Abstract
The specific aim of this study was to assess the faecal shedding of zoonotic enteropathogens by semi-domesticated reindeer (Rangifer tarandus tarandus) to deduce the potential risk to human health through modern reindeer herding. In total, 2,243 faecal samples of reindeer from northern regions of Finland and Norway were examined for potentially enteropathogenic bacteria (Campylobacter species, Enterococcus species, Escherichia coli, Salmonella species and Yersinia species) and parasites (Cryptosporidium species) in accordance with standard procedures. Escherichia coli were isolated in 94.7%, Enterococcus species in 92.9%, Yersinia species in 4.8% of the samples and Campylobacter species in one sample only (0.04%). Analysis for virulence factors in E. coli and Yersinia species revealed no pathogenic strains. Neither Salmonella species nor Cryptosporidium oocysts were detected. The public health risk due to reindeer husbandry concerning zoonotic diseases included in this study has to be considered as very low at present but a putative epidemiological threat may arise when herding conditions are changed with respect to intensification and crowding.
Similar content being viewed by others
Introduction
Zoonotic organisms such as viruses, bacteria or parasites can possess the potential to cause severe diseases both in humans and animals. Free-ranging animals with sporadic or indirect contact to domestic livestock and humans may serve as reservoirs or sentinels for diseases. Transmission of these pathogens can occur directly from a reservoir to the susceptible animal or human being, for example through direct contact with free-ranging animals including cervids [1]. Indirect transmission occurs via vectors, i.e. mosquitos, by contamination of the environment through faecal shedding [2] or by consumption of venison [3]. The epidemiological situation in free-ranging, semi-domesticated animals like reindeer (Rangifer tarandus tarandus), however, is difficult to assess, in contrast to domestic animals. Conclusions derived from studies which focus on domestic animal species in extensive husbandry systems can only be used with caution. But it can be assumed that pathogens are transmitted easily through close animal contact and lead to high animal losses as it is known from intensive husbandry systems. This is of special importance as the crowding of reindeer for winter feeding is becoming more and more common, particularly in southern parts of Northern Finland.
With regard to the occurrence of possibly zoonotic pathogens in reindeer, data is rare. In view of the zoonotic enteropathogens examined in this study, Campylobacter species, Enterococcus species, Escherichia coli, Salmonella species and Yersinia species are among the most important bacteria which cause severe enteric diseases. In detail, various Campylobacter species (C. coli, C. jejuni subspecies jejuni, C. hyointestinalis, C. lari and C. upsaliensis) were isolated from humans with gastro-enteritis [4, 5]. Campylobacter hyointestinalis was detected in healthy Finnish reindeer [6]. In a recent study about Campylobacter species and other bacteria in wild cervids in Norway, no Campylobacter species were detected in wild reindeer [7]. Enterococcus species are known as pyogenic organisms and can cause hospital acquired infections, but are primarily natural inhabitants of the gastrointestinal tract of humans and animals. Enterococcus species in reindeer have not yet been described. The virulence and pathogenesis of the examined bacteria depends on different factors. In E. coli, for instance, the ability to cause severe disease in humans and animals is associated with the occurrence of several virulence factors such as shigatoxins. Therefore, the presence of shigatoxin genes can indicate the virulence of certain strains, also known as shigatoxin-producing E. coli (STEC). In 50 faecal samples from wild Norwegian reindeer, 42 were positive for E. coli, but no shigatoxin-producing strains were identified [7]. Like STEC, Campylobacter species, Salmonella species and Yersinia species are a problem in meat production with a high infection risk for humans who consume contaminated products. Salmonella species have been found to be associated with mortality in reindeer in Finland and Sweden [8]. Among 153 wild Norwegian reindeer, no positive samples for Salmonella species were found [7]. Out of the former eleven known species of Yersinia, Y. pestis as the cause of plague, Y. pseudotuberculosis and certain biotypes of Y. enterocolitica are of great significance for human health. In Europe, the Y. enterocolitica biotypes O:3, O:9 and O:5,27 in particular are associated with human gastro-enteritis [9]. A novel species, Y. aleksiciae, identified by using 16S rRNA gene sequence type analysis and lysine decarboxylase (LDC) and in this points different from members of Y. kristensenii, was described by Sprague & Neubauer [10].
No data is available on the occurrence of zoonotic protozoa in Northern European reindeer such as Cryptosporidium species as a causative agent for heavy diarrhoea in humans and animals.
Materials and methods
Faecal samples
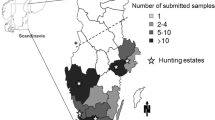
In total, 2,243 faeces samples from healthy reindeer, adults and calves, of both genders were taken over eleven month (June 2001 – April 2002) from eight Finnish and Norwegian free-ranging and corralled reindeer herds, considering parameters such as the degree of intensity of herding, location and season. The origin of samples and further information are given in Table 1. Samples were taken off the ground or per rectum from slaughter animals, sent to the laboratory directly after collection and kept frozen (-4°C) until being processed further within one week.
Examination for Campylobacter species
The examination was carried out by inoculating 1 g faecal material into 9 ml Preston broth (Oxoid). After 24 hours incubation in a microaerophilic atmosphere (5% oxygen, 10% carbon dioxide, 3% hydrogen and 82% nitrogen) at 37°C, a loopful of the enriched suspension was plated on Preston agar (Oxoid, Wesel, Germany) and incubated for 48 hours under the above-mentioned conditions. Campylobacter-like colonies were analysed by Gram-staining, catalase and oxidase tests, and further biochemical reactions (ApiCampy, bioMérieux, Marcy-l'Etoile, France). For all bacteria species, positive controls (ATCC, Manassas, USA) were used to approve the sensitivity of the culture methods.
Examination for Enterococcus species
For the selective enrichment of Enterococcus species, 1 g faecal material was diluted in 9 ml glucose-azide broth (Merck, Darmstadt, Germany) and incubated for 48 hours at 37°C. A loopful of broth was then spread both on kanamycin-aesculin-azide agar (Merck) and Slanetz and Bartley agar (Oxoid). After 48 hours at 37°C suspicious colonies were Gram-stained and their biochemical reactions were analysed further by catalase and oxidase tests.
Examination for Escherichia coli
Escherichia coli was isolated by adding 1 g faeces to 9 ml Gram-negative broth (Becton & Dickinson, Franklin Lakes, USA). After 24 hours of incubation at 37°C a loopful of broth was then plated onto Endo-c agar (Merck) and incubated under the above-mentioned conditions for 24 h. Typical metallic shiny colonies were subcultured on blood agar (Oxoid), incubated for 24 hours at 37°C and tested for their biochemical reactions applying API 20E (bioMérieux). PCR was used to detect the occurrence of shigatoxin1 and 2 genes (stx1, stx2), the intimin gene (eae) and EHEC-haemolysin gene (hly EHEC ) as indicators for the pathogenicity of the isolated strains. Primers were developed with help of the European Molecular Biological Library database and the oligo 6.0 software (Molecular Biology Insight, Cascade, USA) and produced commercially (Invitrogen, Paisley, UK).
Examination for Salmonella species
For the selective enrichment of Salmonella species 1 g faeces was inoculated into 14 ml of tetrathionate broth (Merck) and incubated for 24 hours at 37°C as described by Baird [11]. One ml of this enriched broth was brought into tetrathionate broth the next day and incubated for another 24 hours at 37°C. This enrichment step was repeated one more time. On the fourth day, one loopful of the cultured medium was plated both on Salmonella-Shigella agar (Difco) and Leifson agar (Merck). After 24 hours of incubation at 37°C presumptive Salmonella species colonies were Gram-stained and tested by API 20E (bioMérieux).
Examination for Yersinia species
Cultural examination of Yersinia species was performed by adding 1 g faeces into 9 ml of Gram-negative broth and incubating for 48 hours at 21°C. One loopful of broth was then plated on Yersinia-selective agar (Difco) and incubated for another 48 hours at 21°C. Colonies with the typical bull's-eye appearance were subcultured on blood agar and Gram-stained and biochemical tests were subsequently carried out by use of API 20E (bioMérieux) and Micronaut (Merlin, Bornheim-Hersel, Germany). To detect various Yersinia-genes, crucial for the pathogenicity of the detected strains, PCR was performed using primers to detect the genes encoding 16SrRNA, yadA and v-antigen.
Examination for Cryptosporidium oocysts
For the detection of Cryptosporidium oocysts, immuno-magnetic separation was applied using Dynabeads anti-Cryptosporidium (Dynal Biotech, Oslo, Norway). Twenty μl of the immuno-concentrate were used for a direct immuno-fluorescence test (medac, Wedel, Germany) [12].Cryptosporidium parvum oocysts from a calf (Iowa isolate, Waterborne, USA) served as the positive control. Using a fluorescence microscope at x400-x1000 magnification Cryptosporidium oocysts appear as 6–10 μm in size, round or oval in shape with bright green fluorescence.
For statistical analyses, the data was evaluated with the Statistica 5.0 software (StatSoft Inc., Tulsa, USA), following the instructions of Trampisch & Windeler [13]. For all analyses, differences were considered significant at P ≤ 0.05.
Results
In 2,224 (99.2%) out of the total number of 2,243 faecal samples, one or more of the examined bacteria species were isolated.
Campylobacter species, identified as Campylobacter hyointestinalis, was detected in one faeces sample only (0.04%). Enterococcus species were isolated in 2,084 (92.9%) samples. Escherichia coli were isolated in 2,123 (94.7%) samples. Only few of the isolated E. coli-strains possessed genes encoding stx1, stx2, eae and hly EHEC , as shown in Table 2.
There was no evidence of the occurrence of Salmonella species nor Cryptosporidium species. In total, 108 (4.8%) strains of Yersinia species were isolated, consisting of Y. enterocolitica biogroup 1A (n = 29), Y. intermedia (n = 2), Y. kristensenii (n = 72), Y. mollaretii (n = 3) and Y. rhodei (n = 2).
No significant differences were found for Enterococcus species and E. coli with regard to the degree of intensity of reindeer herding, the season or the geographic origin, whereas the prevalence of Yersinia species differed significantly (p ≤ 0,001): prevalence for Yersinia species in free-ranging reindeer in summer and autumn were significantly higher than in fenced reindeer during winter, as shown in Figure 1. A direct comparison of fenced (n = 100) and free-living (n = 147) reindeer during different seasons was possible in Näkkälä, but with prevalences of 100,00% (fenced) and 65,99% (free-living) for Enterococcus species, 85,00% and 91,63% for E. coli and 1,00% and 0 % for Yersinia species no significant differences could be detected.
Discussion
In reindeer, Enterococcus species and E. coli occurred at very high prevalence, showing the affiliation of these two species to the normal intestinal flora of healthy reindeer. With regard to E. coli, there are few reports of diseases caused by shigatoxin-producing bacteria in ruminants [14, 15]. However, these bacteria are of extreme importance in causing severe diseases in humans [16]. As the genes encoding stx1, eae and hly EHEC were detected in very few of the isolated E. coli-strains, the human health risk due to E. coli excreted by reindeer can be considered very low at present. These results comply with two other studies detecting no E. coli O157:H7 in 1,387 faecal and 421 meat samples from reindeer [17] and no STEC in 50 faecal samples from reindeer [7]. It is known, however, that STEC virulence factors are mobile within bacterial populations [18]. Therefore, an increase in the occurrence of toxin genes in E. coli from reindeer cannot be excluded when influencing parameters such as herding conditions are changed.
Yersinia species were isolated in 108 samples. The identified species Y. intermedia,Y. kristensenii, Y. mollaretii and Y. rhodei had been isolated before from various environmental samples, food, healthy animals and healthy and diseased humans [19, 20]. Even though these species are widely distributed in nature, their actual impact on human health is a matter of controversy. However, as they are isolated from persons with gastrointestinal disorders, the role of these species should not be disregarded [20]. These species seem to be well adapted to warm-blooded animals, including humans. After the identification of the novel species Y. aleksiciae [10], the isolates originally phenotyped as members of Y. kristensenii had to be investigated again (unpublished data). The isolated Y. enterocolitica strains belonged to biogroup 1A, which embraces the non-pathogenic European Y. enterocolitica strains, often isolated from environmental samples, foods, animal and human faeces [21]. Yersinia species, further analysed as Yersinia pseudotuberculosis serotype 1a, had also been isolated from reindeer before [22], but in samples from 35 Norwegian reindeer no Yersinia species were found [23]. The seasonal pattern of Yersinia species with an increase in the summer period can be attributed to the constant accidental incorporation of environmental strains which in this extent is not given in winter with limited survival conditions for bacteria.
Campylobacter hyointestinalis was isolated from one sample only. As the cultivation of Campylobacter species is difficult, the actual prevalence might be higher. Campylobacter hyointestinalis has hitherto been associated only sporadically with human gastrointestinal disorders [5, 24]. Even though the prevalence of Campylobacter species in this study was very low, it shows that reindeer can be carriers. This is supported by a study from Hänninen et al. [6] who detected Campylobacter hyointestinalis at a prevalence of 6% in 399 healthy Finnish reindeer. Lillehaug et al. [7] isolated no Campylobacter species in 150 faecal samples of wild reindeer.
Neither Salmonella species nor Cryptosporidium oocysts were detected in reindeer in this study. Both pathogens had been isolated from the environment, farm animals and humans in Fennoscandia [25, 26]. The occurrence of Salmonella species in Finnish reindeer is described by Kuronen et al. [8]. Regarding Salmonella species, the results of this study and of the study from Lillehaug et al. [7] with negative results for Salmonella species in 153 faecal samples from wild reindeer, indicate that wild cervids do not contract Salmonella-infections to any significant extent from other wildlife.
In Rangifer tarandus, a new genotype of Cryptosporidium closely related to Cr. serpentis, Cr. muris and Cr. andersoni, was isolated from 3 out of 49 caribou examined in Canada [27]. Concerning Northern European reindeer, no bibliographical references exist, but deriving from other species, the prevalence was expected to be higher in younger animals. This could not be supported by this study, identifying no Cryptosporidium oocysts at all.
All bacteria analysed in this study may be found in Northern Europe in the environment in aquatic, terrestrial and animal reservoirs [28] and have been isolated before from the intestinal tract of healthy or diseased ruminants world-wide [29, 30]. Even though most of the isolated bacteria strains do not have the potential to cause severe human or animal health problems, certain strains might be a risk, especially for immuno-supressed, old or very young persons and animals. Therefore, one has to regard the epidemiological impact of transmission of these infectious agents from the environment to reindeer and man and vice versa, depending on a number of local factors. To recapitulate, the excretion risk of pathogens by reindeer has to be considered in the context of the extreme climatic conditions in the research area. Permafrost soils of the tundra and taiga are a domain of psychrophilic and psychrotolerant organisms [31], but as enteropathogens, living in intestines of warm-blooded animals at 37°C, are not adapted to these extreme northern environmental conditions, fast destruction is probable.
In conclusion, the enteropathogens examined were either not detected at all (Salmonella species and Cryptosporidium species), in very small numbers (Campylobacter species) or if detected, their virulence and pathogenicity was very low (E. coli and Yersinia species). The potential human and animal health risk from reindeer excreting various important enteropathogenic bacteria and Cryptosporidium species should be regarded very low at present. No differences could be found in the flora between fenced and free-living animals. However, especially if reindeer are crowded, e.g. for winter feeding, an increased prevalence of enteric pathogens excreted by reindeer and eventually an increased risk to the consumer has to be considered as is already known from other intensive animals husbandry systems worldwide.
Conclusion
With respect to the investigated pathogens, the analysis of faecal samples from Norwegian and Finnish reindeer indicates that the animals do not represent an important source for zoonotic diseases at the moment. Enterococcus species and E. coli belong to the normal intestinal flora of reindeer. Climate conditions in the northern regions are a limiting factor to the survival of enteropathogens in the environment and might be a reason for the low prevalences of the other pathogens examined.
Sammanfattning
Campylobacter spp., Enterococcus spp., Escherichia coli, Salmonella spp., Yersinia spp., og Cryptosporidium oocyster hos semi-domesticerade renar (Rangifer tarandus tarandus) i Finlands och Norges nordliga trakter
Målsättningen med studien var värdering av zoonotiska enteropatogeners fekala utbredning hos semi-domesticerade renar (Rangifer tarandus tarandus) för att bedöma potentialla risker för männsikans hälsa med modern renskötsel. Sammanlagt 2243 träckprover från renar i Finlands och Norges nordliga trakter har analyserats på potentiellt enteropatogena bakterier (Campylobacter species, Enterococcus species, Escherichia coli, Salmonella species och Yersinia species) och parasiter (Cryptosporidium species) med standardiserade metoder. Escherichia coli har isolerats i 94.7%, Enterococcus species i 92.9%, Yersinia species i 4.8% av proverna och Campylobacter species i bara ett prov (0.04%). Analys av virolensfaktorer i E. coli och Yersinia species visade inga patogena egenskaper. Varken Salmonella species eller Cryptosporidium-oocystor har påvisats. Den allmänna hälsorisken gällande zoonotiska sjukdomar på grund av renskötseln bedöms för tillfället som mycket låg men risken kan öka om renskötselns villkor ändras med hänsyn till intensitet och hållande i hägn.
References
Fanning A, Edwards S: Mycobacterium bovis infection in human beings in contact with elk (Cervus elaphus) in Alberta, Canada. Lancet. 1991, 228: 1253-1255. 10.1016/0140-6736(91)92113-G.
Aavitsland P, Hofshagen T: Salmonella utbrudd i Herøy kommune (Salmonella outbreak in the Herøy district). Nytt fra miljø og samfunnsmedisin. 1999, 3: 12-(In Norwegian)
Keene WE, Sazie E, Kok J, Rice DH, Hancock DD, Balan VK, Zhao T, Doyle MP: An outbreak of Escherichia coli O157:H7 infections traced to jerky made from deer meat. JAMA. 1997, 277: 1229-1231. 10.1001/jama.277.15.1229.
Lawson AJ, Logan MJ, O'Neill GL, Desa M, Stanley J: Large-scale survey of Campylobacter species in human gastroenteris by PCR and PCR-enzyme-linked immunosorbent assay. J Clin Microbiol. 1999, 37: 3860-3864.
Gorkiewicz G, Feierl G, Zechner R: Transmission of Campylobacter hyointestinalis from a pig to human. J Clin Microbiol. 2002, 40: 2601-2605. 10.1128/JCM.40.7.2601-2605.2002.
Hänninen ML, Sarelli L, Sukura A, On SL, Harrington CS, Matero P, Hirvelä-Koski V: Campylobacter hyointestinalis subsp. hyointestinalis, a common Campylobacter species in reindeer. J Appl Microbiol. 2002, 92: 717-723. 10.1046/j.1365-2672.2002.01574.x.
Lillehaug A, Bergsjo B, Schau J, Bruheim T, Vikoren T, Handeland K: Campylobacter spp., Salmonella spp., verocytotoxic Escherichia coli, and antibiotic resistance in indicator organisms in wild cervids. Acta Vet Scand. 2005, 46: 23-32.
Kuronen H, Hirvelä-Koski V, Nylund M: Salmonella poronvasoissa (Salmonella in a reindeer calv). Poromies. 1998, 4–5: 54-(In Finnish)
Neubauer H, Hensel A, Aleksic S, Meyer H: Identification of Yersinia enterocolitica within the genus Yersinia. Syst Appl Microbiol. 2000, 23: 58-62.
Sprague LD, Neubauer H: Yersinia aleksiciae sp. nov. Int J Syst Evol Microbiol. 2005, 55: 831-835. 10.1099/ijs.0.63220-0.
Baird RM: Pharmacopoeia of Culture Media for Food Microbiology – Additional Monograph. Mueller-Kauffmann Tetrathionate Broth. Int J Food Microbiol. 1989, 132-134.
Das Graças C, Pereira M, Atwill ER, Jones T: Comparison of sensitivity of immunofluorescent microscopy to that of a combination of immunofluorescent microscopy and immunomagnetic separation for detection of Cryptosporidium parvum oocysts in adult bovine faeces. Appl Environ Microbiol. 1999, 65: 3236-3239.
Trampisch HJ, Windeler J: Medizinische Statistik (Medical statistics). 1997, Berlin, Springer, (In German), 1
Sherwood D, Snodgrass DR, O'Brien AD: Shiga-like toxin production from Escherichia coli associated with calf diarrhea. Vet Rec. 1985, 116: 217.
Mainil J: Shiga/Verocytotoxins and Shiga/Verotoxigenic Escherichia coli in animals. Vet Res. 1999, 30: 235-257.
Griffin PM, Tauxe RV: The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated haemolytic uremic syndrome. Epidemiol Rev. 1991, 13: 60-98.
Lahti E, Hirvelä-Koski V, Honkanen-Buzalski T: Occurrence of Escherichia coli O157 in reindeer (Rangifer tarandus). Vet Rec. 2001, 19: 633-634.
Yamamoto T, Honda T, Miwatani T, Yokota T: A virulence plasmid in Escherichia coli enterotoxigenic for humans: intergenetic transfer and expression. J Infect Dis. 1984, 150: 688-698.
Baier R, Puppel H: Enteritis durch „atypische" Yersinien (Enteritis caused by„atypical" Yersinia). Dtsch Med Wochenschr. 1981, 106: 208-210. (In German)
Sulakvelidze A: Yersiniae other than Y. enterocolitica, Y. pseudotuberculosis, and Y. pestis : the ignored species. Microbes Infect. 2000, 2: 497-513. 10.1016/S1286-4579(00)00311-7.
Bottone EJ: Yersinia enterocolitica : the charisma continues. Clin Microbiol Rev. 1997, 10: 257-276.
Ueshiba H, Kato H, Tohru MA, Tsubokura M, Nagano T, Kanako S, Uchiyama T: Analysis of the superantigen-producing ability of Yersinia pseudotuberculosis strains of various serotypes isolated from patients with systemic or gastroenteric infections, wildlife animals and natural environment. Zentralbl Bakteriol. 1998, 288: 277-291.
Aschfalk A, Kemper N, Höller C: Bacteria of pathogenic importance in faeces from cadavers of free-ranging or corralled semi-domesticated reindeer in Northern Norway. Vet Res Commun. 2003, 27: 93-100. 10.1023/A:1022802918630.
Edmonds P, Patton CM, Ward GE, Griffin PM: Campylobacter hyointestinalis associated with human gastrointestinal disease in the United States. Clin Microbiol. 1987, 25: 251-263.
Refsum T, Heir E, Kapperud G, Vardund T, Holstad G: Molecular epidemiology of Salmonella enterica serovar typhimurium isolates determined by pulsed-field gel electrophoresis: comparison of isolates from avian wildlife, domestic animals, and the environment in Norway. Appl Environ Microbiol. 2002, 68 (11): 5600-6.
Hörman A, Rimhanen-Finne R, Maunula L, von Bonsdorff C, Torvela N, Heikinheimo A, Hänninen ML: Campylobacter spp., Giardia spp., Cryptosporidium spp., noroviruses, and indicator organisms in surface waters in southwestern Finland. Appl Environ Microbiol. 2004, 70: 87-95. 10.1128/AEM.70.1.87-95.2004.
Siefker C, Rickard LG, Pharr GT, Simmons JS, O'Hara TM: Molecular characterisation of Cryptosporidium sp. isolated from Northern Alaskan caribou (Rangifer tarandus). J Parasitol. 2002, 88: 213-216. 10.2307/3285424.
Kapperud G: Survey on the reservoirs of Yersinia enterocolitica and Yersinia enterocolitica -like bacteria in Scandinavia. Acta Pathol Microbiol Scand [B]. 1981, 89: 29-35.
Adesiyun AA, Seepersadsingh N, Inder L, Caesar K: Some bacterial enteropathogens in wildlife and racing pigeons from Trinidad. J Wildl Dis. 1998, 34: 73-80.
Busato A, Lentze T, Hofer D, Burnens A, Hentrich B, Gaillard C: A case control study of potential enteric pathogens for calves raised in cow-calf herds. Zentralbl Veterinarmed B. 1998, 45: 519-528.
Bölter M: Ecophysiology of psychrophilic and psychrotolerant microorganisms. Cell Mol Biol. 2004, 50: 563-573.
Kemper N, Aschfalk A, Höller C: The occurrence and prevalence of potentially zoonotic enteropathogens in semi-domesticated reindeer. Rangifer. 2004, 24: 15-20.
Acknowledgements
This examination was performed in the context of the RENMAN-Project http://www.urova.fi/home/renman/, funded by the EC's 5th framework programme. The authors wish to thank the Nordic Council for Reindeer Husbandry for providing permission to modify data presented at the 11th Arctic Ungulate Conference in Saariselkä, Finland in 2003 [32].
Author information
Authors and Affiliations
Corresponding author
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kemper, N., Aschfalk, A. & Höller, C. Campylobacter spp., Enterococcus spp., Escherichia coli, Salmonella spp., Yersinia spp., and Cryptosporidium oocysts in semi-domesticated reindeer (Rangifer tarandus tarandus) in Northern Finland and Norway. Acta Vet Scand 48, 7 (2006). https://doi.org/10.1186/1751-0147-48-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1751-0147-48-7