Abstract
Background
The discovery of molecular markers associated with various breast cancer subtypes has greatly improved the treatment and outcome of breast cancer patients. Unfortunately, breast cancer cells acquire resistance to various therapies. Mounting evidence suggests that resistance is rooted in the deregulation of the G1 phase regulatory machinery.
Methods
To address whether deregulation of the G1 phase regulatory machinery contributes to radiotherapy resistance, the MCF10A immortalized human mammary epithelial cell line, ER-PR-Her2+ and ER-PR-Her2- breast cancer cell lines were irradiated. Colony formation assays measured radioresistance, while immunocytochemistry, Western blots, and flow cytometry measured the cell cycle, DNA replication, mitosis, apoptosis, and DNA breaks.
Results
Molecular markers common to all cell lines were overexpressed, including cyclin A1 and cyclin D1, which impinge on CDK2 and CDK4 activities, respectively. We addressed their potential role in radioresistance by generating cell lines stably expressing small hairpin RNAs (shRNA) against CDK2 and CDK4. None of the cell lines knocked down for CDK2 displayed radiosensitization. In contrast, all cell lines knocked down for CDK4 were significantly radiosensitized, and a CDK4/CDK6 inhibitor sensitized MDA-MB-468 to radiation induced apoptosis. Our data showed that silencing CDK4 significantly increases radiation induced cell apoptosis in cell lines without significantly altering cell cycle progression, or DNA repair after irradiation. Our results indicate lower levels of phospho-Bad at ser136 upon CDK4 silencing and ionizing radiation, which has been shown to signal apoptosis.
Conclusion
Based on our data we conclude that knockdown of CDK4 activity sensitizes breast cancer cells to radiation by activating apoptosis pathways.
Similar content being viewed by others
Background
Breast cancer is the most common malignancy and the major cause of cancer related deaths of women in industrialized countries [1]. Fortunately, the discoveries of various biomarkers have greatly improved the treatment of breast cancer patients [2]. For example, breast cancer patients that express progesterone receptor (PR) and estrogen receptor (ER) (luminal A breast cancers are ER+PR+HER2- or ER+PR+HER2+, while luminal B are ER+PR+HER2-) are treated with endocrine therapies and/or with the antibody Herceptin, in combination with chemotherapy and/or radiotherapy [3]. Regrettably, the defining characteristic of triple negative breast cancers (a subtype of basal breast cancers) is the absence of ER, PR and HER2, rendering those cancers insensitive to some of the most effective therapies [4]. ER-PR-HER2- and ER-PR-HER2+ patients show decreased survival relative to luminal subtypes [5, 6].
Radiotherapy is one of the cornerstones in the treatment of patients with breast cancer [7, 8]. Radiation greatly reduces the risk of recurrence in women with ductal carcinoma in situ [9, 10] and in breast cancer patients who are lymph node positive [11–13]. The outcomes following chemotherapy vary depending on the subtype: for example, ER-PR-HER2- and ER-PR-HER2+ breast cancers respond better than luminal subtypes to anthracycline-based chemotherapies [14]. In contrast, all subtypes respond equally well to radiation [15]. Regardless of treatment, ER-PR-HER2- or ER-PR-HER2+ breast cancers are more likely to recur upon breast conserving therapies or following mastectomy and radiation [16–19]. Even though radiotherapy shows promise in the treatment of all breast cancer subtypes, radiotherapy is associated with increased risk of cardiovascular disease [20]. In addition, breast tumors can acquire radioresistance [21, 22]. Finding agents that sensitize malignant cells to radiation would increase tumor response while minimizing toxicity to surrounding organs by lowering effective therapeutic doses.
The G1/S phase regulatory machinery includes cyclins and cyclin kinase inhibitors (CKIs) that regulate the activities of the G1 phase CDKs [23, 24] and the Rb/E2F pathway [25, 26]. Besides being distinguished by their ER, PR, or Her2 status, the various breast cancer subtypes exhibit deregulated expression of proteins that ensure progression through the G1/S phase of the cell cycle. ER-PR-HERr2- breast cancers are more likely to overexpress mutant p53, E2F3, p16, and cyclin E and display lower levels of cyclin D1, Bcl2, and Rb relative to other breast cancer subtypes [5, 6, 27, 28]. Cyclin D1 overexpression is more often found in tumors with wild type p53, higher grades of differentiation, and expression of ER or PR [29–32]. Specifically, 53% PR+ and 58% ER+ breast cancer patients overexpress cyclin D1, while a smaller, but significant fraction of ER- and PR- breast cancers (21% and 31% respectively) overexpress cyclin D1 [33].
Several studies have shown that G1/S phase regulatory molecules may drive lower survival rates in patients and resistance to adjuvant therapies. Deregulated expression of cell cycle molecules that specifically modulate CDK2 kinase activity has been associated with poor prognosis of breast cancer patients. For example, high cyclin A expression in metastatic breast cancer correlates significantly with a shorter time to first relapse and is a prognostic factor in early stage ER+ breast tumors [34, 35]. Additionally, high cyclin E expression predicts a poor prognosis in breast cancer [36, 37]. Unregulated CDK2 activity may result in poorer survival due to the modulation of responses to various therapeutic agents. For example, low expression of p27Kip1 and/or cyclin E overexpression predicts early relapse in patients treated with adjuvant therapy that includes tamoxifen [38, 39], doxorubicin, cyclophosphamide [40], and Herceptin [41]. Increased CDK2 kinase activity drives Herceptin resistance in vitro and in vivo [41, 42].
Deregulated CDK4/CDK6 activities have also been associated with decreased survival and resistance to various therapies. Cyclin D1 is overexpressed in over 50% of breast cancers [43, 44]. The oncogenic capacity of cyclin D1/CDK4/CDK6 in experimental models, including mouse models of mammary carcinogenesis has been established [45–47]. However, the role of cyclin D1/CDK4/CDK6 in breast cancers is highly controversial. For example, increased cyclin D1 mRNA and amplification of CCND1 in ER+ breast cancers strongly correlated with increased risk of relapse, local recurrence, metastasis, and death [30, 31, 48], and ER- patients with cyclin D1 overexpression display shorter overall survival [33]. In contrast, other studies showed that patients overexpressing cyclin D1 (including ER+ tumors and unclassified breast tumors not falling within the canonical HER2, PR, ER classification) were less likely to recur following treatment and displayed longer survival [32, 49–51]. Similar to cyclins E and A, there is experimental evidence as to the involvement of cyclin D in resistance or sensitivity to various therapies. Patients within the ER+ subgroup who received endocrine therapy for their primary or recurrent breast cancers showed an association between high cyclin D1 and a shorter response duration [30, 52, 53]. In addition, in vitro studies showed that overexpressed cyclin D1 leads to resistance to antiestrogens [54]. While some studies showed that ectopic cyclin D1 expression is directly involved in radioresistance and the poor prognosis of various carcinomas after radiotherapy [55–59], others found that ectopic expression of cyclin D1 markedly increases cell sensitivity to apoptosis induced by various agents including ionizing radiation [60–63]. In addition, chemical inhibition of CDK4 and CDK6 synergizes with Herceptin and tamoxifen treatments [64].
Although CDK4 and CDK2 are promising targets in cancer therapeutics, their role in the response of ER-PR-HER2+ or ER-PR-HER2- breast cancer cells to ionizing radiation is controversial and not extensively explored. We present data showing that knockdown of CDK4, but not of CDK2, imparts radiosensitivity to breast cancer cells and normal mammary epithelial cells by signaling an apoptotic program.
Results
G1/S phase regulatory molecules are ectopically expressed in radioresistant breast cancer cells
While many deregulated signaling pathways, including PI3K, NFKB, and the MAPK pathways are involved in signaling radiation resistance [65], the role played by the G1/S phase regulatory machinery phase in radiation resistance is unclear. The overall hypothesis of this study is that breast cancer cells are radioresistant because they harbor deregulated G1/S phase cell cycle machinery. The controversy regarding the role played by the G1/S phase regulatory machinery in radioresistance may be due to the use of normal or transformed cells lines, their tissue of origin, or to the dose and duration of radiation treatments [55, 58–60]. Because of this, we tested our hypothesis in a non-transformed mammary epithelial cell line and breast cancer cell lines of various molecular subtypes, irradiated with increasing, single doses of radiation. We first assessed the ability of MCF10A (non-transformed, immortalized mammary epithelial cells lacking the cyclin kinase inhibitors p16INK4A and p15INK4B due to a homozygous deletion), ER-PR-HER2- and ER-PR-HER2+ breast cancer cells to form colonies after increasing single doses of γ-radiation (0, 1, 2, 4, 6, or 8 Gy). Analyses revealed that most of the breast cancer cell lines tested in this study showed resistance to radiation with a statistically significantly increased IC50 comparing to the immortalized MCF10A cells (Table 1).
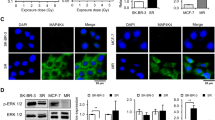
To identify mechanisms resulting in radioresistance, we assessed the relative protein expression of G1/S cell cycle regulatory molecules that have been shown to associate with relapse and to impaired responses to various therapies. We focused on proteins that affect CDK2 or CDK4 activities, including cyclins A, E, D, p21CIP1, and p27KIP1 (Figure 1). Western blot analyses showed that p27KIP1 levels were higher in untreated ER-PR-HER2- breast cancer cells relative to MCF10A cells and showed no change upon irradiation (Figure 1A). Levels of cyclin E were higher in MCF10A cells relative to breast cancer cells irrespective of irradiation. Other molecules, including cyclin A and p21CIP1, were upregulated in both ER-PR-HER2- and MCF10A cells after irradiation, while cyclin D1 levels did not change. We measured cyclin D1 levels in non-irradiated MCF10A, ER-PR-HER2-, and ER-PR- HER2+ cell lines and showed that all breast cancer cell lines overexpressed cyclin D1 (Figure 1B).
G1/S phase regulatory molecules are ectopically expressed in radioresistant breast cancer cells. (A) Non-transformed human mammary epithelial cells (MCF10A) and ER-PR-HER2- human breast cancer cell lines (HCC1806, HS578, MDA-MB-231 & MDA-MB-468) were irradiated at 2 Gy. Western blots of lysates prepared from both the unirradiated (U) and the irradiated (I) cells were immunoblotted with anti-cyclin A, anti-cyclin E, anti-p21CIP1 and anti-p27KIP1 antibodies. (B) Western blot analyses of lysates prepared from various unirradiated human breast cancer cell lines (triple negative and ER-PR-HeR2+ and MCF10A) were immunoblotted with anti-cyclin D1 antibody. (C) Lentivirus containing shRNA constructs specific for CDK2 or CDK4 were used to infect human breast cancer cell lines and MCF10A cells. A pLKO.1 lentiviral vector containing a randomized shRNA sequence was used as control. After puromycin selection, the surviving cells were pooled. The lysates prepared from the infected cells were immunoblotted with anti-CDK2 or anti-CDK4 antibodies to confirm the knockdown efficiency. Equal loading was assessed by immunoblotting with a β-actin antibody.
Because the breast cancer cell lines overexpressed cyclin D1, and cyclin A protein levels were elevated following irradiation, we tested whether knockdowns of their respective kinase partners CDK4 and CDK2 might alter the relative radioresistance of various breast cancer cell lines. Therefore, we generated ER-PR-Her2- and MCF10A cell lines stably expressing shCDK2 or shCDK4. All cell lines showed consistently stable decreases in the protein level of CDK2 or CDK4 (Figure 1C). MCF10A and HCC1806 cells displayed complete downregulation of CDK4, while MDA-MB-468 and MDA-MB-231 cells showed a partial, but almost complete knockdown of CDK4. In addition, HCC1806 and MDA-MB-231 cells displayed complete knockdown of CDK2, while MCF10A and MDA-MB-468 cells displayed a partial, but significant knockdown of CDK2. Importantly, knockdown of CDK2 did not affect CDK4 levels and vice versa, demonstrating that specific knockdowns were achieved. To assess whether silencing of CDK4 altered radioresistance in other breast cancer subtypes, we generated HCC1954 and SKBR3 cells (both ER-PR-HER2+ cell lines) knocked down for CDK4 (Figure 1C). Similarly to ER-PR-HER2- and MCF10A cells, CDK2 levels in ER-PR-HER2+ cells were not affected by knockdown of CDK4.
Downregulated CDK4 sensitizes breast cancer cells to radiation
After cell lines stably silenced for CDK2 and CDK4 were established, all cell lines received ionizing radiation at doses ranging between 0 to 8 Gy. The relative radiosensitivity and the radiation dose that causes a 50% decrease in the number of colonies (IC50) were calculated using colony forming assays (Figure 2A, Table 2). Even though CDK2 has been reported to mediate resistance to various adjuvant therapies against breast cancers, downregulation of CDK2 did not alter the sensitivity (IC50) to radiation in ER-PR-HER2- cells or in MCF10A cells relative to cells expressing control vector (Figure 2B, Table 2). In contrast, all cell lines harboring CDK4 knockdown, including ER-PR-HER2+, ER-PR-HER2-, and MCF10A showed significantly decreased IC50, meaning that the ablation of CDK4 in both the normal and tumor cell lines sensitizes these cells to radiation.
Downregulated CDK4 sensitizes breast cancer cells to radiation. Cells stably expressing pLKO.1, shCDK2 or shCDK4 were irradiated at single doses of 0, 1, 2, 4, 6, or 8 Gy γ-rays. After an average of 8 days, cells were fixed and stained with 1% crystal violet; colonies were counted manually. (A) The figure shows colony formation in MCF10A and ER-PR-HER2- cells at 0 and 6 Gy. (B) Survival curves of various cells transfected with pLKO.1, shCDK2 or shCDK4. ER-PR-Her2+cells were only transfected with pLKO.1 or shCDK4. The y axis represents percentage colony formation relative to unirradiated cells of the respective groups; the x axis represents the various radiation doses. The IC50 and statistical significance are presented in Table 2.
Radiosensitivity imparted by downregulated CDK4 is not caused by cell cycle blocks
Radioresistance can be a consequence of a deregulated cell cycle, altered frequencies of apoptosis, or altered frequencies of DNA repair [58–60]. To address whether radiosensitization occurs because of cell cycle blocks, we irradiated MCF10A and ER-PR-HER2- cells with 2 Gy and assessed percentages of proliferating cells using the BrdU incorporation assay by direct counting using fluorescence microscopy (Additional file 1A) and by flow cytometry (Table 3). Ki-67 was used as an independent marker of proliferation (Additional file 1B). Additional file 1 represents two preliminary experiments with a wider range of timepoints, to establish whether DNA replication was affected up to 48 hours post-irradiation. Neither BrdU nor Ki-67 immunocytochemistry showed differences in proliferation in cells downregulated for CDK4 relative to the control pLKO.1 group. Although cells expressing shCDK2 incorporated BrdU at a lower extent than other groups (Additional file 1A), no differences were found when using Ki67 as an independent marker of proliferation (Additional file 1B). After establishing that DNA replication was not affected, a smaller number of timepoints (0, 6, 12 and 24 hours post-irradiation) were chosen for final analyses. The relative percentage of cells in each phase of the cell cycle was assessed by measuring DNA content using flow cytometry (Figure 3), while a flow cytometry-based BrdU assay measured percentage of replicating cells post-irradiation (Table 3). The BrdU assay did not detect significant differences in the percentage of proliferating cells between control and cells silenced for CDK4 (Table 3). Likewise, flow cytometry did not reveal any major changes in the percentage of cells in G1, S, or G2/M phases of the cell cycle in irradiated or non-irradiated cells when comparing shCDK4 cells relative to pLKO.1 controls (Figure 3). Thus, downregulation of CDK4 does not radiosensitize cells through compromising their proliferation capacity.
Radiosensitivity imparted by downregulated CDK4 is not caused by altering cell cycle progression. Cells stably expressing pLKO.1 or shCDK4 were unirradiated (basal), or irradiated at 2 Gy. Then, cells were collected at different times post-irradiation (0, 6, 12, & 24 hours) stained with 7-AAD and were analyzed using flow cytometry to determine the cell cycle stages; 5000 cells per group were analyzed by the flow cytometer. Flow cytometry and analysis was repeated three times. The columns represent the percentage of cells in the G1, S or G2/M phase and presented as average±standard deviation. The G1/S and G2/M graphs were separated in order to make the G2/M data visible.
To measure if knockdown of CDK4 radiosensitizes cells by signaling mitotic blocks, cells were immunostained for phospho-histone 3 after irradiation. Histone 3 is one of eight core histone proteins that play an important role in transcription regulation in eukaryotes through various modifications, including phosphorylation [66]. Phosphorylation of histone 3 at Ser10, Ser28 and Thr11 is correlated with chromosome condensation during mitosis [67, 68] and is regarded as an indicator of mitosis. Overall, no significant elevations in phospho-histone 3 positive cells were observed after irradiation in pLKO.1 or shCDK4 cells (Table 4). This demonstrates that CDK4 silencing does not radiosensitize cells by causing accumulation of cells in mitosis.
Downregulation of CDK4 does not alter rates of DNA break repair
To establish whether increased DNA repair is responsible for radiation resistance, we measured DNA breaks by detecting γ-H2AX foci. H2A.X is a variant histone which is phosphorylated on Ser139 (γ-H2AX) by ATM, ATR, or DNA PK at sites of DNA damage to recruit DNA repair enzymes, thus representing an important mark of DNA breaks [69–71]. This mark is one of the most sensitive ways to detect active DNA repair foci [70, 72]. Increased frequencies of γ-H2AX foci have been extensively used to measure DNA damage by physical agents, oncogenes, or inactive tumor suppressors [59, 73, 74]. Because phosphorylated γ-H2AX is a major sensor of double strand breaks, clearance of phospho-H2AX foci following irradiation is also used as a measure of the repair of that DNA damage [70, 72]. Additional file 2 represents a preliminary experiment with a wider range of timepoints (up to 48 hours post-irradiation), to establish whether γ-H2AX foci were detected in non-irradiated cells (basal) or following irradiation with 2 Gy. To measure the relative repair capability of various cells, we irradiated cells with 2 Gy and detected γ-H2AX foci at different time points post-irradiation. γ-H2AX foci were clearly detectable upon irradiation (Additional file 2A). Regardless of the cell line, the percentage of γ-H2AX foci decreased gradually post-irradiation in all groups and reached basal levels by 24 hours. These baseline levels were maintained 48 hours post-irradiation. The γ-H2AX results obtained by immunocytochemistry were independently investigated in MCF10A, MDA-MB-231, and MDA-MB-468 cells using flow cytometry. This assay confirmed that no significant differences existed between control and cells silenced for CDK4 (Table 5). The experiments indicate that the double stranded DNA repair capacity of all cells analyzed in this study was intact.
Irradiated breast cancer cell lines silenced for CDK4 display increased frequencies of apoptosis
Another mechanism that can explain the radiosensitization of cells after CDK4 silencing is that its loss potentially enhances frequencies of apoptosis. A pilot experiment established the window of maximal apoptosis following irradiation. Cell lines were either non-irradiated (basal), or received a single dose (2 Gy) of radiation and were collected at different times post-irradiation (0, 6, 12, 24 and 48 hours). Commitment to apoptosis was detected by immunocytochemistry against the apoptotic effector cleaved caspase-3 [75] (Figure 4A). This experiment revealed a gradual increase in the percentage of apoptotic cells, which became maximal at 48 hours (Additional file 3). Silencing of CDK4 caused elevation in apoptosis following irradiation in all cell lines tested, while knockdown of CDK2 did not lead to any changes in frequencies of apoptosis. Based on the results from the initial experiment, cells were irradiated with 0, 2, and 4 Gy, and analyzed 48 hours post-irradiation. Those experiments confirmed a significant elevation of apoptosis in cells silenced for CDK4 (Figure 4B). While MCF10A cells silenced for CDK4 displayed a significant proportion of caspase-3-positive cells under non-irradiated and with 2 Gy radiation, MDA-MB-468 cells silenced for CDK4 displayed a significant elevation in apoptosis only when irradiated at 2 and 4 Gy. Even though the proportion of apoptotic cells in MDA-MB-231 silenced for CDK4 were higher than controls in the basal and the 4 Gy groups, they did not reach statistical significance.
Irradiated breast cancer cell lines knocked down for CDK4 display increased frequencies of apoptosis. (A) Cells stably expressing pLKO.1, shCDK2 or shCDK4 were unirradiated (basal), irradiated at 2 or 4 Gy. Cells were fixed at 48 hours post-irradiation. Cells were subjected to immunostaining with anti-cleaved caspase-3 antibody (arrows), and an Alexa Fluor 488 secondary antibody; DNA was counter-stained with DAPI. Pictures were collected at 20× magnification. (B) The number of cells positively stained with cleaved caspase-3 were counted in a population of at least 287 cells and the results are shown as the proportion of positive cells between treatment and control groups. The experiments were repeated 6 times. Statistical significance was calculated by a Chi-square/Fisher exact test (*=p≤0.05; **=p≤0.01). (C) Cells stably expressing pLKO.1, or shCDK4 were unirradiated (0), irradiated at 2 or 4 Gy. Protein lysates were prepared 48 hours later and were subjected to Western blot with anti-cleaved PARP antibody. β-actin was used as a loading control. (D) The CDK4/6 inhibitor PD0332991 was added to cells stably expressing pLKO.1 (100 nM, 500 nM and 1000 nM for MCF10A, MDA-MB-231 and MDA-MB-468, respectively) and unirradiated (0), irradiated at 2 or 4 Gy. Protein lysates were prepared 48 hours later and were subjected to Western blot with anti-cleaved PARP antibody. β-actin was used as a loading control. Levels were normalized based on β-actin, and are indicated as fold-induction relative to non-irradiated pLKO.1 controls.
To establish whether the increases in apoptosis caused by radiation in cells silenced for CDK4 could be detected with an independent apoptosis marker, cells were subjected to Western blots using cleaved PARP (Figure 4C). In accordance with cleaved caspase-3 immunocytochemisty, higher levels of cleaved PARP were detected in MCF10A (6 to 11 fold changes) cells silenced for CDK4 compared to vector control. Consistent with the small increases in apoptosis with cleaved caspase-3, cleaved PARP levels were slightly elevated in MDA-MB-231 silenced for CDK4 relative to pLKO.1 controls. Likewise, we detected slightly elevated cleaved PARP levels in MDA-MB-468 shCDK4 cells irradiated with 2 Gy relative to MDA-MB-468 pLKO.1 cells. In contrast, no difference was found in cleaved PARP between MDA-MB-468 shCDK4 and MDA-MB-468 pLKO.1 cells upon irradiation with 4 Gy.
To establish whether radiosensitization also occurred through autophagy, we probed Western blots with an antibody against LC3A/3B. In MCF10A cells, only the lower band, which is the indicator of autophagy, was detected with no great difference among samples. Both inactive and active bands were observed in MDA-MB-231 and MDA-MB-468 cells, again with no great differences among samples (Additional file 4A). Finally, to investigate if the CDK4/6 kinase inhibitor, PD0332991, has a similar effect on inducing apoptosis or autophagy, MCF10A, MDA-MB-231 and MDA-MB-468 cells were treated at their respective IC50s [64] of 100 nM, 500 nM, and 1000 nM PD0332991, irradiated after 24 hours, and analyzed after 48 hours for cleaved PARP (Figure 4D). Unlike biological knockdown, levels of cleaved PARP did not increase in MCF10A pLKO.1 cells treated with PD0332991. Instead, high levels of cleaved PARP (2 to 3 fold) were detected in MDA-MB-468 pLKO.1 cells treated with PD0332991 compared to untreated cells. Again, LC3A/3B levels were not changed greatly among any of the cell lines, and did not change in the presence or absence of CDK4 (Additional file 4B). These results demonstrate that knockdown of CDK4 and chemical inhibition of CDK4/CDK6 lead to different outcomes, as the inhibitor is protective of apoptosis in irradiated MCF10A and MDA-MB-231 cells, while promoting apoptosis in MDA-MB-468 cells. Overall, the results also demonstrate that the primary mechanism of cell death in this system is through apoptosis and not autophagy.
Knockdown of CDK4 results in reduced phosphorylation of BAD (ser136) and upregulation of protein phosphatase type 2A (PP2A)
To assess why cells knocked down for CDK4 are prone to apoptosis upon irradiation, we performed a screen of various Bcl family members, with antibodies against the phosphorylated forms of those proteins, as well as total protein levels (Figure 5A). The lysates were obtained from cells pre-treated with Calyculin-A inhibitors, which inhibits phosphatase 1, 2A and 2B; the limitation of such method is that it increases the basal levels of most serine/threonine phosphorylation in cells. This method has been successfully used to detect phosphorylation of CDK2 or CDK4 targets, including NPM [73, 76, 77]. There were no significant changes in the levels of Bcl-2 (ser70) or Mcl-1 (ser159/thr163), or in the total levels of those proteins. However, lower levels of phospho-Bad (ser136) were observed in all irradiated ER-PR-HER2- cells expressing shCDK4, while MCF10A cells had elevated levels of phospho-Bad (ser136). Bad is a proapoptotic member of the Bcl-2 family that promotes cell death by displacing Bax from binding to Bcl-2 and Bcl-xL [78]. Phosphorylation of Bad at ser112 or ser136 via AKT inhibits the apoptotic activity of Bad by promoting binding of Bad to 14-3-3 proteins to prevent an association between Bad with Bcl-2 and Bcl-xl [79]. The dephosphorylation of Bad is mainly accomplished by various protein phosphatases [80–82]. We speculated that Bad ser136 was being dephosphorylated through the increased activity and/or levels of a phosphatase. It has been reported that phospho-ser136 is more efficiently hydrolyzed by PP1? and PP2A relative to PP2B and PP2C [82]. To find out what is responsible for the dephosphorylation of Bad at ser136, we detected the relative protein levels of the protein phosphatases PP1α, PP2A, and PP2B. As shown in Figure 5B, the basal levels of PP2A at time 0 were immediately upregulated in MCF10A and MDA-MB-231 cells lacking CDK4 upon irradiation, and the levels remained upregulated. As for MDA-MB-468 cells, the levels of PP2A increased gradually following irradiation. These experiments suggested that downregulation of CDK4 lead to increases in PP2A, which dephosphorylates phospho-Bad in ser136.
Downregulation of Cdk4 results in impaired phosphorylation of BAD (ser136) and upregulation of protein phosphatase type 2A (PP2A). Cells stably expressing pLKO.1 or shCDK4 were irradiated at 2 Gy. Protein lysates were then prepared at different times (0, 0.5, 1 & 2 hours) post-irradiation and were immunoblotted with anti-Bad(ser136), anti-Mcl-1(ser159/thr163) and anti-Bcl-2(ser70) antibodies, and their counterparts detecting total levels of those proteins. (B) The levels of PP2A and PP2B were further determined by immunoblotting with anti-PP2A and anti-PP2B antibodies. Cells transfected with pLKO.1 were used as control. The β-actin controls for (B) is the same as for (A).
Silencing the catalytic subunit of PP2A does not lead to increased PARP cleavage in irradiated cells silenced for CDK4
To investigate whether silencing of PP2A reverses apoptosis in irradiated cells silenced for CDK4, the PP2A catalytic unit was knocked down using siRNAs (Figure 6). PP2A knock down was efficient in MDA-MB-231 (70%) and MDA-MB-468 cells (50%) and irradiation further decreased PP2A level in both negative control (30% less compared to nonirradiated) and cells silenced for PP2Ac (40-60% less compared to nonirradiated control). Overall, silencing PP2A did not change upregulated cleaved PARP levels in MDA-MB-468 shCDK4. On the other hand, it elevated cleaved PARP in MDA-MB-231 shCDK4 cells level compared to the negative controls. Knockdown PP2A in MCF10A cells silenced for CDK4 under the same conditions was not successful. The autophagy marker LC3A/3B levels were not changed greatly among cell lines and samples (Additional file 4C). This data indicates that downregulation of PP2A does not reverse apoptosis in cells knocked down for CDK4. We conclude that apoptosis in irradiated cells silenced for CDK4 does not involve the PP2A catalytic subunit.
Silencing PP2A does not change cleaved PARP level in Cdk4 silencing cells. A cocktail of siRNAs targeting the PP2A catalytic unit was transiently transfected into cells stably expressing shCDK4 and cells were irradiated at 0, 2 and 4 Gy after 48 hours transfection. Protein lysates were then prepared at 48 hours post-irradiation and were immunoblotted with PP2A, cleaved PARP. β-actin was used as a loading control. Protein levels were normalized based on β-actin, and are indicated as fold-induction relative to scrambled siRNA controls.
Discussion
The G1 phase CDKs, (CDK2, CDK4, and CDK6) display specific functions in terms of development and redundant functions in terms of cell cycle regulation [23, 24, 83]. The redundant nature of the G1 phase CDKs is best illustrated by work showing that cells can proliferate in the absence of CDK2 and CDK4, and that CDK1 activity is required for cell cycle progression [84, 85]. However, injured hepatocytes do not require CDK1 to proliferate [86]. Another important function of the CDKs is in DNA repair. Specifically, cyclin D1, CDK4 [58, 59, 87], cyclin A1, and CDK2 have been implicated in DNA repair [88–92]. In fact, the activities of all CDKs are required for efficient DNA repair [93]. Even though CDK2 and CDK4 are largely dispensable in the entry and progression through S phase, they are important mediators of mammary tumorigenesis in mice, as ablation of either CDK2 or CDK4 can prevent mouse mammary tumorigenesis triggered by HER2/Neu [94, 95]. Also, CDK2 and CDK4 can differentially mediate centrosome amplification depending on the oncogenic or cellular context [96]. While cyclin D1/CDK4 specifically mediates centrosome amplification triggered by H-RasG12D, H-RasG12D&c-Myc [97], and in HER2+ breast cancer cells [76], silencing of cyclin E or CDK2 have no impact on centrosome amplification in the HER2/Ras systems. In contrast, both CDK2 and CDK4 mediate centrosome amplification and chromosome instability in p53 deficient cells [73].
Overall, the ability of CDK2 and CDK4 to mediate various malignant phenotypes that play important roles in cancer biogenesis has prompted the development of CDK2 or CDK4 specific inhibitors. Those inhibitors have proven to be effective in a variety of breast cancer cell lines. For example, ER+ luminal cells that overexpress cyclin D1 and Rb and display downregulated p16 respond well to cell cycle inhibition by the CDK4/CDK6 specific inhibitor PD-0332991, while non-luminal/basal cells are refractive to growth inhibition [64]. It has been reported that the Rb status dictates the early responses to that inhibitor [98]. Even though the CDK4/CDK6 inhibitor has shown promise in preclinical trials, cells acquire resistance through the upregulation of CDK2 activity [98]. A major limitation of PD-0332991 is that while it is effective in extinguishing mammary tumors, its ability to block the cell cycle interferes with chemotherapy [99, 100].
In the present study, we addressed whether the G1/S cell cycle regulatory machinery influences radioresistance in MCF10A, ER-PR-HER2-, and ER-PR-HER2+ cells. There is a precedent for the role of ectopic CDK2 and CDK4 activities in imparting resistance or sensitivity to radiotherapy. For example, knockdown of cyclin E synergizes with doxorubicin to enhance radioresistance in breast cancer cells lines [101]. Induction of cyclin A has previously been observed in cells treated with UV or irradiation [91, 102]. Cyclin A null cells are radiosensitive and display impaired double strand break repair; cyclin A/CDK2 is involved in DNA repair following irradiation by phosphorylating KU-70 [91].
Various mechanisms have been postulated to explain how cyclin D1/CDK4 leads to radioresistance or radiosensitivity of various cells. Inducible expression of cyclin D1 in MCF7 breast cancer cells leads to radiosensitivity through activation of the p53 pathway [60]. Low dose irradiation promotes the free, cytoplasmic cyclin D1 accumulation in human keratinocytes, correlating with radioresistance; in this context, low level radiation disrupted the interaction of cyclin D1 with 14-3-3ζ [55]. Cytoplasmic cyclin D1 then interacts with Bax, suppressing the ability of Bax to induce apoptosis. In another example, long term fractionated irradiation of the human cancer cell lines HepG2 and HeLa induced radioresistance [59]. This radioresistance correlated with upregulation of cyclin D1 due to the stabilization of cyclin D1 by prevention of its proteolysis, achieved by the DNA PK/AKT/GSK3β pathway. In addition, they first proposed that ectopic expression of cyclin D1 leads to increased radioresistance in breast cancer cells by enhancing DNA repair in the radioresistant cells, as downregulation of cyclin D1 in this system abrogated enhanced DNA repair, resulting in radiosensitivity. This group showed using chemical inhibitors that cyclin D1 dependent radioresistance is reversed by AKT or CDK4 chemical inhibitors. A recent report also demonstrated that enhanced cyclin D1 results in enhanced DNA repair and radioresistance [58]. This report postulates that cyclin D1 promotes DNA repair by interacting with RAD51 within DNA repair foci; this interaction is promoted by radiation.
Our studies demonstrate that knockdown of CDK4 acts as a potent radiosensitizer, independently of the breast cancer molecular subtype. In our studies, we tested whether radiosensitization was a consequence of cell cycle inhibition by silenced CDK4, altered DNA break repair, or through the activation of an apoptotic program. We used various assays to show that knockdown of CDK4 did not influence the cell cycle of non-irradiated or irradiated cells. This is consistent with our previous observations that in MCF10A cells expressing control pLKO.1, H-RasG12V, H-RasG12V&c-Myc, or in HER2+ breast cancer cells, knockdown of CDK4 does not greatly influence DNA replication or cell cycle progression [76, 97]. This is also consistent with our data showing that ablation of CDK4 does not alter the cell cycle of p53−/− mouse embryonic fibroblasts [73]. In contrast, co-inhibition of CDK4 and CDK6 using a chemical inhibitor interferes with the cell cycle, leading to protection against chemotherapy [99, 100]. Our studies conducted with the PD0332991 CDK4/CDK6 inhibitor revealed a similar story, as treatment of MCF10A or MDA-MB-231 protected against radiation-induced apoptosis. This is contrary to our studies revealing that knockdown of CDK4 synergizes with radiation to induce apoptosis in breast cancer cells and MCF10A controls. The only cell in which radiation synergized with inhibition of CDK4/CDK6 was MDA-MB-468, a cell that is insensitive to growth inhibition by PD0332991. However, we do not think that insensitivity to growth inhibition is the sole cause of the sensitivity to apoptosis triggered by PD0332991 treatment, as we treated all cell lines exactly at IC50 to prevent complete growth inhibition. One potential explanation for the differences between knockdown of CDK4 and inhibition with PD0332991 is that while the chemical inhibits CDK4/CDK6 activity, the shCDK4 may disrupt the protein:protein interactions of CDK4 with cyclin D, p21, p27, p15, p16, p18 and p19, leading to modulation of the activities of other CDKs.
In contrast to studies showing that increased cyclin D1 activity results in radioresistance through increasing cellular DNA repair capacity, all breast cancer cells tested in this study efficiently repaired DNA breaks irrespective of CDK4 levels, as indicated by the efficient clearance of H2AX foci after radiation. A recent study is consistent with our results, as chemical inhibition of CDK4 and CDK6 in breast cancer cells did not influence double-strand break repair after irradiation [99]. We speculate that the differences between cyclin D1 and CDK4/CDK6 inhibition in relation to DNA repair is because cyclin D1 displays functions that are independent of CDK4. For example, it can bind the promoters of genes involved in chromosome instability [103].
The presence of increased levels of cleaved caspase-3 and of cleaved PARP in all irradiated cells down regulated for CDK4 indicated that apoptosis was a significant driver of radioresistance in mammary epithelial and breast cancer cells. Although apoptosis is high in unirradiated MCF10A cells silenced for CDK4, levels of apoptosis rise significantly in those and in the breast cancer cells silenced for CDK4 upon irradiation. This suggests synergy between downregulation of CDK4 and irradiation in regards to apoptosis. To establish how absence of CDK4 cooperates with radiation to enhance cell death, we screened various Bcl family members. No changes in the levels of phosphorylation of the antiapoptotic members Bcl2 or Mcl1 were observed in the cells. In contrast, levels of phosphorylated Bad ser136 were sharply diminished Even though dephosphorylation of Bad in serine 136 is associated with apoptosis, we observe increased phosphorylation in irradiated shCDK4 MCF10A cells. The regulation of apoptosis is complex, involving various protein/protein interactions, as well as the post-translational modification of numerous proteins. Further studies must be performed in order to establish why MCF10A cells silenced for CDK4 display phosphorylated Bad Ser 136, and still apoptose. Bad ser136 phosphorylation is reversed by various phosphatases, including PP2A and PP1α [81, 82]. In fact, silencing of CDK4 resulted in higher levels of PP2A, suggesting that it specifically dephosphorylates Bad ser136 upon irradiation PP2A is a protein complex with many cellular functions, including the regulation of apoptotic and mitogenic pathways [104–106], as well as the modulation of DNA repair [107]. Inhibition of PP2A by DNA tumor viruses is tightly linked to cellular transformation [108]. Also, the dephosphorylation of Bcl2 by PP2A promotes its anti-apoptotic activity; this event may enhance Bcl2’s oncogenic potential [109]. In wide contrast, PP2A’s proapoptotic activities have been mapped to its ability to dephosphorylate Bad [81, 82] and Bax [110]. We did not observe significant dephosphorylation of Bcl2 in any of the cell lines, suggesting that upregulation of PP2A in breast cancer cells exclusively dephosphorylates Bad ser136 without significantly affecting other Bcl family members. Thus, our observation that downregulation of CDK4 resulted in decreased phosphorylation of Bad ser136 suggests that this event is primarily responsible for apoptosis and radioresistance. Additional experiments are required to address how CDK4 regulates PP2A levels; that regulation may result from increased protein stability, increased degradation, or enhanced de-repression of E2F target genes upon CDK4 silencing. Another important future area of exploration is whether and how the apoptotic program triggered by downregulation of CDK4 in conjunction with radiation is rooted in Rb/E2F dependent transcription.
Unfortunately, we were unable to show that ectopic expression of PP2Ac radiosensitizes breast cancer cells, or that its inhibition modulates radiation induced apoptosis in breast cancer cells. We conclude that apoptosis in cells knocked down for CDK4 is independent of PP2A. Further experiments are required to establish pathways signaling apoptosis in irradiated breast cancer cells knocked down for CDK4.
Conclusion
We conclude that stable knockdown of CDK4 enhances the effects of radiation by enhancing apoptotic pathways. This result has implications for future therapies, as patients can be treated with radiation or chemotherapeutic agents and with doses of CDK4/CDK6 inhibitors that do not block the cell cycle. In that scenario, suppression of CDK4/CDK6 activity would allow cell cycle progression, while synergizing with radiation to increase cell death.
Materials and methods
Cell culture
A human mammary epithelial cell line (MCF10A) and breast cancer cell lines (ER-PR-HER2-:HCC1806, HS578, MDA-MB-231, MDA-MB-468; ER-PR-HER2+: SKBR3 and HCC1954), were purchased from ATCC (Rockville, MD), or obtained through collaborators. The molecular markers and various other characteristics of the breast cancer cells used in this study have been previously described [111]. HCC1806, HCC1954, and SKBR3 cells were cultured in RPMI supplemented with 10% fetal bovine serum (FBS). HS578, MDA-MB-231 and MDA-MB-468 cells were cultured in DMEM supplemented with 10% FBS. MCF10A cells were cultured in DMEM/F12 supplemented 10% FBS, 20 ng/ml EGF, 100 ng/ml cholera toxin, 0.01 mg/ml insulin, and 500 ng/ml hydrocortisone. All cell lines were cultured at 37°C in a humidified atmosphere (5% CO2).
Colony forming assay
Two thousand five hundred cells were plated per well in a 6-well culture plate or one thousand cells were plated per well in a 12-well cell culture plate (Day 0) in triplicates. After 24 hours (Day 1), cells were irradiated at a dose of 0, 1, 2, 4, 6, or 8Gy in an X-RAD 320 irradiator (Precision X-Ray, CA). After an average of 8 days, cells were fixed with 75% ethanol and stained with 1% crystal violet, and colonies containing more than 50 cells (established by microscopy) were counted manually. Plating efficiency was calculated by dividing the number of colonies in the unirradiated group divided by the number of colonies plated and multiplying by 100.
Lentiviral shRNA transduction
The shRNA constructs against CDK2 or CDK4 were cloned into the lentiviral vector pLKO.1, which are commercially available from OPEN Biosystems. Lentiviral infections were done as described in our publications [97]. Briefly, cells were plated in regular medium at 50% confluence on day 1. This was followed by changing into medium containing 5 μg/ml polybrene and infection of cells by adding undiluted shRNA lentiviral particles to the culture on day 2. Stable clones expressing the shRNA were obtained by splitting cells 1:5 and culturing in medium containing puromycin.
Immunocytochemistry assays
Immunofluorescence was performed by following our published protocols [73, 97]. Cells were plated at 4×104 per well into two-well tissue culture chamber slides and received 2 Gy irradiation after cells attached. Cells were then fixed in cold 4% paraformaldehyde at different times postirradiation (from 0 to 48 hr), washed in PBS, permeabilized in a 1% NP-40–PBS solution, and blocked in 10% goat serum. Commitment to apoptosis was detected by using an antibody against cleaved caspase-3 (9661, Cell Signaling). DNA breaks were detected using an antibody against phosphorylated histone 2A variant X (γ-H2AX) (ab2893, Abcam). Cell proliferation was examined using an antibody against Ki-67 (ab15580, Abcam). The BrdU assay was used to detect cells in S phase and was done by pulsing cells with 20 μM BrdU (51-7581KZ; BD Pharmingen) 30 minutes before harvest and detecting with an anti-BrdU antibody (NA61; Calbiochem). To detect mitosis, cells were plated at 1×104 per well into four-well tissue culture chamber slides and received 0, 2 and 4 Gy irradiation 24 hours after plating and incubated 48 hours and phosphor-histone 3 (Ser 28, 9713, Cell Signaling) antibody was used to detect mitosis. Secondary antibodies are conjugated with Alexa Fluor 555 or Alexa Fluor 488. Cells were counterstained with DAPI. Frequencies of the various phenotypes are done by counting the number of antigen-positive cells divided by the total number of cells; unless otherwise indicated, we count at least 200 cells per group. For mitosis, over 300 cells per group were counted.
Western blots
Western blotting was performed according to published protocols [73, 97]. Protein lysates were obtained by incubating cells in lysis buffer (50 mM HEPES, 250 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.1% NP-40, 10% glycerol) containing protease inhibitors (04693124001, Roche) for 30 min at 4°C. Calyculin phosphatase inhibitor (Millipore 19-139v) was added to the culture medium at a concentration of 100 nM, 30 minutes before harvest for the detection of phosphorylated proteins. Samples were denatured at 95°C for 5 min in sodium dodecyl sulfate (SDS) sample buffer, resolved by SDS polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Bio-Rad). The blots were incubated in blocking buffer (5% nonfat dry milk in 1X Tris-buffered saline plus 0.1% Tween 20 [TBS-T]) for 1hr and were then probed overnight at 4°C with the primary antibodies. The blots were then rinsed in 1X TBS-T and incubated with the appropriate horseradish peroxidase conjugated secondary antibodies at room temperature. The blots were then rinsed in 1X TBS-T and the antibody-antigen complex was visualized using Lumigen TMA-6 (RPN2135v1 and RPN2135v2; GE Healthcare).
Antibodies and inhibitors
The antibodies used in the various Western blotting experiments were as follows: CDK2 (sc-163, Santa Cruz), CDK4 (2906, Cell Signaling), p21Waf1 (sc-397, Santa Cruz), p27Kip1 (sc-528, Santa Cruz), β-actin (4970, Cell Signaling), cyclin A (ab38, Abcam), cyclin D1 (2922, Cell Signaling), cyclin E (sc-481, Santa Cruz), Phospho-Bcl-2 ser70 (2827, Cell Signaling), Bcl-2 (OP60, Calbiochem), Phospho-Mcl ser159/thr163 (4579, Cell Signaling), Mcl-1 (sc-819, Santa Cruz), Phospho-Bad ser136 (06–846, Upstate), Bad (1541–1, Epitomic), Phospho-AKT Ser 473 (Cell Signaling), PP1α (2582, Cell Signaling), PP2A (2259, Cell Signaling) and PP2B (2614, Cell Signaling), cleaved PARP (Asp214, 5625, Cell Signaling). The CDK4/6 inhibitor PD0332991 (Axon1505) was purchased from Axon Medchem. When indicated, ImageJ software was used to quantify protein levels of Western blots (NIH).
Flow cytometry
Cells were irradiated at 2 Gy, pulsed with BrdU (added at a final concentration of 10 μM 30 min prior to harvest) and harvested at various time points (6, 12, and 24 hours). Timepoints were chosen based on our preliminary and supplementary data. A unirradiated control was also collected. 1×106 cells were transferred to 5ml round bottom tubes (352235, BD), and cell staining was performed according to the protocol included with the “Apoptosis, DNA damage and Cell Proliferation kit” (562253, BD) or stained with 7-AAD (559925, BD Biosciences). BrdU was detected using PerCP-Cy™5.5 Mouse Anti-BrdU (51–9007682, BD), PARP cleavage was identified with PE Mouse Anti-Cleaved PARP (Asp214) Antibody (51–9007684, BD) and γH2AX was distinguished using Alexa Fluor® 647 Mouse Anti-H2AX (pS139) (51–9007683, BD). A LSRII flow cytometer (BD) was used to read each sample. Compensation was performed using Anti-Mouse Ig, κ/Negative Control (FBS) Compensation Particles Set (BD, 552843) and samples were analyzed using Flowjo v7.6.5 and v10.4 (Tree Star).
Statistical methods
For the colony forming assay, a dose response analysis was conducted, in which the dose response relationship was assumed to be the commonly used four parameter logistic function [112]. The data was normalized relative to the baseline dose level=0 Gy (no radiation), and the percentage of control was used as the primary outcome in the analysis. IC50, the radiation dose that is required for 50% reduction from baseline dose level, was the parameter of interest and compared accordingly. The higher value of IC50, the higher the resistance. A proper data transformation, Box- Cox transformation, [113] was applied to assure the fitted model meeting the statistical assumptions. All data management and statistical analyses were done in R [114]. T-tests were done in Excel. The statistical significance level of all tests was set at 0.05.
For the flow cytometry assays (DNA content and BrdU) the data was aligned by the six-hour gates. The two measurements (percentages of positive cells) per each run are averaged at each time point and the averaged measurement at each of 6 hours, 12 hours and 24 hours after radiation is divided by the averaged measurement at baseline. The mean percentage of the positive cells relative to no radiation is used for analyses, named fold-change to no radiation. The main goal of this data analysis is to compare the fold change between control and treatment groups at each time point, within each cell line, biomarker, and gate. The fold change is compared between control group and treatment group (pLKO.1 vs. shCDK4) at each time point within each cell line, biomarker, and gate using ANOVA (Analysis of Variance) to estimate the mean and standard error for the fold change to non-radiation. For the cleaved caspase-3 assay, observations (number of positive or negative cells) are combined within each radiation dose of 2 Gy, 4 Gy or no radiation treated cells for each control group and treatment group and then the Chi-square test or Fisher’s exact test is used to compare the proportions of positive and negative cells between treatment and control groups. Analysis unit is a cell. The significance levels are set at 0.05 for all tests. The SAS statistical package V9.3 (SAS Institute, Inc., Cary, North Carolina) is used for data managements and analyses.
Ethical approval
No research on humans or animals were conducted. All human cell lines were purchased from the publically available American Type Culture Collection (ATCC), or obtained through collaborators.
Abbreviations
- CDK:
-
Cyclin-dependent kinase
- Gy:
-
Gray.
References
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ: Cancer statistics, 2008. CA Cancer J Clin 2008, 58: 71–96. 10.3322/CA.2007.0010
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de-Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL: Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001, 98: 10869–10874. 10.1073/pnas.191367098
De-Laurentiis M, Cianniello D, Caputo R, Stanzione B, Arpino G, Cinieri S, Lorusso V, De-Placido S: Treatment of triple negative breast cancer (TNBC): current options and future perspectives. Cancer Treat Rev 2010,36(Suppl 3):S80-S86.
Irvin WJ Jr, Carey LA: What is triple-negative breast cancer? Eur J Cancer 2008, 44: 2799–2805. 10.1016/j.ejca.2008.09.034
Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, Deming SL, Geradts J, Cheang MC, Nielsen TO, Moorman PG, Earp HS, Millikan RC: Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 2006, 295: 2492–2502. 10.1001/jama.295.21.2492
Lund MJ, Trivers KF, Porter PL, Coates RJ, Leyland-Jones B, Brawley OW, Flagg EW, O’Regan RM, Gabram SG, Eley JW: Race and triple negative threats to breast cancer survival: a population-based study in Atlanta, GA. Breast Cancer Res Treat 2009, 113: 357–370. 10.1007/s10549-008-9926-3
Bartelink H, Horiot JC, Poortmans PM, Struikmans H, Van den-Bogaert W, Fourquet A, Jager JJ, Hoogenraad WJ, Oei SB, Warlam-Rodenhuis CC, Pierart M, Collette L: Impact of a higher radiation dose on local control and survival in breast-conserving therapy of early breast cancer: 10-year results of the randomized boost versus no boost EORTC 22881–10882 trial. J Clin Oncol 2007, 25: 3259–3265. 10.1200/JCO.2007.11.4991
Ragaz J, Olivotto IA, Spinelli JJ, Phillips N, Jackson SM, Wilson KS, Knowling MA, Coppin CM, Weir L, Gelmon K, Le N, Durand R, Coldman AJ, Manji M: Locoregional radiation therapy in patients with high-risk breast cancer receiving adjuvant chemotherapy: 20-year results of the British Columbia randomized trial. J Natl Cancer Inst 2005, 97: 116–126. 10.1093/jnci/djh297
Correa C, McGale P, Taylor C, Wang Y, Clarke M, Davies C, Peto R, Bijker N, Solin L, Darby S: Overview of the randomized trials of radiotherapy in ductal carcinoma in situ of the breast. J Natl Cancer Inst Monogr 2010, 2010: 162–177.
Solin LJ: The impact of adding radiation treatment after breast conservation surgery for ductal carcinoma in situ of the breast. J Natl Cancer Inst Monogr 2010, 2010: 187–192. 10.1093/jncimonographs/lgq020
Kim SI, Park S, Park HS, Kim YB, Suh CO, Park BW: Comparison of Treatment Outcome Between Breast-Conservation Surgery with Radiation and Total Mastectomy without Radiation in Patients with One to Three Positive Axillary Lymph Nodes. Int J Radiat Oncol Biol Phys 2010,1;80(5):1446–1452.
Kuru B: Prognostic factors for locoregional recurrence and survival in stage IIIC breast carcinoma: impact of adjuvant radiotherapy. Singapore Med J 2011, 52: 289–298.
Masinghe SP, Faluyi OO, Kerr GR, Kunkler IH: Breast radiotherapy for occult breast cancer with axillary nodal metastases–does it reduce the local recurrence rate and increase overall survival? Clin Oncol (R Coll Radiol) 2011, 23: 95–100. 10.1016/j.clon.2010.10.001
Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, Ollila DW, Sartor CI, Graham ML, Perou CM: The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res 2007, 13: 2329–2334. 10.1158/1078-0432.CCR-06-1109
Wilder RB, Curcio LD, Khanijou RK, Eisner ME, Kakkis JL, Chittenden L, Agustin J, Lizarde J, Mesa AV, Macedo JC, Ravera J, Tokita KM: Results with accelerated partial breast irradiation in terms of estrogen receptor, progesterone receptor, and human growth factor receptor 2 status. Int J Radiat Oncol Biol Phys 2010, 78: 799–803. 10.1016/j.ijrobp.2009.08.081
Gabos Z, Thoms J, Ghosh S, Hanson J, Deschenes J, Sabri S, Abdulkarim B: The association between biological subtype and locoregional recurrence in newly diagnosed breast cancer. Breast Cancer Res Treat 2010, 124: 187–194. 10.1007/s10549-010-1135-1
Nguyen PL, Taghian AG, Katz MS, Niemierko A, Abi Raad RF, Boon WL, Bellon JR, Wong JS, Smith BL, Harris JR: Breast cancer subtype approximated by estrogen receptor, progesterone receptor, and HER-2 is associated with local and distant recurrence after breast-conserving therapy. J Clin Oncol 2008, 26: 2373–2378. 10.1200/JCO.2007.14.4287
Wang SL, Li YX, Song YW, Wang WH, Jin J, Liu YP, Liu XF, Yu ZH: Triple-Negative or HER2-Positive Status Predicts Higher Rates of Locoregional Recurrence in Node-Positive Breast Cancer Patients After Mastectomy. Int J Radiat Oncol Biol Phys 2010.
Zaky SS, Lund M, May KA, Godette KD, Beitler JJ, Holmes LR, O’Regan RM, Yu ES, Yu DS, Landry JC: The Negative Effect of Triple-Negative Breast Cancer on Outcome after Breast-Conserving Therapy. Ann Surg Oncol 2011,18(10):2858–2865. 10.1245/s10434-011-1669-4
Hooning MJ, Botma A, Aleman BM, Baaijens MH, Bartelink H, Klijn JG, Taylor CW, van Leeuwen FE: Long-term risk of cardiovascular disease in 10-year survivors of breast cancer. J Natl Cancer Inst 2007, 99: 365–375. 10.1093/jnci/djk064
Jameel JK, Rao VS, Cawkwell L, Drew PJ: Radioresistance in carcinoma of the breast. Breast 2004, 13: 452–460. 10.1016/j.breast.2004.08.004
Pearce AG, Segura TM, Rintala AC, Rintala-Maki ND, Lee H: The generation and characterization of a radiation-resistant model system to study radioresistance in human breast cancer cells. Radiat Res 2001, 156: 739–750. 10.1667/0033-7587(2001)156[0739:TGACOA]2.0.CO;2
Malumbres M, Barbacid M: Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009, 9: 153–166.
Satyanarayana A, Kaldis P: Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 2009, 28: 2925–2939. 10.1038/onc.2009.170
Chen HZ, Tsai SY, Leone G: Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer 2009, 9: 785–797. 10.1038/nrc2696
Iaquinta PJ, Lees JA: Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol 2007, 19: 649–657. 10.1016/j.ceb.2007.10.006
Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, Delaloge S, Hortobagyi GN, Symmans WF, Lazar V, Pusztai L: Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res 2009, 15: 441–451. 10.1158/1078-0432.CCR-08-1791
Gauthier ML, Berman HK, Miller C, Kozakeiwicz K, Chew K, Moore D, Rabban J, Chen YY, Kerlikowske K, Tlsty TD: Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell 2007, 12: 479–491. 10.1016/j.ccr.2007.10.017
Bukholm IK, Berner JM, Nesland JM, Borresen-Dale AL: Expression of cyclin Ds in relation to p53 status in human breast carcinomas. Virchows Arch 1998, 433: 223–228. 10.1007/s004280050240
Kenny FS, Hui R, Musgrove EA, Gee JM, Blamey RW, Nicholson RI, Sutherland RL, Robertson JF: Overexpression of cyclin D1 messenger RNA predicts for poor prognosis in estrogen receptor-positive breast cancer. Clin Cancer Res 1999, 5: 2069–2076.
Roy PG, Pratt N, Purdie CA, Baker L, Ashfield A, Quinlan P, Thompson AM: High CCND1 amplification identifies a group of poor prognosis women with estrogen receptor positive breast cancer. International journal of cancer. Journal international du cancer 2010, 127: 355–360.
Utsumi T, Yoshimura N, Maruta M, Takeuchi S, Ando J, Mizoguchi Y, Harada N: Correlation of cyclin D1 MRNA levels with clinico-pathological parameters and clinical outcome in human breast carcinomas. Int J Cancer 2000, 89: 39–43. 10.1002/(SICI)1097-0215(20000120)89:1<39::AID-IJC7>3.0.CO;2-T
Umekita Y, Ohi Y, Sagara Y, Yoshida H: Overexpression of cyclinD1 predicts for poor prognosis in estrogen receptor-negative breast cancer patients. Int J Cancer 2002, 98: 415–418. 10.1002/ijc.10151
Poikonen P, Sjostrom J, Amini RM, Villman K, Ahlgren J, Blomqvist C: Cyclin A as a marker for prognosis and chemotherapy response in advanced breast cancer. Br J Cancer 2005, 93: 515–519. 10.1038/sj.bjc.6602735
Strand C, Ahlin C, Bendahl PO, Fjallskog ML, Hedenfalk I, Malmstrom P, Ferno M: Combination of the proliferation marker cyclin A, histological grade, and estrogen receptor status in a new variable with high prognostic impact in breast cancer. Breast Cancer Res Treat 2011. Epub 2011
Potemski P, Kusinska R, Pasz-Walczak G, Piekarski JH, Watala C, Pluciennik E, Bednarek AK, Kordek R: Prognostic relevance of cyclin E expression in operable breast cancer. Med Sci Monit 2009, 15: MT34-MT40.
Voduc D, Nielsen TO, Cheang MC, Foulkes WD: The combination of high cyclin E and Skp2 expression in breast cancer is associated with a poor prognosis and the basal phenotype. Hum Pathol 2008, 39: 1431–1437. 10.1016/j.humpath.2008.03.004
Filipits M, Rudas M, Heinzl H, Jakesz R, Kubista E, Lax S, Schippinger W, Dietze O, Greil R, Stiglbauer W, Kwasny W, Nader A, Stierer M, Gnant MF: Low p27 expression predicts early relapse and death in postmenopausal hormone receptor-positive breast cancer patients receiving adjuvant tamoxifen therapy. Clin Cancer Res 2009, 15: 5888–5894. 10.1158/1078-0432.CCR-09-0728
Span PN, D e-Mulder PH, Sweep FC: Re: p27(Kip1) and cyclin E expression and breast cancer survival after treatment with adjuvant chemotherapy. J Natl Cancer Inst 2007, 99: 738.
Porter PL, Barlow WE, Yeh IT, Lin MG, Yuan XP, Donato E, Sledge GW, Shapiro CL, Ingle JN, Haskell CM, Albain KS, Roberts JM, Livingston RB, Hayes DF: p27(Kip1) and cyclin E expression and breast cancer survival after treatment with adjuvant chemotherapy. J Natl Cancer Inst 2006, 98: 1723–1731. 10.1093/jnci/djj467
Scaltriti M, Eichhorn PJ, Cortes J, Prudkin L, Aura C, Jimenez J, Chandarlapaty S, Serra V, Prat A, Ibrahim YH, Guzman M, Gili M, Rodriguez O, Rodriguez S, Perez J, Green SR, Mai S, Rosen N, Hudis C, Baselga J: Cyclin E amplification/overexpression is a mechanism of trastuzumab resistance in HER2+ breast cancer patients. Proc Natl Acad Sci USA 2011, 108: 3761–3766. 10.1073/pnas.1014835108
Nahta R, Takahashi T, Ueno NT, Hung MC, Esteva FJ: P27(kip1) down-regulation is associated with trastuzumab resistance in breast cancer cells. Cancer Res 2004, 64: 3981–3986. 10.1158/0008-5472.CAN-03-3900
Buckley MF, Sweeney KJ, Hamilton JA, Sini RL, Manning DL, Nicholson RI, deFazio A, Watts CK, Musgrove EA, Sutherland RL: Expression and amplification of cyclin genes in human breast cancer. Oncogene 1993, 8: 2127–2133.
Gillett C, Fantl V, Smith R, Fisher C, Bartek J, Dickson C, Barnes D, Peters G: Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res 1994, 54: 1812–1817.
Jiang W, Kahn SM, Zhou P, Zhang YJ, Cacace AM, Infante AS, Doi S, Santella RM, Weinstein IB: Overexpression of cyclin D1 in rat fibroblasts causes abnormalities in growth control, cell cycle progression and gene expression. Oncogene 1993, 8: 3447–3457.
Wang TC, Cardiff RD, Zukerberg L, Lees E, Arnold A, Schmidt EV: Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature 1994, 369: 669–671. 10.1038/369669a0
Zhou P, Jiang W, Zhang YJ, Kahn SM, Schieren I, Santella RM, Weinstein IB: Antisense to cyclin D1 inhibits growth and reverses the transformed phenotype of human esophageal cancer cells. Oncogene 1995, 11: 571–580.
Seshadri R, Lee CS, Hui R, McCaul K, Horsfall DJ, Sutherland RL: Cyclin DI amplification is not associated with reduced overall survival in primary breast cancer but may predict early relapse in patients with features of good prognosis. Clin Cancer Res 1996, 2: 1177–1184.
Bostrom P, Soderstrom M, Palokangas T, Vahlberg T, Collan Y, Carpen O, Hirsimaki P: Analysis of cyclins A, B1, D1 and E in breast cancer in relation to tumour grade and other prognostic factors. BMC research notes 2009, 2: 140. 10.1186/1756-0500-2-140
Huo D, Ikpatt F, Khramtsov A, Dangou JM, Nanda R, Dignam J, Zhang B, Grushko T, Zhang C, Oluwasola O, Malaka D, Malami S, Odetunde A, Adeoye AO, Iyare F, Falusi A, Perou CM, Olopade OI: Population differences in breast cancer: survey in indigenous African women reveals over-representation of triple-negative breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 2009, 27: 4515–4521. 10.1200/JCO.2008.19.6873
Reis-Filho JS, Savage K, Lambros MB, James M, Steele D, Jones RL, Dowsett M: Cyclin D1 protein overexpression and CCND1 amplification in breast carcinomas: an immunohistochemical and chromogenic in situ hybridisation analysis. Mod Pathol 2006, 19: 999–1009. 10.1038/modpathol.3800621
Jirstrom K, Stendahl M, Ryden L, Kronblad A, Bendahl PO, Stal O, Landberg G: Adverse effect of adjuvant tamoxifen in premenopausal breast cancer with cyclin D1 gene amplification. Cancer Res 2005, 65: 8009–8016.
Rudas M, Lehnert M, Huynh A, Jakesz R, Singer C, Lax S, Schippinger W, Dietze O, Greil R, Stiglbauer W, Kwasny W, Grill R, Stierer M, Gnant MF, Filipits M: Cyclin D1 expression in breast cancer patients receiving adjuvant tamoxifen-based therapy. Clin Cancer Res 2008, 14: 1767–1774. 10.1158/1078-0432.CCR-07-4122
Hui R, Finney GL, Carroll JS, Lee CS, Musgrove EA, Sutherland RL: Constitutive overexpression of cyclin D1 but not cyclin E confers acute resistance to antiestrogens in T-47D breast cancer cells. Cancer Res 2002, 62: 6916–6923.
Ahmed KM, Fan M, Nantajit D, Cao N, Li JJ: Cyclin D1 in low-dose radiation-induced adaptive resistance. Oncogene 2008, 27: 6738–6748. 10.1038/onc.2008.265
Chang AR, Wu HG, Park CI, Jun YK, Kim CW: Expression of epidermal growth factor receptor and cyclin D1 in pretreatment biopsies as a predictive factor of radiotherapy efficacy in early glottic cancer. Head Neck 2008, 30: 852–857. 10.1002/hed.20788
Higuchi E, Oridate N, Homma A, Suzuki F, Atago Y, Nagahashi T, Furuta Y, Fukuda S: Prognostic significance of cyclin D1 and p16 in patients with intermediate-risk head and neck squamous cell carcinoma treated with docetaxel and concurrent radiotherapy. Head Neck 2007, 29: 940–947. 10.1002/hed.20632
Jirawatnotai S, Hu Y, Michowski W, Elias JE, Becks L, Bienvenu F, Zagozdzon A, Goswami T, Wang YE, Clark AB, Kunkel TA, van Harn T, Xia B, Correll M, Quackenbush J, Livingston DM, Gygi SP, Sicinski P: A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature 2011, 474: 230–234. 10.1038/nature10155
Shimura T, Kakuda S, Ochiai Y, Nakagawa H, Kuwahara Y, Takai Y, Kobayashi J, Komatsu K, Fukumoto M: Acquired radioresistance of human tumor cells by DNA-PK/AKT/GSK3beta-mediated cyclin D1 overexpression. Oncogene 2010, 29: 4826–4837. 10.1038/onc.2010.238
Coco Martin JM, Balkenende A, Verschoor T, Lallemand F, Michalides R: Cyclin D1 overexpression enhances radiation-induced apoptosis and radiosensitivity in a breast tumor cell line. Cancer Res 1999, 59: 1134–1140.
Han EK, Begemann M, Sgambato A, Soh JW, Doki Y, Xing WQ, Liu W, Weinstein IB: Increased expression of cyclin D1 in a murine mammary epithelial cell line induces p27kip1, inhibits growth, and enhances apoptosis. Cell Growth Differ 1996, 7: 699–710.
Niu MY, Menard M, Reed JC, Krajewski S, Pratt MA: Ectopic expression of cyclin D1 amplifies a retinoic acid-induced mitochondrial death pathway in breast cancer cells. Oncogene 2001, 20: 3506–3518. 10.1038/sj.onc.1204453
Zhou Q, Fukushima P, DeGraff W, Mitchell JB, Stetler Stevenson M, Ashkenazi A, Steeg PS: Radiation and the Apo2L/TRAIL apoptotic pathway preferentially inhibit the colonization of premalignant human breast cells overexpressing cyclin D1. Cancer Res 2000, 60: 2611–2615.
Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, Los G, Slamon DJ: PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast cancer research: BCR 2009, 11: R77. 10.1186/bcr2419
Begg AC, Stewart FA, Vens C: Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer 2011, 11: 239–253.
Cheung P, Allis CD, Sassone-Corsi P: Signaling to chromatin through histone modifications. Cell 2000, 103: 263–271. 10.1016/S0092-8674(00)00118-5
Goto H, Tomono Y, Ajiro K, Kosako H, Fujita M, Sakurai M, Okawa K, Iwamatsu A, Okigaki T, Takahashi T, Inagaki M: Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J Biol Chem 1999, 274: 25543–25549. 10.1074/jbc.274.36.25543
Preuss U, Landsberg G, Scheidtmann KH: Novel mitosis-specific phosphorylation of histone H3 at Thr11 mediated by Dlk/ZIP kinase. Nucleic Acids Res 2003, 31: 878–885. 10.1093/nar/gkg176
Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A: Genomic instability in mice lacking histone H2AX. Science 2002, 296: 922–927. 10.1126/science.1069398
Mah LJ, El-Osta A, Karagiannis TC: gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia 2010, 24: 679–686. 10.1038/leu.2010.6
Rogakou EP, Boon C, Redon C, Bonner WM: Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 1999, 146: 905–916. 10.1083/jcb.146.5.905
Mah LJ, Orlowski C, Ververis K, Vasireddy RS, El-Osta A, Karagiannis TC: Evaluation of the efficacy of radiation-modifying compounds using gammaH2AX as a molecular marker of DNA double-strand breaks. Genome Integr 2011, 2: 3. 10.1186/2041-9414-2-3
Adon AM, Zeng X, Harrison MK, Sannem S, Kiyokawa H, Kaldis P, Saavedra HI: Cdk2 and Cdk4 regulate the centrosome cycle and are critical mediators of centrosome amplification in p53-null cells. Mol Cell Biol 2010, 30: 694–710. 10.1128/MCB.00253-09
Felsher DW, Bishop JM: Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci USA 1999, 96: 3940–3944. 10.1073/pnas.96.7.3940
Olsson M, Zhivotovsky B: Caspases and cancer. Cell Death Differ 2011,18(9):1441–1449. 10.1038/cdd.2011.30
Harrison Pitner MK, Saavedra HI: Cdk4 and nek2 signal binucleation and centrosome amplification in a her2+ breast cancer model. PLoS One 2013, 8: e65971. 10.1371/journal.pone.0065971
Saavedra HI, Maiti B, Timmers C, Altura R, Tokuyama Y, Fukasawa K, Leone G: Inactivation of E2F3 results in centrosome amplification. Cancer Cell 2003, 3: 333–346. 10.1016/S1535-6108(03)00083-7
Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ: Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell 1995, 80: 285–291. 10.1016/0092-8674(95)90411-5
Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ: Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14–3-3 not BCL-X(L). Cell 1996, 87: 619–628. 10.1016/S0092-8674(00)81382-3
Ayllon V, Cayla X, Garcia A, Roncal F, Fernandez R, Albar JP, Martinez C, Rebollo A: Bcl-2 targets protein phosphatase 1 alpha to Bad. J Immunol 2001, 166: 7345–7352.
Chiang CW, Kanies C, Kim KW, Fang WB, Parkhurst C, Xie M, Henry T, Yang E: Protein phosphatase 2A dephosphorylation of phosphoserine 112 plays the gatekeeper role for BAD-mediated apoptosis. Mol Cell Biol 2003, 23: 6350–6362. 10.1128/MCB.23.18.6350-6362.2003
Klumpp S, Selke D, Krieglstein J: Protein phosphatase type 2C dephosphorylates BAD. Neurochem Int 2003, 42: 555–560. 10.1016/S0197-0186(02)00174-2
Berthet C, Kaldis P: Cell-specific responses to loss of cyclin-dependent kinases. Oncogene 2007, 26: 4469–4477. 10.1038/sj.onc.1210243
Berthet C, Kaldis P: Cdk2 and Cdk4 cooperatively control the expression of Cdc2. Cell division 2006, 1: 10. 10.1186/1747-1028-1-10
Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, Caceres JF, Dubus P, Malumbres M, Barbacid M: Cdk1 is sufficient to drive the mammalian cell cycle. Nature 2007, 448: 811–815. 10.1038/nature06046
Diril MK, Ratnacaram CK, Padmakumar VC, Du T, Wasser M, Coppola V, Tessarollo L, Kaldis P: Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc Natl Acad Sci USA 2012, 109: 3826–3831. 10.1073/pnas.1115201109
Pagano M, Theodoras AM, Tam SW, Draetta GF: Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev 1994, 8: 1627–1639. 10.1101/gad.8.14.1627
Deans AJ, Khanna KK, McNees CJ, Mercurio C, Heierhorst J, McArthur GA: Cyclin-dependent kinase 2 functions in normal DNA repair and is a therapeutic target in BRCA1-deficient cancers. Cancer Res 2006, 66: 8219–8226. 10.1158/0008-5472.CAN-05-3945
Jirawatnotai S, Aziyu A, Osmundson EC, Moons DS, Zou X, Kineman RD, Kiyokawa H: Cdk4 is indispensable for postnatal proliferation of the anterior pituitary. J Biol Chem 2004, 279: 51100–51106. 10.1074/jbc.M409080200
Lee J, Desiderio S: Cyclin A/CDK2 regulates V(D)J recombination by coordinating RAG-2 accumulation and DNA repair. Immunity 1999, 11: 771–781. 10.1016/S1074-7613(00)80151-X
Muller-Tidow C, Ji P, Diederichs S, Potratz J, Baumer N, Kohler G, Cauvet T, Choudary C, van der Meer T, Chan WY, Nieduszynski C, Colledge WH, Carrington M, Koeffler HP, Restle A, Wiesmuller L, Sobczak-Thepot J, Berdel WE, Serve H: The cyclin A1-CDK2 complex regulates DNA double-strand break repair. Mol Cell Biol 2004, 24: 8917–8928. 10.1128/MCB.24.20.8917-8928.2004
Neganova I, Vilella F, Atkinson SP, Lloret M, Passos JF, von Zglinicki T, O’Connor JE, Burks D, Jones R, Armstrong L, Lako M: An important role for CDK2 in G1 to S checkpoint activation and DNA damage response in human embryonic stem cells. Stem Cells 2011, 29: 651–659. 10.1002/stem.620
Cerqueira A, Santamaria D, Martinez-Pastor B, Cuadrado M, Fernandez-Capetillo O, Barbacid M: Overall Cdk activity modulates the DNA damage response in mammalian cells. J Cell Biol 2009, 187: 773–780. 10.1083/jcb.200903033
Ray D, Terao Y, Christov K, Kaldis P, Kiyokawa H: Cdk2-null mice are resistant to ErbB-2-induced mammary tumorigenesis. Neoplasia 2011, 13: 439–444.
Reddy HK, Mettus RV, Rane SG, Grana X, Litvin J, Reddy EP: Cyclin-dependent kinase 4 expression is essential for neu-induced breast tumorigenesis. Cancer Res 2005, 65: 10174–10178. 10.1158/0008-5472.CAN-05-2639
Harrison MK, Adon AM, Saavedra HI: The G1 phase Cdks regulate the centrosome cycle and mediate oncogene-dependent centrosome amplification. Cell division 2011, 6: 2. 10.1186/1747-1028-6-2
Zeng X, Shaikh FY, Harrison MK, Adon AM, Trimboli AJ, Carroll KA, Sharma N, Timmers C, Chodosh LA, Leone G, Saavedra HI: The Ras oncogene signals centrosome amplification in mammary epithelial cells through cyclin D1/Cdk4 and Nek2. Oncogene 2010, 29: 5103–5112. 10.1038/onc.2010.253
Dean JL, Thangavel C, McClendon AK, Reed CA, Knudsen ES: Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene 2010, 29: 4018–4032. 10.1038/onc.2010.154
Dean JL, McClendon AK, Knudsen ES: Modification of the DNA damage response by therapeutic CDK4/6 inhibition. J Biol Chem 2012, 287: 29075–29087. 10.1074/jbc.M112.365494
Roberts PJ, Bisi JE, Strum JC, Combest AJ, Darr DB, Usary JE, Zamboni WC, Wong KK, Perou CM, Sharpless NE: Multiple roles of cyclin-dependent kinase 4/6 inhibitors in cancer therapy. J Natl Cancer Inst 2012, 104: 476–487. 10.1093/jnci/djs002
Liang Y, Gao H, Lin SY, Goss JA, Brunicardi FC, Li K: siRNA-based targeting of cyclin E overexpression inhibits breast cancer cell growth and suppresses tumor development in breast cancer mouse model. PLoS One 2010, 5: e12860. 10.1371/journal.pone.0012860
Boothman DA, Meyers M, Odegaard E, Wang M: Altered G1 checkpoint control determines adaptive survival responses to ionizing radiation. Mutat Res 1996, 358: 143–153. 10.1016/S0027-5107(96)00115-7
Casimiro MC, Crosariol M, Loro E, Ertel A, Yu Z, Dampier W, Saria EA, Papanikolaou A, Stanek TJ, Li Z, Wang C, Fortina P, Addya S, Tozeren A, Knudsen ES, Arnold A, Pestell RG: ChIP sequencing of cyclin D1 reveals a transcriptional role in chromosomal instability in mice. J Clin Invest 2012, 122: 833–843. 10.1172/JCI60256
Garcia A, Cayla X, Guergnon J, Dessauge F, Hospital V, Rebollo MP, Fleischer A, Rebollo A: Serine/threonine protein phosphatases PP1 and PP2A are key players in apoptosis. Biochimie 2003, 85: 721–726. 10.1016/j.biochi.2003.09.004
Mumby M: PP2A: unveiling a reluctant tumor suppressor. Cell 2007, 130: 21–24. 10.1016/j.cell.2007.06.034
Westermarck J, Hahn WC: Multiple pathways regulated by the tumor suppressor PP2A in transformation. Trends Mol Med 2008, 14: 152–160. 10.1016/j.molmed.2008.02.001
Wang Q, Gao F, Wang T, Flagg T, Deng X: A nonhomologous end-joining pathway is required for protein phosphatase 2A promotion of DNA double-strand break repair. Neoplasia 2009, 11: 1012–1021.
Sablina AA, Hahn WC: SV40 small T antigen and PP2A phosphatase in cell transformation. Cancer Metastasis Rev 2008, 27: 137–146. 10.1007/s10555-008-9116-0
Simizu S, Tamura Y, Osada H: Dephosphorylation of Bcl-2 by protein phosphatase 2A results in apoptosis resistance. Cancer Sci 2004, 95: 266–270. 10.1111/j.1349-7006.2004.tb02214.x
Xin M, Deng X: Protein phosphatase 2A enhances the proapoptotic function of Bax through dephosphorylation. J Biol Chem 2006, 281: 18859–18867. 10.1074/jbc.M512543200
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW: A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10: 515–527. 10.1016/j.ccr.2006.10.008
Ritz C, JC S: Bioassay Analysis using R. J. Statistics Software. 2005, 12.
Box G a, a. C. C,DR: An analysis of transformations. Journal of the Royal Statistical Society Series B 1964, 26: 211–252.
Team RDC: R: A language and environment for statistical computing. 2009.
Acknowledgements
We would like to thank Dr. Rita Nahta for various Her2+ breast cancer cell lines, and Dr. Jing Chen for 293T cells and helper plasmids. We thank Dr. Sungjin Kim from the Biostatistics and Bioinformatics Shared Resource at Winship Cancer Institute for statistical analysis. This research project was supported by R01 CA151521 from the National Institutes of Health, the Georgia Cancer Coalition, The Sindab Triple Negative Breast Cancer Foundation, and by the Emory University Integrated Cellular Imaging Microscopy Core of the Winship Cancer Institute comprehensive cancer center grant, P30CA138292.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare they have no competing interests.
Authors’ contributions
KRH: Performed the radiation and flow cytometry experiments to detect DNA proliferation, apoptosis and DNA breaks. Performed caspase immunoflourecent staining assays. Was involved in finalizing the writing of the manuscript. XZ: Performed western blots and caspase, BrdU, Ki67 and γ-H2AX preliminary and supplementary experiments in the manuscript. Participated in early drafts of the manuscripts. MYL Performed Western blots for cleaved PARP and LC3A/3B in the presence of shCDK4 and the CDK4/CDK6 inhibitor, immunocytochemistry for phospho-histone 3 Contributed and modified figures and was involved in manuscript editing. STK: Performed the analysis showing that breast cancer cell lines are resistant to radiation. Showed that silencing CDK4 radiosensitized breast cancer cells. MKHP: Developed cell lines stably silenced for CDK4 and performed Western blots showing downregulation of CDK4. Edited the manuscript. SSZ: Developed the irradiation protocols for the radiation sensitivity and colony formation experiments. YL: Biostatistician that performed many of the statistical assays in the manuscript. RMO: Designed some of the experiments and provided various cell lines. XD: Provided antibodies and designed the experiments that lead to the Western blot analyses of various apoptotic proteins. HIS: Designed the experiments in the manuscript, edited the manuscript, and established all collaborations in the manuscript. All authors read and approved the final manuscript.
Katie R Hagen, Xiangbin Zeng, Mi-Young Lee, Shannon Tucker Kahn contributed equally to this work.
Electronic supplementary material
13008_2013_181_MOESM1_ESM.pdf
Additional file 1: Radiosensitivity imparted by downregulated Cdk4 is not caused by impaired proliferation. (A) Cells stably expressing control pLKO.1, shCDK2 or shCDK4 were unirradiated (Basal) or irradiated at 2 Gy. Cells were pulse-labeled with BrdU following different times post-irradiation (0, 6, 12, 24, & 48 hours), and were subjected to immunostaining with an anti-BrdU antibody, and an Alexa Fluor secondary antibody. Nuclei were counter-stained with DAPI. The number of cells positively stained with BrdU was counted in 200 cells per group, and the results are shown as the average±standard deviation from two independent experiments. (B) Cells stably expressing pLKO.1, shCDK2 or shCDK4 were unirradiated (Basal), or irradiated at 2 Gy. Cells collected from different time points (0, 6, 12, 24, & 48 hours) post-irradiation were subjected to immunostaining with anti-Ki67 antibody, an Alexa Fluor secondary and the DNA was counter-stained with DAPI. The number of cells positively stained with Ki67 was counted in 200 cells per group, and the results are shown as the average±standard deviation from two experiments. (PDF 19 KB)
13008_2013_181_MOESM2_ESM.pdf
Additional file 2: Downregulation of Cdk4 does not alter rates of DNA break repair. Non-infected cells and cells stably expressing pLKO.1, shCDK2 or shCDK4 were irradiated at 2 Gy. The basal levels were determined in unirradiated cells. Cells were fixed at different times (0, 6, 12, 24, & 48 hours) post-irradiation and were subjected to immunostaining with anti-H2A.X antibody (γ-H2AX) and an Alexa Fluor 555 secondary antibody; DNA was counter-stained with DAPI (A). Blue cells represent nuclei, while the red cells (arrows) represent cells expressing γ-H2AX. Pictures were taken at a 65× magnification. (B) The number of cells positively stained with γ-H2AX was counted in 200 cells per group, and the results are shown as the average±standard deviation from two experiments. (PDF 316 KB)
13008_2013_181_MOESM3_ESM.pdf
Additional file 3: Silencing of Cdk4 promotes apoptosis. Cells stably expressing pLKO.1, shCDK2 or shCDK4 were irradiated at 2 Gy. The basal levels were determined in unirradiated cells. Cells were fixed at different times (0, 6, 12, 24, & 48 hours) post-irradiation and were subjected to immunostaining with anti-cleaved caspase-3 antibody and an Alexa Fluor 555 secondary antibody; DNA was counter-stained with DAPI. The number of cells positively stained with cleaved caspase-3 was counted in 200 cells per group, and the results are shown as the average±standard deviation from two experiments. (PDF 27 KB)
13008_2013_181_MOESM4_ESM.pdf
Additional file 4: CDK4 silencing did not change degree of autophagy. (A) Cells stably expressing control pLKO.1 and shCDK4 were irradiated at 0, 2 and 4 Gy. Protein lysates were prepared after 48 hours post irradiation and were subjected to Western blot with an anti-LC3A/3B antibody. β-actin was used as a loading control. (B) Cells stably expressing control pLKO.1 were treated with the CDK4/6 inhibitor PD0332991 and irradiated at 0, 2 and 4 Gy. Protein lysates were prepared after 48 hours post irradiation and were subjected to Western blot with an anti-LC3A/3B antibody. β-actin was used as a loading control. (C) Cells stably expressing shCDK4 was transfected with siRNA targeting the PP2A catalytic unit for 48 hours and irradiated at 0, 2 and 4 Gy. Protein lysates were prepared after 48 hours post irradiation and were subjected to Western blot with an anti-LC3A/3B antibody. β-actin was used as a loading control. (PDF 324 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Hagen, K.R., Zeng, X., Lee, MY. et al. Silencing CDK4 radiosensitizes breast cancer cells by promoting apoptosis. Cell Div 8, 10 (2013). https://doi.org/10.1186/1747-1028-8-10
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1747-1028-8-10