Abstract
Background
AATD is one of the most common inherited disorders in the World. However, it is generally accepted that AATD in North African populations is not a risk factor for lung and/or liver disease, based on a number of small studies. We therefore planned a screening study for detection of AATD in patients with OLD in a cohort of patients from Kairouan in central Tunisia. Methods: One hundred twenty patients with OLD (asthma, emphysema, COPD) were enrolled in the screening programme. Laboratory diagnosis for AATD was performed according to current diagnostic standards.
Results
We found that 6/120 OLD patients carried an AAT deficient allele, 1 PI*MZ, 1 PI*MPlowel, 3 PI*MMmalton, 1 PI*MMwurzburg.
Conclusion
this pilot study demonstrated that alleles related to deficiency of AAT are not absent in the Tunisian population, and that rare AATD variants prevailed over commonest PI*Z variant. These results would support a larger scale screening for AATD in Tunisia.
Similar content being viewed by others
Background
Alpha 1 antitrypsin (AAT) is an acute phase glycoprotein predominantly derived from the liver, and its major biological function is to inhibit neutrophil elastase [1]. AAT, a highly polymorphic protein with more than 120 variants known to date [2] is coded by a gene, called SERPINA1, located on chromosome 14q31-32.3 within the SERPIN cluster [3]. The SERPINA1 PI*M alleles code for the commonest normal AAT variants, whereas PI*S and PI*Z are the most common deficiency alleles associated with reduced concentrations of plasma AAT. Nevertheless, there are at least 30 SERPINA1 alleles rarely detected, other than the PI*Z and the PI*S alleles, which are associated with significantly reduced AAT levels [4–6].
Inherited deficiency of AAT (AATD) is one of the most common genetic disorders in the world, and is associated with an increased risk of developing lung and, to a lesser extent, liver disease [7]. Individuals homozygous for the PI*Z allele usually have an AAT level in the region of 0.35 g/L [3] and are at high risk for the development of emphysema [8], asthma [9], chronic bronchitis and bronchiectasis [10]. However, heterozygous individuals with PI*S and PI*Z mutation (or rare mutations) showing a more protective AAT level (> 0,8 g/L) [3] can also be at increased risk for lung disease, depending on multiple environmental factors, such as smoking, occupational exposure, and environmental exposure.
Epidemiology studies showed that highest prevalence of PI*ZZ related AATD is recorded among Northern Europeans and populations with North European background [11]. Notwithstanding, during the last few years, based on estimates from allele frequencies obtained in available cohort studies, it has been suggested that the Z variant is not only common in Caucasians, but also among other ethnic groups worldwide [12, 13].
A total of 30 cohorts have been investigated for AATD in the African continent. Twenty four cohorts, having a total of 4,718 individuals, were in Sub-Saharian Africa [14], 6 cohorts with a total sample size of 1,735 have been investigated in the north African populations [15–20], Three of them in Tunisia: in the Tunisian population, the PI*Z allele, previously considered as virtually absent [15, 16], has then been detected once on the heterozygous state in a total cohort of 1,168 individuals (allele frequency 0.04%) [17]. However, none of the previous surveys in Tunisia has looked at rare, non-S and non-Z SERPINA1 variants, which have been hypothesised to be particularly frequent in the Mediterrenean area, where PI*Z allele frequency is reduced [4]. We therefore aimed this paper at investigating AATD variants in a Tunisia area, targeting a cohort of obstructed individuals, in which the diagnosis of AATD has been recommended accoding to the ATS/ERS document [21].
Methods
Study subjects
Upon approval by the local Ethical Committee, patients with obstructive lung disease (OLD: asthma, emphysema, COPD), referred to the pulmonary disease department in Kairouan regional hospital (central Tunisia) from June 2006 to September 2006, were enrolled in the screening programme. Diagnosis of obstructive lung disease was obtained by symptoms, radiology, lung function examinations and allergy history.
Quantitative determination of AAT level
AAT concentration measurement was performed on plasma samples with a rate immune turbidimetric method (Konelab 20, Thermo Clinical Labsystem, Finland), using a polyclonal anti-human AAT antibody (Thermo Electron Corporation, Finland). A calibrator (Specical, Thermo Electron Corporation, Finland) with an assigned AAT value was used as a standard. A control (Specitrol, Thermo Electron Corporation, Finland) with an assigned AAT value was used after a predefined reaction number. Plasma samples for the turbidimetric assay are diluted by the instrument 1:10 (AAT reading range, 0.23–4.4 g/L). The turbidimetry of antigen-antibody complex was measured at 360 nm. Values obtained are regressed according to the calibration curve, and expressed as g/L.
Molecular diagnosis of AATD
DNA extracted from the white blood cells of all subjects by standard methods, was submitted to genotyping for Z and S SERPINA1 alleles, by PCR-RFLP, as previously described [22]. According to published diagnostic algorythm [23], samples negative for Z and S alleles, but with inconsistent AAT level/genotype were submitted to sequencing of SERPINA1 exons II, III, IV, and V, as previously described [24].
Results
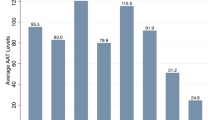
One hundred twenty subjects with OLD were enroled in this study. There were 115 males and 5 females, mean age (SD) 49.2 (9.5) years (range 34–81). The large majority of patients were smokers (84.1%). As far as the OLD phenotype was concerned, there were 49 COPD (40.8%), 48 emphysema (40%), and 23 asthma patients (19.2%). Mean (SD) plasma AAT level was 1.74 g/L (0.63) (range 0.81 to 3.3). Plasma AAT level distribution is shown in figure 1.
SERPINA1 S and Z allele genotyping, performed in all OLD subjects, allowed the detection of one PI*MZ individual (plasma AAT level: 0.81 g/l, consistent with the detected genotype).
Nevertheless, some subjects negative for PI*S and PI*Z allele detection, displayed plasma AAT levels consistent with intermediate AATD; in particular, 4 subjects displayed plasma AAT level < 1 g/L [mean (SD) 0.91 g/L (0.02)]. According to our protocol for detection of rare AATD variants, such samples with inconsistent AAT level/genotype were suitable for sequencing [4, 24]. In the absence of reference values of plasma AAT in the general population from Tunisia, we decided to submit to sequencing, DNA samples from subjects with plasma AAT levels < 1.5 g/L, rather than the usual cut-off of 1.13 g/L [25]. We therefore sequenced 24 DNA samples (plasma AAT level range: 0.89–1.48). We found that 5 subjects were heterozygous for rare deficiency variants: 3 subjects carried the PI* MMmalton, 1 the PI*MPlowell, and 1 the PI*MMwurzburg genotypes. Figure 2 summarises the results of genotyping/sequencing.
Discussion
Meta-analysis of available data showed that SERPINA 1 PI*S and PI*Z deficiency alleles are extremely rare in North African populations [12, 13]. Recent investigations in the Tunisian population confirmed the low frequency of AATD common deficiency variants in the general population and healthy subjects (1078 subjects investigated: 15 PI*MS detected (PI*S: 0.7%; PI*Z: 0%)) [15–17], as well as in COPD patients (90 subjects investigated: 1 PI*SS and 1 PI*MZ detected (PI*S: 1%; PI*Z: 0.5%)) [17]. The present study, performed in 120 patients with OLD, confirms these data: PI*S allele was not seen, whereas PI*Z was detected only once on the heterozygous state (frequency 0.4%).
In this study, we extended the SERPINA1 gene investigation to variants not detectable by rapid PI*S and PI*Z genotyping: by this strategy, we found 5 more deficiency variants (frequency as a whole 2%) detected on the heterozygous state. Such a condition of a relatively higher frequency of rare than common AATD variants, is shared by Central – Southern regions of Italy, in which PI*Mmalton and PI*Mprocida variants seem to prevail over PI*Z [4]. Interestingly, the PI*Mmalton variant, detected in three unrelated individuals in this paper, is the commonest AATD variant in Sardinia, where the PI*Z variant is detected very rarely [4, 26]. A novel SERPINA1 Null mutation, first described in an Egyptian family, and for that reason called Q0*cairo [27] has been repeatedly detected in unrelated individuals from regions of Southern Italy. Population admixture due to migration occurred in ancestral periods, as well as contacts for commercial purposes in more recent centuries are likely to be responsible for dissemination of rare AATD variants in the Southern Mediterrenean basin. Table 1 summarizes the rare variants reported in the Mediterranean basin, as well as the mutations types, the cellular defect and the related clinical data [4, 28–30].
This study strengthens the concept that for a correct laboratory diagnosis of AATD, there is a need of a combination of biochemical and biomolecular methods [24], otherwise rare AATD variants will be missed [5]. These preliminary data also confirm the usefulness of enrolling patients with OLD in a screening programme for AATD. According to the ATS/ERS statement's evidence based recommendations, all subjects with COPD and asthma should be submitted to diagnostic testing for AATD [21]. In this paper, although no subjects with severe AATD deficiency were detected, we found six out of 120 subjects (5%) carrying the so called "intermediate deficiency", that means heterozygosity with one normal PI*M allele and one severe AATD allele [mean (SD) plasma AAT level: 0.93 g/L (0.12)]. These findings are consistent with the hypothesis that intermediate AATD, such as PI*MZ or equivalent genotypes, represent a risk factor for developing COPD [20]. Recent meta-analysis [31] reported the increase in risk of COPD in PI*MZ heterozygous individuals (OR for PI*MZ versus PI*MM (normal genotype) was 2.31 (95% CI 1.60 to 3.35)).
Severe AATD diagnosis in the North African countries might be beneficial to the treatment of patients with the introduction of AAT replacement therapy in these populations. Furthermore, AATD carrier diagnosis might provide genetic counseling to persons who are planning a pregnancy or are in the prenatal period [21]. In addition, awareness of carrying a gene that may increase the susceptibility to COPD may be an additional factor for a successful enrolment of patients in smoking cessation programmes [32].
Conclusion
This investigation, performed for the first time with current diagnostic standards in a North African population, highlights the implication of AATD in development of OLD in this area, whereas previous data excluded a role for AATD in the African continent. This would suggest that, similarly to what happens in the Caucasians, also in Northern Africans AATD is an underdiagnosed condition, and therefore investigations in larger sample sizes would be advisable. It seems also likely that targeted detection strategy to identify affected individuals produces a rate of detecting disease higher than the population-based screening programs. Such approach may also contribute to change the widespread concept concerning the AAT deficiency epidemiology in North African and others populations.
References
Lomas DA, Parfrey H: Alpha 1 Antitrypsin deficiency 4: molecular pathophysiology. Thorax. 2004, 59: 529-535. 10.1136/thx.2003.006528.
DeMeo DL, Silverman EK: Alpha 1 antitrypsin deficiency. 2: genetic aspects of alpha 1 antitrypsin deficiency: phenotypes and genetic modifiers of emphysema risk. Thorax. 2004, 59: 259-264. 10.1136/thx.2003.006502.
Crystal RG: α1-antitrypsin Deficiency, emphysema, and liver disease genetic basis and strategies for therapy. J Clin Invest. 1990, 85: 1343-1352. 10.1172/JCI114578.
Ferrarotti I, Baccheschi J, Zorzetto M, Tinelli C, Corda L, Balbi B: Prevalence and phenotype of subjects carrying rare variants in the Italian registry for alpha1-antitrypsin deficiency. J Med Genet. 2005, 42: 282-287. 10.1136/jmg.2004.023903.
Prins J, Mejiden van der BB, Kraaijenhagen RJ, Wielders JPM: Inherited obstructive pulmonary disease: new selective sequencing workup for α1-antitrypsin deficiency identified two previously unidentified Null allels. Clin Chem. 2008, 51: 101-107.
Fregonese L, Stolk J, Frants RR, Veldhuisen B: Alpha1 antitrypsin Null mutations and severity of emphysema. Respir Med. 2008, 102: 876-884. 10.1016/j.rmed.2008.01.009.
Stolk J, Seersholm N, Kalsheker N: Alpha1-antitrypsin deficiency: current perspective on research, diagnosis, and management. Int J COPD. 2006, 1: 151-160. 10.2147/copd.2006.1.2.151.
Laurell CB, Eriksson S: The electrophoretic alpha 1-globulin pattern of serum in alpha 1-antitrypsin deficiency. Scand J Clin Lab Invest. 1963, 15: 132-140. 10.3109/00365516309051324.
Eden E, Hammel J, Rouhani FN, Brantly ML, Barker AF, Buist AS: Asthma features in severe alpha1-antitrypsin deficiency: experience of the National Heart, Lung, and Blood Institute Registry. Chest. 2003, 123: 765-771. 10.1378/chest.123.3.765.
Dowson LJ, Guest PJ, Stockley RA: The relationship of chronic sputum expectoration to physiologic, radiologic, and health status characteristics in alpha(1)-antitrypsin deficiency (Pi Z). Chest. 2002, 122: 1247-1255. 10.1378/chest.122.4.1247.
Luisetti M, Seersholm N: Alpha1-antitrypsin Deficiency. 1: Epidemiology of Alpha1-antitrypsin deficiency. Thorax. 2004, 59: 164-169. 10.1136/thorax.2003.006494.
de Serres FJ: Worldwide racial and ethnic distribution of α1-antitrypsin deficiency. Summary of an analysis of published genetic epidemiology surveys. Chest. 2002, 122: 1818-1829. 10.1378/chest.122.5.1818.
de Serres FJ, Blanco I, Fernández-Bustillo E: PI S and PI Z Alpha-1 antitrypsin deficiency worldwide. A review of existing genetic epidemiological data. Monaldi Arch Chest Dis. 2007, 67: 184-208.
de Serres FJ, Blanco I, Fernández-Bustillo E: Health implications of alpha1-antitrypsin deficiency in Sub-Sahara African countries and their emigrants in Europe and the New World. Genet Med. 2005, 7: 175-184.
Makni S, Zitouni M, Ayed K, Mhirii S, Azabi S, Cherif F: Absence of the alpha 1 antitrypsin PiZ allele in Tunisia substantiates the particular genetic structure of African populations. Am J Hum Biol. 1997, 9: 223-224. 10.1002/(SICI)1520-6300(1997)9:2<223::AID-AJHB8>3.0.CO;2-0.
Chaabani H, Martin JP, Frants RR, Lefranc G: Genetic study of Tunisian Berber. Alpha 1 antitrypsin (Pi) polymorphism. Report of a new allele (Pi S berber). Exp Clin Immunogenet. 1984, 1: 19-24.
Denden S, Haj Khelil A, Perrin P, Daimi H, Leban N, Ouaja A: Alpha 1 antitrypsin polymorphism in the Tunisian population with special reference to pulmonary disease. Pathol Biol. 2008, 56: 106-110. 10.1016/j.patbio.2007.05.003.
Ezzikouri S, El Feidi AE, El Kihal L, Afifi R, Bennazzouz R, Hassar M: Prevalence of common HFE and SERPINA1 mutations in patients with hepatocellular carcinoma in a Moroccan population. Arch Med Res. 2008, 39: 236-241. 10.1016/j.arcmed.2007.09.006.
Harich N, Esteban E, Chafic A, López-Alomar A, Vona G, Moral P: Classical polymorphisms in Berbers from Moyen Atlas (Morocco): genetics, geography, and historical evidence in the Mediterranean peoples. Ann Hum Biol. 2002, 29: 473-487. 10.1080/03014460110104393.
Sebetan IM: a new allele in the alpha-1-antitrypsin system. Hum Hered. 1992, 42: 206-208. 10.1159/000154070.
American Thoracic Society/European Respiratory Society Statement: Standards for the Diagnosis and Management of Individuals with Alpha-1 Antitrypsin Deficiency. Am J Respir Crit Care Med. 2003, 168: 818-900. 10.1164/rccm.168.7.818.
Ferrarotti I, Zorzetto M, Scabini R, Mazzola P, Campo I, Luisetti M: A novel method for rapid genotypic identification of alpha1-antitrypsin variants. Diagn Mol Pathol. 2004, 12: 160-163. 10.1097/01.pdm.0000128700.22128.80.
Zorzetto M, Russi E, Senn O, Imboden M, Ferrarotti I, Tinelli C: SERPINA1 gene variants in individuals from the general population with reduced α1-antitrypsin concentrations. Clin Chem. 2008, 54: 1331-1338. 10.1373/clinchem.2007.102798.
Ferrarotti I, Scabini R, Campo I, Ottaviani S, Zorzetto M, Gorrini M: Laboratory diagnosis of alpha1-antitrypsin deficiency. Transl Res. 2007, 150: 267-274. 10.1016/j.trsl.2007.08.001.
Gorrini M, Ferrarotti I, Lupi A, Bosoni T, Mazzola P, Scabini R: Validation of a rapid, simple method to measure α1-antitrypsin in human dried blood spots. Clin Chem. 2006, 52: 899-901. 10.1373/clinchem.2005.062059.
Orrù G, Faa G, Pillai S, Pilloni L, Montaldo C, Pusceddu G: Rapid PCR real-time genotyping of M-Malton alpha1-antitrypsin deficiency alleles by molecular beacons. Diagn Mol Pathol. 2005, 14: 237-242. 10.1097/01.pas.0000178221.44474.b3.
Zorzetto M, Ferrarotti I, Campo I, Balestrino A, Nava S, Gorrini M: Identification of a novel alpha1-antitrypsin null variant (Q0cairo). Diagn Mol Pathol. 2005, 14: 121-124. 10.1097/01.pas.0000155023.74859.d6.
Jardi R, Rodriguez F, Miravitlles M, Vidal R, Cotrina M, Pascual C: Identification and molecular characterisation of the new alpha-1-antitrypsin deficient allele PI Ybarcelona (Asp256→Val and Pro391→His). Hum Mutation #174 1-6 (1998) Online.
Faber JP, Poller W, Weidinger S, Kirchgesser M, Schwaab R, Olek K: Sequence data of fifteen new α1-antitrypsin variants including two PI*Q0 and one deficient PI*M allele. Am J Hum Genet. 1994, 55: 1113-1121.
Jardi R, Rodriguez F, Lopez-Talavera JC, Miravitlles M, Cotrina M, Costa X: Characterization of the New Alpha-1-Antitrypsin-Deficient PI M-Type Allele, PI Mvall d'hebron (Pro369→Ser). Hum Hered. 2000, 50: 320-321. 10.1159/000022935.
Hersh CP, Dahl M, Ly NP, Berkey CS, Nordestgaard BG, Silverman EK: Chronic obstructive pulmonary disease in a1-antitrypsin PI MZ heterozygotes: a meta-analysis. Thorax. 2004, 59: 843-849. 10.1136/thx.2004.022541.
Ferrarotti I, Gorrini M, Scabini I, Ottaviani S, Mazzola P, Campo I: Secondary outputs of alpha1-antitrypsin deficiency targeted detection programme. Respir Med. 2008, 102: 354-358. 10.1016/j.rmed.2007.10.015.
Acknowledgements
SD was supported by a Short Term Training Fellowship (n° 721) from European Respiratory Society. The diagnostic center for AATD in Pavia gratefully acknoweledges the support from Fondazione IRCCCS Policlinico San Matteo, Ricerca Corrente programs, and from Talecris Biotherapeutics, Germany.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SD, MZ, IF and IC designed the study and drafted the Manuscript. FA and JK participated in data collection. SO, RS and MG participated in laboratory investigations. JBC, AHK and ML supervised the study process, revised and edited the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Denden, S., Zorzetto, M., Amri, F. et al. Screening for Alpha 1 antitrypsin deficiency in Tunisian subjects with obstructive lung disease: a feasibility report. Orphanet J Rare Dis 4, 12 (2009). https://doi.org/10.1186/1750-1172-4-12
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1750-1172-4-12