequency differences in cases and controls. The top 136 SNPs showing the greatest allele frequency differences (8% difference between cases and controls) and satisfying the above filtering criteria were put forward for potential individual genotyping. SNPs that had previously been flagged as being in a cluster were not included.
3) "Combined Z-test"
The third method was based on p-values estimated using the following statistic which combines experimental and sampling errors, a general description of which has previously been presented [11, 36, 38]:
This statistic combines:
-
a)
chi-square statistic T for testing differences between two proportions (allele frequencies) in cases and in controls accounting for the sampling variance:
where is the mean of the allele frequencies over n k pool replicates, is the binomial sampling variance and N k is number of controls and cases respectively (k = 1,2).
-
b)
Z-statistics for testing the difference in mean allele frequencies between cases and controls:
where is the square of the standard error due to experimental error.
Thus we have taken into account the two single available sources of error: sampling error and experimental error in a simple way which is equivalent to a simplified version of the complex regression model suggested by MacGregor[36].
Excluding 36 SNPs already selected by the allele frequency (25 SNPs) or cluster (11 SNPs) methods, and any filtered ones, 79 of the highest-ranking SNPs identified by the combined Z-test method were put forward for individual genotyping.
Individual Genotyping
The 273 SNPs chosen by the three methods, (in addition to 5 SNPs at the APOE locus which had been identified by all three methods) were presented to the Sequenom Assay Design 3.1 software (Sequenom, San Diego, CA), which selected 130 for genotyping. We deliberately presented about twice as many SNPs to the software, as we wanted to type, in order to maximise the chance of producing well-performing panels.
Genotyping was performed with the MassARRAY and iPlex systems (Sequenom, San Diego, CA) according to the manufacturer's recommendations. Assays were optimized in 30 reference CEU parent – offspring trios, which have been used extensively in the HapMap project. All sample plates contained cases, controls, blanks, CEU and duplicate samples. Quality control (QC) measures included independent double genotyping blind to sample identity and blind to the other rater, and comparison of our CEU genotypes to those in the HapMap database http://www.hapmap.org. SNPs showing deviation from Hardy-Weinberg equilibrium (p-value < 0.001 in controls) or with genotyping success rates of less than 90%, or showing differences in genotypes in the CEU samples from those in the HapMap database were excluded from analysis. Individual DNA samples for whom the genotyping success rate across all SNPs was < 75% were also excluded (40 cases and 49 controls).
Of the 130 SNPs chosen from the pooling GWA study, 114 passed QC. Of these 5 were located within 32 kb of APOE, 33 were selected from the cluster method, 30 were identified solely by the combined Z-test method and 46 were chosen based on the allele frequency difference.
Calculation of allele frequencies, genotype counts, tests for departure from HWE, allelic association and odds ratio were carried out using PLINK (vers 0.99s, http://pngu.mgh.harvard.edu/purcell/plink/, [39].
Linkage Disequilibrium analysis was conducted using Haploview version 4.0 http://www.broad.mit.edu/mpg/haploview/, [40].
Additional control genotype data were obtained from (http://www.b58cgene.sgul.ac.uk/, accessed September 2007) which contains genotyping data on the British 1958 Birth Cohort DNA Collection, deposited by the Wellcome Trust Sanger Institute.
Abbreviations
- AD:
-
Alzheimer's Disease
- APOE:
-
Apolipoprotein E
- DNA:
-
Deoxyribonucleic Acid
- GWA:
-
Genome-Wide Association
- Kb:
-
kilobase pairs
- LD:
-
Linkage Disequilibrium
- LOAD:
-
Late-Onset Alzheimer's Disease
- LRAT:
-
lecithin retinol acyltransferase
- MRC:
-
Medical Research Council
- SNP:
-
Single Nucleotide Polymorphism.
References
Ferri CP, Prince M, Brayne C, Brodaty H, Fratiglioni L, Ganguli M, Hall K, Hasegawa K, Hendrie H, Huang Y, et al: Global prevalence of dementia: a Delphi consensus study. Lancet. 2005, 366 (9503): 2112-2117. 10.1016/S0140-6736(05)67889-0.
Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL: Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006, 63 (2): 168-174. 10.1001/archpsyc.63.2.168.
Daw EW, Payami H, Nemens EJ, Nochlin D, Bird TD, Schellenberg GD, Wijsman EM: The number of trait loci in late-onset Alzheimer disease. Am J Hum Genet. 2000, 66 (1): 196-204. 10.1086/302710.
Wang WY, Barratt BJ, Clayton DG, Todd JA: Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005, 6 (2): 109-118. 10.1038/nrg1522.
Eberle MA, Ng PC, Kuhn K, Zhou L, Peiffer DA, Galver L, Viaud-Martinez KA, Lawley CT, Gunderson KL, Shen R, et al: Power to detect risk alleles using genome-wide tag SNP panels. PLoS Genet. 2007, 3 (10): 1827-1837. 10.1371/journal.pgen.0030170.
Risch NJ: Searching for genetic determinants in the new millennium. Nature. 2000, 405 (6788): 847-856. 10.1038/35015718.
Cardon LR, Bell JI: Association study designs for complex diseases. Nat Rev Genet. 2001, 2 (2): 91-99. 10.1038/35052543.
MacGregor S, Zhao ZZ, Henders A, Nicholas MG, Montgomery GW, Visscher PM: Highly cost-efficient genome-wide association studies using DNA pools and dense SNP arrays. Nucleic Acids Res. 2008, 36 (6): e35-10.1093/nar/gkm1060.
Kirov G, Nikolov I, Georgieva L, Moskvina V, Owen MJ, O'Donovan MC: Pooled DNA genotyping on Affymetrix SNP genotyping arrays. BMC Genomics. 2006, 7 (1): 27-10.1186/1471-2164-7-27.
Docherty SJ, Butcher LM, Schalkwyk LC, Plomin R: Applicability of DNA pools on 500 K SNP microarrays for cost-effective initial screens in genomewide association studies. BMC Genomics. 2007, 8: 214-10.1186/1471-2164-8-214.
Sham P, Bader JS, Craig I, O'Donovan M, Owen M: DNA Pooling: a tool for large-scale association studies. Nat Rev Genet. 2002, 3 (11): 862-871. 10.1038/nrg930.
Norton N, Williams NM, Williams HJ, Spurlock G, Kirov G, Morris DW, Hoogendoorn B, Owen MJ, O'Donovan MC: Universal, robust, highly quantitative SNP allele frequency measurement in DNA pools. Hum Genet. 2002, 110 (5): 471-478. 10.1007/s00439-002-0706-6.
Pearson JV, Huentelman MJ, Halperin RF, Tembe WD, Melquist S, Homer N, Brun M, Szelinger S, Coon KD, Zismann VL, et al: Identification of the genetic basis for complex disorders by use of pooling-based genomewide single-nucleotide-polymorphism association studies. Am J Hum Genet. 2007, 80 (1): 126-139. 10.1086/510686.
Saunders AM, Schmader K, Breitner JC, Benson MD, Brown WT, Goldfarb L, Goldgaber D, Manwaring MG, Szymanski MH, McCown N, et al: Apolipoprotein E epsilon 4 allele distributions in late-onset Alzheimer's disease and in other amyloid-forming diseases. Lancet. 1993, 342 (8873): 710-711. 10.1016/0140-6736(93)91709-U.
Farrer LA, Cupples LA, van Duijn CM, Kurz A, Zimmer R, Muller U, Green RC, Clarke V, Shoffner J, Wallace DC, et al: Apolipoprotein E genotype in patients with Alzheimer's disease: implications for the risk of dementia among relatives. Ann Neurol. 1995, 38 (5): 797-808. 10.1002/ana.410380515.
Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, Zismann VL, Beach TG, Leung D, Bryden L, et al: A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J Clin Psychiatry. 2007, 68 (4): 613-618.
Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M, et al: Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol . 2008, 65 (1): 45-53. 10.1001/archneurol.2007.3.
Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007, 447 (7145): 661-678. 10.1038/nature05911.
Colhoun HM, McKeigue PM, Davey Smith G: Problems of reporting genetic associations with complex outcomes. Lancet. 2003, 361 (9360): 865-872. 10.1016/S0140-6736(03)12715-8.
Goodman AB, Pardee AB: Evidence for defective retinoid transport and function in late onset Alzheimer's disease. Proc Natl Acad Sci USA. 2003, 100 (5): 2901-2905. 10.1073/pnas.0437937100.
Goodman AB: Retinoid receptors, transporters, and metabolizers as therapeutic targets in late onset Alzheimer disease. J Cell Physiol. 2006, 209 (3): 598-603. 10.1002/jcp.20784.
Corcoran JP, So PL, Maden M: Disruption of the retinoid signalling pathway causes a deposition of amyloid beta in the adult rat brain. Eur J Neurosci. 2004, 20 (4): 896-902. 10.1111/j.1460-9568.2004.03563.x.
Morgan AR, Turic D, Jehu L, Hamilton G, Hollingworth P, Moskvina V, Jones L, Lovestone S, Brayne C, Rubinsztein DC, et al: Association studies of 23 positional/functional candidate genes on chromosome 10 in late-onset Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet. 2007, 144B (6): 762-770. 10.1002/ajmg.b.30509.
McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM: Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984, 34 (7): 939-944.
Holmes C, Cairns N, Lantos P, Mann A: Validity of current clinical criteria for Alzheimer's disease, vascular dementia and dementia with Lewy bodies. Br J Psychiatry. 1999, 174: 45-50. 10.1192/bjp.174.1.45.
Foy CM, Nicholas H, Hollingworth P, Boothby H, Willams J, Brown RG, Al-Sarraj S, Lovestone S: Diagnosing Alzheimer's disease – non-clinicians and computerised algorithms together are as accurate as the best clinical practice. Int J Geriatr Psychiatry. 2007, 22 (11): 1154-1163. 10.1002/gps.1810.
Folstein MF, Folstein SE, McHugh PR: "Mini-mental state". A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975, 12 (3): 189-198. 10.1016/0022-3956(75)90026-6.
Roth M, Tym E, Mountjoy CQ, Huppert FA, Hendrie H, Verma S, Goddard R: CAMDEX. A standardised instrument for the diagnosis of mental disorder in the elderly with special reference to the early detection of dementia. Br J Psychiatry. 1986, 149: 698-709. 10.1192/bjp.149.6.698.
Blessed G, Tomlinson BE, Roth M: The association between quantitative measures of dementia and of senile change in the cerebral grey matter of elderly subjects. Br J Psychiatry. 1968, 114 (512): 797-811. 10.1192/bjp.114.512.797.
Bucks RS, Ashworth DL, Wilcock GK, Siegfried K: Assessment of activities of daily living in dementia: development of the Bristol Activities of Daily Living Scale. Age Ageing. 1996, 25 (2): 113-120. 10.1093/ageing/25.2.113.
Webster DD: Critical analysis of the disability in Parkinson's disease. Mod Treat. 1968, 5 (2): 257-282.
Reisberg B, Ferris SH, de Leon MJ, Crook T: Global Deterioration Scale (GDS). Psychopharmacol Bull. 1988, 24 (4): 661-663.
Alexopoulos GS, Abrams RC, Young RC, Shamoian CA: Cornell Scale for Depression in Dementia. Biol Psychiatry. 1988, 23 (3): 271-284. 10.1016/0006-3223(88)90038-8.
Cummings JL: The Neuropsychiatric Inventory: assessing psychopathology in dementia patients. Neurology. 1997, 48 (5 Suppl 6): S10-16.
Sheikh JI, Yesavage JA: A knowledge assessment test for geriatric psychiatry. Hosp Community Psychiatry. 1985, 36 (11): 1160-1166.
Macgregor S: Most pooling variation in array-based DNA pooling is attributable to array error rather than pool construction error. Eur J Hum Genet. 2007, 15 (4): 501-504. 10.1038/sj.ejhg.5201768.
Hoogendoorn B, Norton N, Kirov G, Williams N, Hamshere ML, Spurlock G, Austin J, Stephens MK, Buckland PR, Owen MJ, et al: Cheap, accurate and rapid allele frequency estimation of single nucleotide polymorphisms by primer extension and DHPLC in DNA pools. Hum Genet. 2000, 107 (5): 488-493. 10.1007/s004390000397.
Kirov G, Zaharieva I, Georgieva L, Moskvina V, Nikolov I, Cichon S, Hillmer A, Toncheva D, Owen MJ, O'Donovan MC: A genome-wide association study in 574 schizophrenia trios using DNA pooling. Mol Psychiatry. 2008
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007, 81 (3): 559-575. 10.1086/519795.
Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005, 21 (2): 263-265. 10.1093/bioinformatics/bth457.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1755-8794/1/44/prepub
Acknowledgements
We would like to thank the patients and their families who participated in this study. We acknowledge our collaborators John Powell and Simon Lovestone, Institute of Psychiatry, London and both Carol Brayne and David Rubinsztein at the Cambridge Institute for Medical Research, University of Cambridge for the supervision of sample collection.
This research was supported by funding from the Alzheimer's Research Trust (grant to GK, JW, MJO and MCO'D: ART/PPG2006B/5), the Medical Research Council (UK), and the Welsh Assembly Government.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
All authors read and approved the final manuscript. GK conceived of and designed the study, analysed pooled genotyping and contributed to writing of manuscript. LG, RS and AM assisted with DNA pool construction and individual genotyping. KD and PH were responsible for patient/control diagnosis and sample collection. SC and AH conducted pooled genome-wide genotyping. VM analysed pooled genotype data, designing combined Z-test and conducted test for association using 1958 control group. MO'D contributed to experimental design and writing of manuscript. JW and MO supervised and contributed to sample collection, interpretation of data and writing of manuscript. RA took the lead in DNA pool construction, writing the manuscript, conducted and analysed individual genotyping.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Abraham, R., Moskvina, V., Sims, R. et al. A genome-wide association study for late-onset Alzheimer's disease using DNA pooling. BMC Med Genomics 1, 44 (2008). https://doi.org/10.1186/1755-8794-1-44
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1755-8794-1-44