Abstract
Background
The emerging 3q13.31 microdeletion syndrome appears to encompass diverse neurodevelopmental conditions. However, the 3q13.31 deletion is rare and few adult cases have yet been reported. We examined a cohort with schizophrenia (n = 459) and adult control subjects (n = 26,826) using high-resolution microarray technology for deletions and duplications at the 3q13.31 locus.
Results
We report on the extended adult phenotype associated with a 3q13.31 microdeletion in a 41-year-old male proband with schizophrenia and a nonverbal learning disability. He was noted to have a speech impairment, delayed motor skills, and other features consistent with the 3q13.31 microdeletion syndrome. The 2.06 Mb deletion overlapped two microRNAs and seven RefSeq genes, including GAP43, LSAMP, DRD3, and ZBTB20. No overlapping 3q13.31 deletions or duplications were identified in control subjects.
Conclusions
Later-onset conditions like schizophrenia are increasingly associated with rare copy number variations and associated genomic disorders like the 3q13.31 microdeletion syndrome. Detailed phenotype information across the lifespan facilitates genotype-phenotype correlations, accurate genetic counselling, and anticipatory care.
Similar content being viewed by others
Background
Preliminary evidence suggests that individuals with microdeletions at chromosome 3q13.31 (OMIM #615433) may be predisposed to a broad spectrum of neurodevelopmental/neuropsychiatric conditions including global developmental delay/intellectual disability (DD/ID), speech delay, autism spectrum disorder (ASD), and attention deficit hyperactivity disorder (ADHD) [1–5]. Inconsistent molecular and cytogenetic techniques as well as variable genomic breakpoints have made pinpointing the responsible gene(s) within the 3q13.31 region challenging. In a large case series, Molin et al. defined a 580 kb shortest region of overlap (SRO) that includes five RefSeq genes: DRD3, ZNF80, TIGIT, MIR568, and ZBTB20[1]. Those authors proposed that haploinsufficiency of DRD3 (OMIM #126451), which codes for the D3 subunit of the dopamine receptor, and ZBTB20 (OMIM #606025), a DNA transcription repressor expressed in hippocampal neurons, may contribute to the neurodevelopmental features of the 3q13.31 microdeletion syndrome [1, 6]. Recently, a 1.3 Mb deletion at 3q13.31 downstream of this proposed SRO and encompassing just two genes (LSAMP, GAP43) was identified in a 7-year-old female with ADHD, hypotonia, and postnatal growth above the mean [3]. LSAMP (OMIM #603241) encodes a limbic system-associated membrane protein (LAMP) and has been shown to regulate anxiety-like phenotypes in mice [7]. GAP43 (OMIM #162060) is almost exclusively expressed in neuronal tissue and is a candidate gene for ASD [8].
Later-onset conditions, including neurodevelopmental diseases like schizophrenia, may also be associated with rare copy number variations (CNVs) [9]. To date, few adult cases with 3q13.31 deletions have been described in the literature, highlighting the paucity of phenotypic data across the lifespan needed to inform genetic counselling and anticipatory care. Here we provide a description of the extended adult phenotype associated with a 3q13.31 microdeletion [9] that encompasses all four of the previously proposed neurodevelopmental candidate genes at this locus: DRD3, ZBTB20, LSAMP, and GAP43.
Case presentation
Clinical description
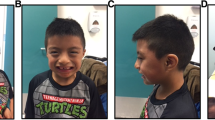
The male proband (Figure 1) was conceived naturally to non-consanguineous parents of European ancestry, a 29-year-old mother and 36-year-old father. There was no known family history of major developmental or neuropsychiatric conditions. The pregnancy and term delivery were unremarkable; birth weight was 3,941 g (75th-90th percentile). There was no evidence of hypotonia. The patient was noted to have delayed motor developmental milestones and “clumsiness” prior to age 3 years, when he was hospitalized for suspected viral meningoencephalitis with decreased level of consciousness and dystonic movements. A speech impairment and enuresis was noted thereafter and persisted into adulthood. At age 9 years, fine motor and perceptual motor skills were noted to be under-developed. A head CT (computed tomography) scan at age 15 years showed focal hypoplasia of the superior cerebellar vermis; a repeat CT scan at age 33 years was read as normal.
Physical features of an adult male with a 3q13.31 microdeletion. Anthropometric data demonstrated a post-natal growth pattern above the mean: at 31 years height was 181.5 cm (75th-90th percentile), weight was 90.9 kg (BMI 27.6), and OFC was 60.9 cm (97th percentile). Findings noted on physical examination as an adult included a broad neck and slight kyphosis. Craniofacial features included facial asymmetry, strabismus, narrow palpebral fissures, a long philtrum, a narrow high arched palate, flat occiput, and frontal and occipital hair whorls. He also had bilateral cubitus valgus, short 4th and 5th metacarpals bilaterally, as well as narrow feet with mild clinodactyly bilaterally of all toes and camptodactyly bilaterally of 4th and 5th toes.
He was enrolled in a special education program beginning at age 7 years, with particular difficulties noted in mathematics and writing. At age 33 years, clinical neuropsychological testing revealed a full-scale IQ in the borderline range. Research-based testing at age 39 years using the WASI (Wechsler Abbreviated Scale of Intelligence) demonstrated a full-scale IQ of 75, with a marked difference between verbal (90) and performance (62) scores consistent with a nonverbal learning disability.
The patient developed schizophrenia and was first treated with standard antipsychotic medications at age 24 years. Overall, he has had an unremarkable course of illness (details available upon request); of note, aggressive behaviour at age 28 years prompted a change in his medication regimen. His past medical history also includes type 2 diabetes mellitus treated with insulin and hypercholesterolemia diagnosed in his 20′s, mild hypocalcemia first identified at age 31 years, bilateral L5-S1 disc protrusion requiring surgery at 34 years, and hypertension diagnosed at age 34 years. At time of last contact, he was 41 years old and living in a supported situation in the community.
Molecular studies
The patient was recruited into a longitudinal study of the genetics of schizophrenia [9] at age 31 years. Research chromosomal microarray analysis using the Affymetrix® Genome-Wide Human SNP Array 6.0 ultimately demonstrated a deletion at cytoband 3q13.31 (chr3:115,308,450–117,370,859, hg18) [9], which was clinically confirmed at a CLIA (Clinical Laboratory Improvement Amendments) approved laboratory and reported back to the patient. All stringent genome-wide CNV calls in this individual are included in Additional file 1: Table S1 with only the 3q13.31 deletion predicted to be pathogenic [10]. As is common with adult-onset disorders like schizophrenia, both parents were unavailable for testing (deceased). The deletion overlaps two microRNAs (miRNAs) and seven RefSeq genes (Figure 2), including the four promising neurodevelopmental gene candidates highlighted by Gimelli et al. (GAP43 and LSAMP) and Molin et al. (DRD3 and ZBTB20) [1, 3]. The 3q13.31 region was inspected for CNVs in the DECIPHER (http://decipher.sanger.ac.uk) database and 10 patients were found to have deletions overlapping the one presented in this case report (Figure 2) [11]. In the 26,826 adult control subjects with genome-wide CNV data available to our colleagues at The Centre for Applied Genomics (TCAG), no similar 3q13.31 deletions or duplications were identified using a 50% reciprocal overlap criterion (Additional file 2: Table S2) [9, 12].
3q13.31 deletions and duplications overlapping neuropsychiatric candidate genes DRD3 , ZBTB20 , LSAMP, and GAP43. The image was modified from the Database of Genomic Variants (http://dgv.tcag.ca), NCBI Build 36 (hg 18) [13]. Only deletions and duplications <5 Mb in size and overlapping at least one of DRD3, GAP43, ZBTB20, and LSAMP are shown. Deletions and duplications are represented by red and blue bars, respectively. All genes (purple) and miRNAs (orange) within the 3q13.2-q13.31 region are shown; splice variants are not included. The grey bar outlines the coordinates for the 580 kb (chr3:115,335,356-115,916,848) shortest region of overlap (SRO) defined by Molin et al. [1]. The purple bar outlines the boundaries for the Gimelli deletion (chr3:116,640,577- 118,002,810, hg 18) that overlapped GAP43 and LSAMP[3]. The 4.5 Mb deletion reported by Wisniowiecka-Kowalnik [14], the 4.57 Mb deletion in DECIPHER Case 258860, and the 3.4 Mb duplication by Vuillaume [4] extend beyond the boundary of the figure and are represented by line breaks. Where necessary, all deletion and duplication coordinates (i.e., excluding the Molin et al. SRO and the case described in this report) were converted from hg 19 to hg 18 using The UCSC Genome LiftOver tool (http://genome.ucsc.edu).
Discussion
3q13.31 deletion expression in adults
Including the present case, four adults (≥18 years) with 3q13.31 microdeletions have been reported in the literature to date (Table 1) [1, 2]. Several features present in this patient, including DD/ID, speech delay, enuresis, postnatal growth above the mean, structural brain abnormalities, high arched palate, and skeletal anomalies, are consistent with previous reports of adults with 3q13.31 microdeletions (Table 1) [1, 2]. Of all forty-one cases previously in the literature, the 3q13.31 deletion was reported as occurring de novo in thirty-eight (92.7%) and of unknown inheritance in three (7.3%) [1–4, 14, 15].
Neuropsychiatric phenotypes (i.e., ADHD and ASD) have been previously associated with the 3q13.31 microdeletion; however, this is the first report of schizophrenia in an individual with a 3q13.31 microdeletion. Moreover, the patient described in this report had a >25 point discrepancy between his performance and verbal IQ scores, consistent with a nonverbal learning disability [16]. Interestingly, the 42 year old female (Case 6) reported in Shuvarikov et al. [2] demonstrated a similar trend in IQ scores (Table 1). Raw IQ scores were not given for many of the paediatric cases, however Vuillaume et al. [4] described an affected 16 year old female (Case 1) as “being able to read and write but educational learning was difficult,” potentially describing a nonverbal learning disability. Detailed neuropsychological phenotyping of additional cases will help determine if nonverbal learning disabilities are part of the emerging 3q13.31 microdeletion syndrome.
Candidate genes for neuropsychiatric expression
The four genes (GAP43, DRD3, LSAMP, and ZBTB20) that have been posited as contributing to the brain phenotype of the 3q13.31 microdeletion syndrome are all overlapped by this patient's deletion (Figure 2) [1, 2]. Regarding these genes and their potential role in schizophrenia and related neuropsychiatric conditions, non-synonymous point mutations in GAP43 were recently identified in two unrelated schizophrenia cases in a next-generation sequencing study [17]. DRD3 has been a longstanding candidate gene for schizophrenia, largely based on its affinity to bind antipsychotic drugs and its localization in limbic brain structures [18]. Further, LSAMP and ZBTB20 have each been implicated in various brain regions associated with schizophrenia. In a post-mortem study of schizophrenia, LSAMP expression was noted to be increased by ~20% in the dorsolateral prefrontal cortex of individuals with schizophrenia compared to controls [19]. In mice, ZBTB20 knockdowns were noted to have faulty hippocampal cytoarchitecture and selective ablation of ZBTB20 in mature hippocampal CA1 neurons led to disruptions in learning and memory [20, 21].
In addition to genic haploinsufficiency, other molecular mechanisms may contribute to the phenotype of the 3q13.31 microdeletion syndrome. In particular, recent reports suggest miRNAs may play a role in mediating the risk for neurodevelopmental disorders [22, 23]. Both miRNAs overlapped by the 3q13.31 deletion in this patient (miR-4796, miR-568) have predicted targets that are additional candidate genes for schizophrenia and related disorders, including SHANK2 and FMR1[24, 25]. The individual and collective influence of the above mentioned genes and miRNAs on the neuropsychiatric expression of the 3q13.31 microdeletion is deserving of further study. Of interest with respect to genotype-phenotype correlations are the few reports to date of the reciprocal 3q13.31 duplication, in which individuals appear to share some (i.e., DD/ID, hypotonia) but not all of the same clinical features as the 3q13.31 microdeletion [4, 26]. The fact that neither deletions nor duplications at 3q13.31 were identified in 26,826 controls suggest that copy number aberrations in this region, rather than haploinsufficiency alone, may be associated with the deleterious phenotypic consequences.
Implications for clinical practice
Cytogenetic anomalies may be found in up to 5-8% of cases with schizophrenia, suggesting a potential future role for clinical microarray testing in this population [9]. The occurrence of schizophrenia in the patient reported here could be unrelated to the 3q13.31 microdeletion. However, multiple lines of evidence, including the variable expression of many other recurrent large, rare CNVs, suggest a genetically-related neuropsychiatric spectrum of disease that includes both DD/ID and schizophrenia [9, 27, 28]. This is, to our knowledge, the first report of a 3q13.31 deletion discovered in a schizophrenia cohort. This attests to the overall rarity of these variants, and to the relative paucity of available data (almost all of which are research-based) compared with diseases like DD/ID where clinical microarray testing is now the first-tier diagnostic test [9, 29]. More data are needed to delineate the role of pathogenic CNVs in the dual-diagnosis (schizophrenia and premorbid ID) population to which this patient would belong, where the yield may be significantly higher and where clinical testing is already indicated [30].
Conclusions
In conclusion, we have identified an adult male with schizophrenia and a 3q13.31 deletion overlapping four promising neurodevelopmental candidate genes: DRD3, ZBTB20, GAP43, and LSAMP. Later-onset conditions like schizophrenia are increasingly associated with rare CNVs. Detailed phenotypic information across the lifespan facilitates genotype-phenotype correlations, accurate genetic counselling, and anticipatory care.
Consent
Written informed consent was obtained from the patient for publication of this Case report and any accompanying images. A copy of the written consent is available for review through the Editor-in-Chief of this journal.
Abbreviations
- DD/ID:
-
Developmental delay/intellectual disability
- ASD:
-
Autism spectrum disorder
- ADHD:
-
Attention deficit hyperactivity disorder
- SRO:
-
Shortest region of overlap
- CNV:
-
Copy number variation
- CT:
-
Computed tomography
- IQ:
-
Intelligence quotient
- WASI:
-
Wechsler Abbreviated Scale of Intelligence
- CLIA:
-
Clinical Laboratory Improvement Ammendments
- miRNA:
-
microRNA
- TCAG:
-
The Centre for Applied Genomics.
References
Molin AM, Andrieux J, Koolen DA, Malan V, Carella M, Colleaux L, Cormier-Daire V, David A, de Leeuw N, Delobel B, Duban-Bedu B, Fischetto R, Flinter F, Kjaergaard S, Kok F, Krepischi AC, Le Caignec C, Ogilvie CM, Maia S, Mathieu-Dramard M, Munnich A, Palumbo O, Papadia F, Pfundt R, Reardon W, Receveur A, Rio M, Ronsboro Darling L, Rosenberg C, Sa J, et al.: A novel microdeletion syndrome at 3q13.31 characterised by developmental delay, postnatal overgrowth, hypoplastic male genitals, and characteristic facial features. J Med Genet 2012,49(2):104–109. 10.1136/jmedgenet-2011-100534
Shuvarikov A, Campbell IM, Dittwald P, Neill NJ, Bialer MG, Moore C, Wheeler PG, Wallace SE, Hannibal MC, Murray MF, Giovanni MA, Terespolsky D, Sodhi S, Cassina M, Viskochi D, Moghaddam B, Herman K, Brown CW, Beck CR, Gambin A, Cheung SW, Patel A, Lamb AN, Shaffer LG, Ellison JW, Ravnan JB, Stankiewicz P, Rosenfeld JA: Recurrent HERV-H-mediated 3q13.2-q13.31 deletions cause a syndrome of hypotonia and motor, language, and cognitive delays. Hum Mutat 2013,34(10):1415–1423. 10.1002/humu.22384
Gimelli S, Leoni M, Di Rocco M, Caridi G, Porta S, Cuoco C, Gimelli G, Tassano E: A rare 3q13.31 microdeletion including GAP43 and LSAMP genes. Mol Cytogenet 2013,6(1):52. 10.1186/1755-8166-6-52
Vuillaume ML, Delrue MA, Naudion S, Toutain J, Fergelot P, Arveiler B, Lacombe D, Rooryck C: Expanding the clinical phenotype at the 3q13.31 locus with a new case of microdeletion and first characterization of the reciprocal duplication. Mol Genet Metab 2013,110(1–2):90–97.
Shimojima K, Saito K, Yamamoto T: A de novo 1.9-Mb interstitial deletion of 3q13.2q13.31 in a girl with dysmorphic features, muscle hypotonia, and developmental delay. Am J Med Genet A 2009,149A(8):1818–1822. 10.1002/ajmg.a.32963
Nielsen JV, Thomassen M, Mollgard K, Noraberg J, Jensen NA: Zbtb20 defines a hippocampal neuronal identity through direct repression of genes that control projection neuron development in the isocortex. Cereb Cortex 2013. (Epub ahead of print)
Innos J, Koido K, Philips MA, Vasar E: Limbic system associated membrane protein as a potential target for neuropsychiatric disorders. Front Pharmacol 2013, 4: 32.
Allen-Brady K, Miller J, Matsunami N, Stevens J, Block H, Farley M, Krasny L, Pingree C, Lainhart J, Leppert M, MacMahon WM, Coon H: A high-density SNP genome-wide linkage scan in a large autism extended pedigree. Mol Psychiatry 2009,14(6):590–600. 10.1038/mp.2008.14
Costain G, Lionel AC, Merico D, Forsythe P, Russell K, Lowther C, Yuen T, Husted J, Stavropoulos DJ, Speevak M, Chow EW, Marshall CR, Scherer SW, Bassett AS: Pathogenic rare copy number variants in community-based schizophrenia suggest a potential role for clinical microarrays. Hum Mol Genet 2013,22(22):4485–4501. 10.1093/hmg/ddt297
Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST, Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee: American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med 2011,13(7):680–685. 10.1097/GIM.0b013e3182217a3a
Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, Carter NP: DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet 2009,84(4):524–533. 10.1016/j.ajhg.2009.03.010
Lionel AC, Crosbie J, Barbosa N, Goodale T, Thiruvahindrapuram B, Rickaby J, Gazzellone M, Carson AR, Howe JL, Wang Z, Wei J, Stewart AF, Roberts R, McPherson R, Fiebig A, Franke A, Schreiber S, Zwaigenbaum L, Fernandez BA, Roberts W, Arnold PD, Szatmari P, Marshall CR, Schachar R, Scherer SW: Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci Transl Med 2011,3(95):95ra75.
Macdonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW: The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 2014,42(1):D986-D992.
Wisniowiecka-Kowalnik B, Kastory-Bronowska M, Bartnik M, Derwinska K, Dymczak-Domini W, Szumbarska D, Ziemka E, Szczaluba K, Sykulski M, Gambin T, Shaw CA, Mazurczak T, Obersztyn E, Bocian E, Stankiewicz P: Application of custom-designed oligonucleotide array CGH in 145 patients with autistic spectrum disorders. Eur J Hum Genet 2013,21(6):620–625. 10.1038/ejhg.2012.219
Materna-Kiryluk A, Kiryluk K, Burgess KE, Bieleninik A, Sanna-Cherchi S, Gharavi AG, Latos-Bielenska A: The emerging role of genomics in the diagnosis and workup of congenital urinary tract defects: a novel deletion syndrome on chromosome 3q13.31–22.1. Pediatr Nephrol 2014,29(2):257–267. 10.1007/s00467-013-2625-2
Rourke BP, Ahmad SA, Collins DW, Hayman-Abello BA, Hayman-Abello SE, Warriner EM: Child clinical/pediatric neuropsychology: some recent advances. Annu Rev Psychol 2002, 53: 309–339. 10.1146/annurev.psych.53.100901.135204
Need AC, McEvoy JP, Gennarelli M, Heinzen EL, Ge D, Maia JM, Shianna KV, He M, Cirulli ET, Gumbs CE, Zhao Q, Campbell CR, Hong L, Rosenquist P, Putkonen A, Hallikainen T, Repo-Tiihonen E, Tiihonen J, Levy DL, Meitzer HY, Goldstein DB: Exome sequencing followed by large-scale genotyping suggests a limited role for moderately rare risk factors of strong effect in schizophrenia. Am J Hum Genet 2012,91(2):303–312. 10.1016/j.ajhg.2012.06.018
Cho DI, Zheng M, Kim KM: Current perspectives on the selective regulation of dopamine D(2) and D(3) receptors. Arch Pharm Res 2010,33(10):1521–1538. 10.1007/s12272-010-1005-8
Behan AT, Byrne C, Dunn MJ, Cagney G, Cotter DR: Proteomic analysis of membrane microdomain-associated proteins in the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder reveals alterations in LAMP, STXBP1 and BASP1 protein expression. Mol Psychiatry 2009,14(6):601–613. 10.1038/mp.2008.7
Ren A, Zhang H, Xie Z, Ma X, Ji W, He DZ, Yuan W, Ding YQ, Zhang XH, Zhang WJ: Regulation of hippocampus-dependent memory by the zinc finger protein Zbtb20 in mature CA1 neurons. Physiol 2012,590(Pt 19):4917–4932.
Xie Z, Ma X, Ji W, Zhou G, Lu Y, Xiang Z, Wang YX, Zhang L, Hu Y, Ding YQ, Zhang WJ: Zbtb20 is essential for the specification of CA1 field identity in the developing hippocampus. Proc Natl Acad Sci U S A 2010,107(14):6510–6515. 10.1073/pnas.0912315107
Brzustowicz LM, Bassett AS: miRNA-mediated risk for schizophrenia in 22q11.2 deletion syndrome. Front Genet 2012, 3: 291.
Mellios N, Sur M: The emerging role of microRNAs in schizophrenia and autism spectrum disorders. Front Psychiatry 2012, 3: 39.
Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, Handsaker RE, McCarroll SA, O’Donnovan MC, Owen MJ, Kirov G, Sullivan PF, Hultman CM, Sklar P, Purcell SM: De novo mutations in schizophrenia implicate synaptic networks. Nature 2014,506(7487):179–184. 10.1038/nature12929
TargetScanHuman: prediction of microRNA targets http://targetscan.org/
Karavitakis E, Kitsiou-Tzeli S, Xaidara A, Kosma K, Makrythanasis P, Apazidou E, Kanavakis E, Tzetis M: Microduplication 3q13.2q13.31 identified in a male with dysmorphic features and multiple congenital anomalies. Am J Med Genet A 2013,164(3):666–670.
Costain G, Bassett AS: Clinical applications of schizophrenia genetics: genetic diagnosis, risk, and counseling in the molecular era. App Clin Genet 2012, 5: 1–18.
Bassett AS, Scherer SW, Brzustowicz LM: Copy number variations in schizophrenia: critical review and new perspectives on concepts of genetics and disease. Am J Psychiatry 2010,167(8):899–914. 10.1176/appi.ajp.2009.09071016
Sahoo T, Theisen A, Rosenfeld JA, Lamb AN, Ravnan JB, Schultz RA, Torchia BS, Neill N, Casci I, Bejjani BA, Shaffer LG: Copy number variants of schizophrenia susceptibility loci are associated with a spectrum of speech and developmental delays and behavior problems. Genet Med 2011,13(10):868–880. 10.1097/GIM.0b013e3182217a06
Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucette WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg CM, Scherer SW, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS, et al.: Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010,86(5):749–764. 10.1016/j.ajhg.2010.04.006
Acknowledgements
The authors thank the patient for his participation, research assistants and staff at Saint John Community Mental Health Services and the Saint John Regional Hospital, and staff at The Centre for Applied Genomics (TCAG). The authors would also like to thank the anonymous reviewers for their valuable feedback. This study was supported by Canadian Institutes of Health Research (CIHR) grants (MOP-111238 and MOP-89066 to A.S.B.). A.S.B. holds the Canada Research Chair in Schizophrenia Genetics and Genomic Disorders, and the Dalglish Chair in 22q11.2 Deletion Syndrome.
Control datasets were obtained, along with permission for use, from the database of Genotypes and Phenotypes (dbGaP) found at http://ncbi.nlm.nih.gov/gap through accession numbers phs000143.v1.p1 (Starr County Health Studies’ Genetics of Diabetes Study), phs000091.v2.p1 (GENEVA NHS/HPFS Diabetes study), phs000169.v1.p1 (Whole Genome Association Study of Visceral Adiposity in the HABC Study), phs000303.v1.p1 (Genetic Epidemiology of Refractive Error in the KORA Study) and phs000404.v1.p1 (COGEND; The Genetic Architecture of Smoking and Smoking Cessation). The Starr County Health Studies Genetics of Diabetes Study was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and the NIDDK Central Repositories. Support for the GWAS of Gene and Environment Initiatives in Type 2 Diabetes was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01HG004399). The human subjects participating in the GWAS derive from The Nurses’ Health Study and Health Professionals’ Follow-up Study and these studies are supported by National Institutes of Health (NIH) grants CA87969, CA55075 and DK58845. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the Gene Environment Association Studies, GENEVA Coordinating Center (U01 HG004446) and the National Center for Biotechnology Information. Support for genotyping, which was performed at the Broad Institute of MIT and Harvard, was provided by the NIH GEI (U01HG004424). Support for the ‘CIDR Visceral Adiposity Study’ was provided through the Division of Aging Biology and the Division of Geriatrics and Clinical Gerontology, National Institute on Aging. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by Health ABC Study (HABC) Investigators. The KORA dataset was obtained from the NEI Refractive Error Collaboration (NEIREC) Database, support for which was provided by the National Eye Institute. Support for genotyping of the COGEND samples, which was performed at the Center for Inherited Disease Research (CIDR), was provided by 1X01 HG005274-01. Assistance with genotype cleaning of the COGEND samples, as well as with general study coordination, was provided by the Gene Environment Association Studies (GENEVA) Coordinating Center (U01HG004446). Support for the collection of COGEND datasets and samples were provided by the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392) and the University of Wisconsin Transdisciplinary Tobacco Use Research Center (P50 DA019706, P50 CA084724).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
SWS is on the Scientific Advisory Board of Population Diagnostics, Inc. and is a co-founder of YouNique Genomics. The other authors declare no conflicts of interest.
Authors’ contributions
CL organized the clinical data and drafted and revised the manuscript. RM organized the clinical data for the manuscript. DJS carried out the clinical cytogenetic studies. ACL, CRM, and SWS carried out the molecular studies. GC carried out the study and helped draft and revise the manuscript. ASB conceived, designed, carried out the study, reviewed the detailed phenotypic data, and helped draft the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
13039_2013_370_MOESM1_ESM.docx
Additional file 1: Table S1: Genome-wide copy number variation detected by Affymetrix 6.0 microarray in the proband with a 3q13.31 deletion described in this report. (DOCX 25 KB)
13039_2013_370_MOESM2_ESM.docx
Additional file 2: Table S2: Overview and negative results of 13 control datasets (total n=26,826) searched for 3q13.31 deletions and duplications. (DOCX 15 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Lowther, C., Costain, G., Melvin, R. et al. Adult expression of a 3q13.31 microdeletion. Mol Cytogenet 7, 23 (2014). https://doi.org/10.1186/1755-8166-7-23
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1755-8166-7-23