Abstract
Background
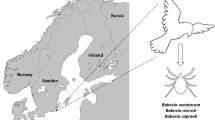
Bovine babesiosis is regarded as a limited health problem for Norwegian cows, and the incidence has decreased markedly since the 1930s. Rare cases of babesiosis in splenectomised humans from infection with Babesia divergens and B.venatorum have been described. The objective of this study was to determine whether birds can introduce Babesia-infected ticks. There are between 30 and 85 million passerine birds that migrate to Norway every spring.
Methods
Passerine birds were examined for ticks at four bird observatories along the southern Norwegian coast during the spring migrations of 2003, 2004 and 2005. The presence of Babesia was detected in the nymphs of Ixodes ricinus by real-time PCR. Positive samples were confirmed using PCR, cloning and phylogenetic analyses.
Results
Of 512 ticks examined, real-time PCR revealed five to be positive (1.0%). Of these, four generated products that indicated the presence of Babesia spp.; each of these were confirmed to be from Babesia venatorum (EU1). Two of the four B. venatorum-positive ticks were caught from birds having an eastern migratory route (P< 0.001).
Conclusions
Birds transport millions of ticks across the North Sea, the Skagerrak and the Kattegat every year. Thus, even with the low prevalence of Babesia-infected ticks, a substantial number of infected ticks will be transported into Norway each year. Therefore, there is a continuous risk for introduction of new Babesia spp. into areas where I. ricinus can survive.
Similar content being viewed by others
Background
Ticks have limited mobility [1], but can be transported over long distances when feeding on their vertebrate hosts. In particular, avian hosts can efficiently transport ticks across geographical barriers such as oceans and deserts [2–6].
Norway is in the northernmost range for the distribution of Ixodes ricinus, which is a vector for many human and animal pathogens [7]. In a review by Gorenflot [8], 83% of the 28 European cases of human babesiosis occured in splenectomised individuals, and 76% were reported to be due to Babesia divergens. Apicomplexans of the genus Babesia have greater vertebrate host specificity than other tick-borne pathogens, such as Borrelia spp. and Anaplasma phagocytophilum. None of the described Babesia species can have both bird and mammal hosts [9, 10]. The first report of Babesia canis in Norway occurred recently; the tick was found on a dog that had not been abroad [11]. The main vector for B. canis is Dermacentor reticulatus, which is not native to Norway. However, a larva of Dermacentor sp. was recovered on a northward-migrating willow warbler (Phylloscopus trochilus) in Akeröya in 2005 [6]. B. divergens, which causes redwater disease in cattle, is widespread in Europe [12]. This pathogen was first described in Norway in 1901 [13] and has been recovered and confirmed in Norway, using molecular tools (Ø. Øines unpublished results). A comparison of the data of Thambs-Lyche from the 1930s [14] with data from the Norwegian Dairy Herd Recording System (NDHRS) indicates that the incidence of babesiosis in Norwegian cattle has decreased, although the infection remains highly prevalent in some coastal locations in Norway such as Lista (longitude E 6.7°) (47% of 34 tested cows had antibodies against B. divergens) and the island Jomfruland (longitude E 9.6°) (68%, N = 25) [15]. Lundsett tested 439 flagged ticks along the southern Norwegian coast (although not specifically in cow-pasturing areas) and found only a single tick that was positive for B. divergens by polymerase chain reaction (PCR) analysis [16]. Radzijevskaja and colleagues [17] found no Babesia sp. in 91 ticks (16 adults and 75 nymphs) collected from Jomfruland in Norway. There are no data to support the presence of other Babesia species related to B. divergens in Norway. However, in this study, we report the presence of B. venatorum (Babesia species, EU1) in ticks found on migratory birds. This species was first characterised by Herwaldt in 2003 [18] and has been recovered from roe deer in Slovenia [19] and France [20] and from I. ricinus[21, 22]. Three cases documenting infection with this species in splenectomised humans [18, 23] have raised concern for B. venatorum becoming an emerging tick-borne human disease.
The purpose of this study was to assess the prevalence of Babesia on ticks collected from northward migratory birds arriving in Norway and to document the potential for spread of this parasite across the sea. This is the first report finding Babesia on ticks from birds. In comparison, a Polish study [24] did not detect Babesia in 442 tick specimens collected from birds.
Materials and methods
Four Norwegian bird observatories along 300 km of the southern Norwegian coast were involved in the study; from west to east, these included Lista, Jomfruland, Store Færder and Akerøya [6]. At Akerøya, six hours of flagging yielded five ticks, and at Store Færder, two hours of flagging did not yield any ticks. Although the yield of ticks may vary through the day and through the season, our experience is that ticks are detectable at any time during spring, summer and autumn at localities where they occur. Therefore, it is questionable whether these two islands have any local tick populations. By contrast, ticks are common on Jomfruland and on the mainland locality of Lista [6]. There are pasturing cattle in Jomfruland and Lista, but there are only pasturing sheep at Akerøya or Store Færder, and no cattle or cervid animals. Altogether, 9,768 passerine birds on northward migration were caught with mist nets during the spring migrations of 2003, 2004 and 2005. The birds were examined for ticks around the eyes, beak and ear openings. The ticks were picked off with tweezers and placed in 70% ethanol for subsequent examination. One vial was used for each bird, and tweezers were sterilised with a flame between uses.
The ticks were examined using a stereomicroscope for species identification and for estimation of feeding status. In total, 1,440 nymphs and 517 larvae were found on 713 birds. Five species of ticks were found [6]. However, only Ixodes ricinus was included in this study. At the sites in Jomfruland and Lista, it was necessary to minimise the inclusion of ticks from resident birds. Therefore, for these two locations, we analysed only fully or almost fully engorged tick nymphs collected from birds caught before or during the peak migratory arrival time for each species, as engorgement is not evident during the first 24 hours following attachment [25]. To avoid including unengorged specimens, only obviously thickened nymphs were recorded as engorged. Because there were very few ticks on Akerøya and Store Færder, we examined all nymphs collected from birds at these two sites for pathogens. Larvae were not included in this study, as there was considerable uncertainty as to whether the reduced amounts of Babesia DNA obtained from larvae, in comparison to that from nymphs, would alter the sensitivity of the test. In total, 512 ticks were selected for examination. Of these, 332 were engorged and were from all the locations and 180 were unengorged or slightly engorged and were from the two locations assumed not to have resident tick populations.
The specimens were crushed with the tip of a glass rod, which was discarded after each use, and DNA was isolated using a spin-column method, using the DNeasy Blood & Tissue Kit (Qiagen), according to the manufacturer's protocol. To avoid interspecimen contamination, the ticks were handled with tweezers that were flame-decontaminated after each use. Engorged and unengorged ticks were treated using the same methods. A contract laboratory, Telelab (now Unilabs Telelab, Skien, Norway), performed the initial screening of the samples using real-time PCR to detect B. divergens DNA published by Radzijevskaja et al. [17]. The PCR mix consisted of 5 μL BdiF (3 pmol/μL), 5 μL BdiR (9 pmol/μL), 2 μL BdiT (5 pmol/μL), 8 μL PCR water and 25 μL TaqMan Universal PCR Mastermix (Applied Biosystems, Foster City, CA, USA) in a total volume of 45 μL per sample, with 5 μL of sample DNA or control(dH2O). After heat activation at 93°C for 10 minutes, the two-step PCR protocol consisting of the following steps ran for 40 cycles: denaturation at 94°C for 15 seconds and annealing/extension at 60°C for 60 seconds. Signals were read using an Applied Biosystems AB7000 sequence-detection system and were calculated against a dilution series of synthetic amplicon DNA supplied by Applied Biosystems and standards of B. divergens DNA (30,000, 3,000, 300 and 30 DNA copies). Only signals above the threshold line (ΔRn = 1e-1) at or before cycle (Ct) 37 were interpreted as a positive. DNA from the five positive samples were then used for the two separate nested PCR reactions described by Zintl and collagues [26]. PCRs were performed with two positive controls (B. divergens and B. gibsoni) in each run. The multiple PCR products were cloned using One Shot® TOP-10 E.coli TA Cloning® by Invitrogen (Carlsbad, USA) and pCR®2.1 plasmid. Transformed bacteria were grown overnight at 37°C on LB agar plates containing 75 μg/ml ampicillin and X-gal. From each PCR sample that was cloned, 16 to 24 white colonies were selected and used as templates for a PCR reaction using M13-reverse and T7 primers. Several of these PCR products were sequenced using these primers at Macrogen Europe (http://www.macrogen.com). Chromatograms were edited, trimmed for vector contamination and assembled using Contig Express in Vector Nti Advance® 11.5 (Invitrogen, Carlsbad, USA). An initial BLASTN search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) for several of the sequence assemblages made in Contig Express revealed matches for sequences from Ixodes, uncultured eucaryotes and other non-Babesia targets. However, many of the sequence assemblages produced matches for Babesia. Cloned sequences from one of the five samples did not match any Babesia entries in BLASTN searches even after repeated PCR and cloning. The remaining four samples, however, contained cloned sequences matching Babesia 18S DNA from the databases. Each of the nested PCR products were assembled using high-quality chromatograms into a single sequence. Sequences extending beyond the nested PCR primers were deleted to avoid any confounding errors introduced by the primers. The two nested PCRs reported by Zintl and colleagues [26] target two regions of 18S DNA in separate PCRs (5' and the 3') and produce two non-overlapping sequences due to deletion of regions protruding from the annealing sites of the primers. The 472 bp 5' and 534 bp 3' sequences from each clone were found to contain no more than 3 nucleotide ambiguities (substitutions, deletions or polymorphisms) when compared to the sequence of Babesia EU1 GQ888709. This corresponded to a maximum difference of 0.6% from this 'backbone' sequence.
It is possible that the infrequent substitutions found in the individual chromatogrammes were due to errors from the Taq-polymerase or basecalling or that they may represent intra-host nucleotide differences from Babesia present in each sample. However, when applying the principle of the majority rule to the chromatograms, these substitutions would be eliminated hence producing sequences identical to the GQ888709 Genbank entry for B. venatorum (EU1). The two non-overlapping sequences from the separate nested PCR's were included in an alignment that consisted of 18S sequences from each of the following Babesia species: B. gibsoni (AB118032), B. odocolei (AY237638), B. divergens (AY572456), B. capreoli (AY726009), B. canis canis (AY072926), B canis vogeli (AY371198), B. canis rossi (DQ111760), B. kiwiensis (EF551335), B occultans (EU376017), B. bovis (AY150059), B. equi (AY150062), B. caballi (AY309955), B. motasi (AY533147), B. coco (AY618928) and two sequences from Babesia venatorum (EU1) (GQ888709 and HQ830266). The alignment was exported to MEGA 5 [27], and a Neighbor-joining tree was constructed using 500 bootstrap replicates with pairwise deletion of gaps and missing data. The nucleotides in positions 505-992 and 1090-1626, relative to the complete alignment, were included in a single phylogenetic analysis which used the sequence data produced by the two nested PCRs.
The National Board of Animal Experimentation approved the field collecting and handling of the birds.
Results
Thirty-three species of birds carrying I. ricinus nymphs were collected. Of the 512 nymphs investigated, 114 (22.2%), 98 (19.1%), 54 (10.5%), 40 (7.8%), 38 (7.4%), 31 (6.1%), 23 (4.5%), 22 (4.3%) and 14 (2.7%) were found on Erithacus rubecula, Turdus merula, Turdus philomelos, Phoenicurus phoenicurus, Prunella modolaris, Phylloscopus trochilus, Sylvia curruca, Turdus iliacus and Sylvia atricapilla, respectively. Only two ticks were found on Luscinia svecica and one on Phylloscopus trochiloides, and these are the only bird species in this material that have an eastern migratory route. Initial real time analysis revealed five specimens to be positive for the presence of Babesia sp. Subsequent PCR and cloning confirmed four specimens, collected from a robin (Erithacus rubecula) in Akerøya (cycle thresold: Ct = 30), a greenish warbler (Phylloscopus trochiloides) in Jomfruland (Ct = 24) and a bluethroat (Luscinia svecica) (Ct = 24) and a dunnock (Prunella modularis) from Store Færder (Ct = 27), to be positive. The disproportionality between Babesia-positive ticks caught from birds with an eastern versus a western migratory route was highly siginficant, i.e., P< 0.001 (Fisher's exact test for count data, run in the software R [28]).
Phylogenetic analysis of the sequences obtained after cloning of the samples using a Neighbor-joining tree indicated a very high support for the unknown sequence to belong to Babesia venatorum (EU1) (GQ888709) (Bootstrap > 99) (Figure 1).
Neighbour-joining tree of aligned 18S sequences after a bootstrap test with 500 replicates made using the Jukes-Cantor model [33]. The tree is based on the final dataset that included 1,025 base pair positions (472+534) of the 18S Babesia sequences. The percentage of replicate trees, in which the associated taxa clustered together in the bootstrap test (500 replicates), are shown next to the branches (only values above 50 are displayed). Sequences from the unknown Babesia sample, 'Sample x', cluster with B. venatorum (EU1) sequences that were included in the alignment. Evolutionary analyses were conducted in MEGA 5 [27].
Discussion
The real-time PCR used for screening was initially reported to be specific for B. divergens[17]. However, in this study we report the detection of B. venatorum using this method. In addition, the primer set and probe is capable of generating positive signals with DNA from other Babesia species, such as B. canis canis and B. gibsoni (Ø. Øines unpublished results). The fifth sample that was not confirmed to contain Babesia was only marginally positive in the real-time PCR, as it produced a signal close to the threshold line at 37 cycles, i.e., Ct = 36. This could indicate that either any Babesia DNA may have been present in this sample at concentrations that were too low to allow for verification using the procedures described previously, or, it could indicate that the cycle threshold (Ct) was set too high when selecting for positive samples. Hence positive signals would be false positives, due to unspecific products amplified in the real-time analysis.
Some bird species are partly migratory, with only a fraction of the individuals wintering in Norway [29]. Therefore, for a conservative approach, we only used engorged nymphs caught before or during the peak migratory arrival time in localities where ticks are endemic. In addition, three of the four bird species on which B. venatorum was found are not known to winter in Norway [29], and three Babesia samples were collected from birds on Akerøya and Store Færder (Table 1) where it seems to be no I. ricinus ticks [6]. Thus, it appears reasonable to conclude that the ticks infected by B. venatorum were brought to Norway by the birds they were feeding on. The low number of Babesia-positive nymphs was insufficient for analyses comparing differences in the prevalence of these nymphs by bird species. The robin and dunnock belong to the great majority of birds that migrate to Norway via the Atantic coast of continental Europe [29]. The greenish warbler does not breed west of Gothland, Sweden [30] and is very rarely encountered in Norway, and the bluethroat is one of the few birds that migrate to Norway via an eastern migratory route [29]. Thus, two of the four B. venatorum-positive ticks were collected from the minority of birds that came from Eastern Europe, indicating that B. venatorum may be more common in Eastern Europe.
B. venatorum (Babesia EU1) had previously not been reported in Norway. Thus, we did not expect to find this parasite and not B. divergens, which is a widespread and common parasite of cattle [12, 15]. Duh et al. found B. venatorum in 11 and B. divergens in 28 of 51 roe deers that were tested [19]. The host range for B. venatorum is poorly understood, and apart from splenectomised humans, a literature search on the Internet (http://www.scholar.google.com) revealed no natural hosts besides roe deer or any other tick vector than I. ricinus for B. venatorum.
Few B. venatorum-positive ticks were found on migratory birds in this study. There are 30-85 million passerine birds that migrate to Norway every spring [31], which may carry 0.2 ticks on average (nymphs and larvae) across the sea during their spring migration [6]. B. divergens has been shown to be transmitted transovarially and transstadially in the tick [32]. Therefore, larvae are as likely as nymphs to carry the pathogen. Assuming a prevalence of infection less than 1%, as was obtained in this study, birds could introduce on the order of 100,000 Babesia-infected ticks per year. Therefore, the spring migration represents an important component in the mechanism of the spread of tick-borne pathogens. Although theoretically possible, birds have not been shown to be responsible for the introduction of new tick-borne pathogens. This study shows that such introduction of B. venatorum is possible. Evidence suggests that the transport of ticks by migratory birds into Norway has increased over the last four decades [6], and it is possibile that B. canis, recently described for the first time in Norway [11], may have been introduced by ticks on migratory birds.
References
Korch GW: Geographic dissemination of tick-borne zoonoses. 1994, New York, Oxford: Oxford University Press
Hoogstraal H, Kaiser MN, Traylor MA, Gaber S, Guindy E: Ticks (Ixodoidea) on birds migrating from Africa to Europe and Asia. Bull World Health Organ. 1961, 24: 197-212.
Mehl R, Michaelsen J, Lid G: Ticks (Acari, Ixodides) on migratory birds in Norway. Fauna Norv Ser B. 1984, 31: 46-58.
Olsén B, Jaenson T, Bergstrom S: Prevalence of Borrelia burgdorferi sensu lato-infected ticks on migrating birds. Applied and Environmental Microbiology. 1995, 61: 3082-
Poupon M, Lommano E, Humair P, Douet V, Rais O, Schaad M, Jenni L, Gern L: Prevalence of Borrelia burgdorferi sensu lato in ticks collected from migratory birds in Switzerland. Applied and Environmental Microbiology. 2006, 72: 976-10.1128/AEM.72.1.976-979.2006.
Hasle G, Bjune G, Edvardsen E, Jakobsen C, Linnehol B, Røer J, Mehl R, Røed K, Pedersen J, Leinas H: Transport of ticks by migratory passerine birds to Norway. J Parasitol. 2009, 95: 1342-1351. 10.1645/GE-2146.1.
Estrada-Peña A, Jongejan F: Ticks feeding on humans: a review of records on human-biting Ixodoidea with special reference to pathogen transmission. Experimental and Applied Acarology. 1999, 23: 685-715. 10.1023/A:1006241108739.
Gorenflot A, Moubri K, Precigout E, Carcy B, Schetters TP: Human babesiosis. Annals of Tropical Medicine and Parasitology. 1998, 92: 489-501. 10.1080/00034989859465.
Levine N: Taxonomy of the piroplasms. Transactions of the American Microscopical Society. 1971, 90: 2-33. 10.2307/3224894.
Peirce M: A taxonomic review of avian piroplasms of the genus Babesia Starcovici, 1893 (Apicomplexa: Piroplasmorida: Babesiidae). Journal of Natural History. 2000, 34: 317-332. 10.1080/002229300299507.
Øines Ø, Storli K, Brun-Hansen H: First case of babesiosis caused by Babesia canis canis in a dog from Norway. Vet Parasitol. 2010, 171: 350-353. 10.1016/j.vetpar.2010.03.024.
Zintl A, Mulcahy G, Skerrett HE, Taylor SM, Gray JS: Babesia divergens, a bovine blood parasite of veterinary and zoonotic importance. Clin Microbiol Rev. 2003, 16: 622-636. 10.1128/CMR.16.4.622-636.2003.
Stuen S: Anaplasma phagocytophilum (formerly Ehrlichia phagocytophila) infection in sheep and wild ruminants in Norway. 2002, Oslo: Norwegian School of Veterinary Science
Thambs-Lyche H: Ixodes ricinus og piroplasmosen I Norge. Norsk Veterinærtidsskrift. 1943, 60: 337-366.
Hasle G, Bjune GA, Christensson D, Roed KH, Whist AC, Leinaas HP: Detection of Babesia divergens in southern Norway by using an immunofluorescence antibody test in cow sera. Acta Vet Scand. 2010, 52: 55-10.1186/1751-0147-52-55.
Lundsett AL: Flåtten Ixodes ricinus som sykdomsvektor i Sør-Norge. MSc dissertation. 2004, Telemark University College
Radzijevskaja J, Paulauskas A, Rosef O: Prevalence of Anaplasma phagocytophilum and Babesia divergens in Ixodes ricinus ticks from Lithuania and Norway. Int J Med Microbiol. 2008, 298: 218-221.
Herwaldt BL, Cacciò S, Gherlinzoni F, Aspöck H, Slemenda SB, Piccaluga PP, Martinelli G, Edelhofer R, Hollenstein U, Poletti G: Molecular characterization of a non-Babesia divergens organism causing zoonotic babesiosis in Europe. Emerging Infectious Diseases. 2003, 9: 943-
Duh D, Petrovec M, Bidovec A, Avsic-Zupanc T: Cervids as babesiae hosts, Slovenia. Shouxi. 2005, 11:
Bonnet S, Jouglin M, L'Hostis M, Chauvin A: Babesia sp. EU1 from Roe Deer and Transmission within Ixodes ricinus. Emerging Infectious Diseases. 2007, 13:
Duh D, Petrovec M, Avsic-Zupanc T: Molecular characterization of human pathogen Babesia EU1 in Ixodes ricinus ticks from Slovenia. Journal of Parasitology. 2005, 91: 463-465. 10.1645/GE-394R.
Casati S, Sager H, Gern L, Piffaretti J: Presence of potentially pathogenic Babesia sp. for human in Ixodes ricinus in Switzerland. Annals of agricultural and environmental medicine. 2006, 13: 65-70.
Häselbarth K, Tenter AM, Brade V, Krieger G, Hunfeld KP: First case of human babesiosis in Germany-Clinical presentation and molecular characterisation of the pathogen. Int J Med Microbiol. 2007, 297: 197-204. 10.1016/j.ijmm.2007.01.002.
Skotarczak B, Rymaszewska A, Wodecka B, Sawczuk M, Adamska M, Maciejewska A: PCR detection of granulocytic Anaplasma and Babesia in Ixodes ricinus ticks and birds in west-central Poland. Annals of agricultural and environmental medicine. 2006, 13: 21-23.
Gray J, Stanek G, Kundi M, Kocianova E: Dimensions of engorging Ixodes ricinus as a measure of feeding duration. Int J Med Microbiol. 2005, 295: 567-572. 10.1016/j.ijmm.2005.05.008.
Zintl A, Finnerty EJ, Murphy TM, de Waal T, Gray JS: Babesias of red deer (Cervus elaphus) in Ireland. Vet Res. 2011, 42:
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S: MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution. Molecular Biology and Evolution. 2011
R: A language and environment for statistical computing. [http://www.R-project.org]
Bakken V, Runde O, Tjørve E: Norwegian bird ringing atlas. 2006, Stavanger: Stavanger Museum
Svensson L, Grant PJ, Mullarney K, Zetterström D: Fågelguiden. 1999, Stockholm: Bonniers förlag
Gjershaug JO, Thingstad PG, Eldøy S, Byrkjeland S: Norsk fugleatlas Klæbu. 1994, Norway: Norsk Ornitologisk Forening
Bonnet S, Jouglin M, Malandrin L, Becker C, Agoulon A, L'Hostis M, Chauvin A: Transstadial and transovarial persistence of Babesia divergens DNA in Ixodes ricinus ticks fed on infected blood in a new skin-feeding technique. Parasitology. 2007, 134: 197-207. 10.1017/S0031182006001545.
Jukes TH, Cantor CR: Evolution of protein molecules. 1969, New York: Academic Press
Acknowledgements
Thanks to PhD-student Lise Heier for statistical advice.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
GH organised the fieldwork and wrote the main part of the paper. KHR led the DNA-isolation part of the laboratory work. ØØ performed the nested PCR, cloning and phylogeny of the positive samples from the real-time PCR screening. ØØ, KHR and HPL provided valuable and significant contributions to the writing of the paper. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Hasle, G., Leinaas, H.P., Røed, K.H. et al. Transport of Babesia venatorum-infected Ixodes ricinus to Norway by northward migrating passerine birds. Acta Vet Scand 53, 41 (2011). https://doi.org/10.1186/1751-0147-53-41
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1751-0147-53-41