Abstract
Aims
The goal of our study is to investigate the associations between 18 candidate genetic markers and overweight/obesity.
Methods
A total of 72 eligible articles were retrieved from literature databases including PubMed, Embase, SpingerLink, Web of Science, Chinese National Knowledge Infrastructure (CNKI), and Wanfang. Meta-analyses of 18 genetic markers among 56,738 controls and 48,148 overweight/obese persons were done by Review Manager 5.0.
Results
Our results showed that SH2B1 rs7498665 polymorphism was significantly associated with the risk of overweight/obesity (overall odds ratio (OR) = 1.21, 95% confidence interval (CI) = 1.09-1.34, P = 0.0004). Increased risk of overweight/obesity was also observed in FAIM2 rs7138803 polymorphism (overall OR = 1.11, 95% CI = 1.01-1.22, P = 0.04).
Conclusion
Our meta-analyses have shown the important role of 2 polymorphisms (SH2B1 rs7498665 and FAIM2 rs7138803) in the development of overweight/obesity. This study highlighted the importance of above two candidate genes (SH2B1 and FAIM2) in the risk of overweight/obesity.
Virtual slides
The virtual slide(s) for this article can be found here: http://www.diagnosticpathology.diagnomx.eu/vs/2785487401176182.
Similar content being viewed by others
Introduction
Overweight/obesity as a metabolic disorder is closely associated with diabetes mellitus and cardiovascular disease, which are chronic diseases influencing the average life expectancy [1, 2]. In 2008, world health organization (WHO) has reported that a large portion of adults (>20 yr) were overweight (35%) and obese (12%) [3]. The overweight/obesity will become an epidemic [4] and cause a huge economic burden of society [4] in the near future.
The occurrence and the development of obesity are influenced by both environmental and genetic factors [5, 6]. Environmental factors, such as poor nutritional state and a lack of physical exercise, have an impact on the development of overweight/obesity [7, 8] through the epigenetic modifications such as gene methylation [9]. Genetic polymorphisms can confer the susceptibility of overweight/obesity and obesity-related morbidities [10]. Recent genome-wide association studies (GWAS) have identified a handful of candidate genetic markers to the risk of overweight/obesity [11].
In the present study, we performed a systematic search for eligible studies in the meta-analyses. Our results identified 18 polymorphisms among 16 genes that were all the candidate genes of obesity. Among these genes, GNB3 encodes β3-subunit protein which is involved in the process of hypertension and obesity [12]. MTHFR gene encodes methylenetetrahydrofolate reductase that is shown to be associated with increased fasting homocysteine [13]. MTHFR polymorphism is shown to be associated with lipid metabolism in the elderly women [14]. CNR1 is shown to regulate the endocannabinoid system that might stimulate the metabolism of lipogenesis through central and peripheral mechanisms [15, 16]. CNR1 is associated with low HDL dyslipidemia and a common haplotype of CNR1 could be a protective factor of obesity-related dyslipidemia [17]. BDNF is shown to play an important role in the development of several neuronal systems [18]. As an effector on energy homeostasis through MC4R signaling pathway, BDNF has an effect on the glucose and lipid metabolism in obese diabetic animals [19, 20]. FAAH gene encodes fatty acid amide hydrolase [21] and plays an important role in the development of obesity [22]. ADRB1 is shown to mediate in lipolysis and thus is important for obesity [23]. Rat study identifies that ADRB1 mediates the sympathetic nervous system (SNS) stimulation of thermogenesis in brown adipose tissue [24]. SH2B1 is able to bind leptin to its receptor, and thus increases the JAK2 activation which is involved in the insulin and leptin signaling [25, 26]. PCSK1 encodes prohormone convertase 1/3 that is a vital enzyme in the regulation of a majority of neuroendocrine body weight control [27]. A novel homozygous missense mutation in PCSK1 leads to early-onset obesity [28]. NPY2R is a presynaptic receptor [29] playing an inhibitory role in the control of appetite regulation [30], and thus influences the development of obesity [31]. FAIM2 (Fas apoptotic inhibitory molecule 2) is an anti-apoptotic gene [32]. Mutations of FAIM2 which interferes with Fas-mediated cell death confer risk for obesity [33]. SERPINE1 encodes a member of serine proteinase inhibitor which influences plasma PAI-1 activity with relation to obesity [34]. Serum paraoxonase-1 (PON1) encoded by PON1 as an enzyme associated with HDL-C could be a protector against oxidative damage in obesity [35]. CETP protein product transfers cholesterylesters from HDL to pro-atherogenic apoB-lipoproteins and thus has an impact on the lipid and HDL metabolism [36, 37]. UCP1 encodes uncoupling protein 1 that is mediated by long-chain fatty acids (LCFAs) from brown adipose tissue [38]. UCP1 expression in adipose tissue has an impact on regulating the thermogenesis and lipolysis [39, 40]. Mitochondrial uncoupling by UCP1 has demonstrated to be a target in antiobesity therapies [41]. ABCA1 gene product mediates the transport of cholesterol, phospholipids, and other metabolites [42]. Exercise has an impact on ABCA1 expression along with increased HDL levels in obese boys [43]. APOE plays a fundamental role with ligand-receptor in uptaking lipoproteins, and thus participates in the lipid metabolism [44]. In addition, APOE correlates with inflammation in adipose tissue in high-fat diet-induced obesity [45].
Meta-analysis is a systematic evaluation by combining the results from collected studies [46, 47]. The major advantages of meta-analysis are to improve the precision and accuracy by pooling up the data from multiple sources, and to analyze and quantify the inconsistency of results and the publish bias [48]. In the present study, we conducted comprehensive meta-analyses to identify the contribution of 18 polymorphisms to overweight/obesity.
Materials and methods
Literature search and data extraction
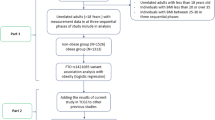
We performed the literature research using related databases such as PubMed, Embase, SpingerLink, Web of Science, Chinese National Knowledge Infrastructure (CNKI), and Wanfang. The combination of keywords in the literature search was obesity or overweight together with polymorphism or mutation or variant or single nucleotide polymorphism (SNP). The studies excluded in the meta-analysis met the following criteria: (1) the study had been included in the previous meta-analysis; (2) the study was not involved with genetic testing; (3) the study was not a case–control study. The criteria for overweight or obesity in adolescents and children were defined by WHO [49, 50]. Finally, we harvested 18 polymorphisms of 16 genes in the current meta-analysis. These included GNB3 rs5443, MTHFR rs1801133, CNR1 rs806381, BDNF rs6265, FAAH rs324420, ADRB1 rs1801253, SH2B1 rs7498665, PCSK1 rs6232 and rs6235, NPY2R rs1047214, FAIM2 rs7138803, SERPINE1 rs1799768, PON1 rs854560 and rs662, CETP TaqIB, UCP1 rs1800592, ABCA1 rs2230806 and APOE ϵ2/ϵ3/ϵ4.
Statistical analysis
Meta-analysis was performed by using Statistical software Review Manager 5.0 [51]. Forest plots included the ORs with the corresponding 95% CIs, cochran’s Q and the inconsistency index (I2). If there were no significant heterogeneity (I2 < 50%, P > 0.05) of the studies in the meta-analysis, we used a fixed-effect model for the analysis. Otherwise, a random-effect model was used for the meta-analysis with large heterogeneity (I2 > 50%, P < 0.05). The weight of each involved study was calculated whatever in fixed-effect or random-effect model in forest plots by Review Manager 5.0. Two tailed P value < 0.05 was treated as significant. Power analyses were calculated by Power and Sample Size Calculation software (v3.0.43) [52].
Results
An initial search returned a total of 7,750 literatures from databases including PubMed, Embase, SpingerLink, Web of Science, Chinese National Knowledge Infrastructure (CNKI), and Wanfang. After a systematic filtration, 72 eligible articles, including 64 English, 6 Chinese, 1 German and 1 Spanish articles, were left for the meta-analyses (Additional file 1: Table S1). The detailed information for the retrieved studies was shown in Tables 1 and 2.
Heterogeneity is an important indicator to identify if there is difference in the collected studies. According to the extent of heterogeneity, we categorized the meta-analyses into three groups that have minimal (I2 = 0), moderate (I2 < 50%), and significant heterogeneity (I2 ≥ 50%), respectively. As shown in Figure 1, minimal heterogeneity (I2 = 0) was found for the meta-analyses of 10 polymorphisms that included MTHFR rs1801133, CNR1 rs806381, ADRB1 rs1801253, SH2B1 rs7498665, PCSK1 rs6235, NPY2R rs1047214, FAIM2 rs7138803, CETP TaqIB and ABCA1 rs2230806. Moderate heterogeneity was found for 5 polymorphisms, including BDNF rs6265 (I2 = 46%), PCSK1 rs6232 (I2 = 34%), GNB3 rs5443 (I2 = 42%), PON1 rs854560 (I2 = 31%), PON1 rs662 (I2 = 18%), and SERPINE1 rs1799768 (I2 = 39%). Significant heterogeneity was found for UCP1 rs1800592 (I2 = 60%) and FAAH rs324420 (I2 = 79%). Moreover, As shown in Figure 2, various heterogeneities were shown in the meta-analyses of APOE ϵ2/ϵ3/ϵ4 polymorphism under the seven genetic models (ϵ2/ϵ3 versus ϵ3/ϵ3: I2 = 48%; ϵ2/ϵ4 versus ϵ3/ϵ3: I2 = 0%; ϵ3/ϵ4 versus ϵ3/ϵ3: I2 = 28%; ϵ4/ϵ4 versus ϵ3/ϵ3: I2 = 63%; ϵ2/ϵ3 versus ϵ3/ϵ3: I2 = 0%; ϵ2 versus ϵ3: I2 = 23%; ϵ4 versus ϵ3: I2 = 65%). No obvious publication bias was observed based on their funnel plots (Figures 3 and 4).
Our results showed that SH2B1 rs7498665 was significantly associated with the risk of overweight/obesity among 6,142 cases and 4,345 controls from four studies (overall OR = 1.21, 95% CI = 1.09-1.34, P = 0.0004, Figure 1). Increased risk of overweight/obesity was also observed in rs7138803 of FAIM2 among 3,477 cases and 4,676 controls from five studies (overall OR = 1.11, 95% CI = 1.01-1.22, P = 0.04, Figure 1). No evidence of association was observed for the meta-analyses of the rest 16 variants (Figures 1 and 3). For the meta-analyses with large heterogeneity, we further performed subgroup meta-analyses by ethnicity. No significant association of UCP1 rs1800592 with overweight/obesity was observed in Caucasian (P = 0.13, I2 = 62%), and Asian (P = 0.59, I2 = 0%, Additional file 2: Figure S1). And the subgroup meta-analysis of APOE ϵ2/ϵ3/ϵ4 polymorphism by excluding the study of Srivastava et al. [53] didn’t produce any significant association of APOE ϵ2/ϵ3/ϵ4 with overweight/obesity (Additional file 3: Figure S2). There was no visual publication bias in all the above meta-analyses (Additional file 4: Figure S3).
Discussion
Current meta-analyses were performed among 48,148 cases and 56,738 controls from 72 studies, covering a total of 6 populations, including Caucasian, Asian, Japanese-American, European-American, African-American, South American, and African. Among the tested 18 polymorphisms, there were two (SH2B1 rs7498665 and FAIM2 rs7138803) with significant association results (P < 0.05). Power analysis also showed large power existed in our meta-analyses of two significant polymorphisms including SH2B1 rs7498665 (100%) and FAIM2 rs7138803 (100%).
SH2B1 encodes an adaptor protein associated with leptin and insulin signaling in the lipid metabolism [54]. SH2B1 is an enhancer that may influence the phenotype of obesity through JAK-STAT pathway [55], which is important in the development and function of adipocytes [56]. SH2B1 acts as a mediator through PI3-kinase pathway which is correlated with the biological actions of leptin [26]. Many animal studies have shown that SH2B1 is involved in the development of obesity. SH2B1 through its participation in the regulation of leptin sensitivity, energy metabolism and body weight [57]. SH2B1 has been identified to be related to obesity through genome-wide association studies (GWAS) [55]. Our meta-analysis of SH2B1 rs7498665 was performed among 6,652 cases and 4,814 controls with four studies. Among the tested populations, no heterogeneity was observed (I2 = 0). Our results confirmed the relationship between SH2B1 and the risk of overweight/obesity (overall OR = 1.21, 95% CI = 1.09-1.34, P = 0.0004, Figure 1).
FAIM2 is an anti-apoptotic gene that provides protection from Fas-mediated cell death [32] that is associated with extreme overweight by GWAS [58]. FAIM2 rs7138803 polymorphism is associated with increased risk of obesity in Japanese [59]. But there is no relationship between FAIM2 rs7138803 and obesity in Chinese [60]. Minor allele frequency of rs7138803 in Chinese populations ranges from 0.28 to 0.29, while FAIM2 rs7138803 is monomorphic in Japanese and Caucasian populations. Our meta-analysis among 3477 cases and 4676 controls demonstrated that FAIM2 rs7138803 was associated with the risk of overweight/obesity (overall OR = 1.11, 95% CI = 1.01-1.22, P = 0.04, Figure 1).
Although meta-analysis is an important method to improve the precision and accuracy, to analyze and quantify the published results [61–63], some disadvantages exist in the meta-analysis. For the current meta-analyses, several limitations need to be taken with cautions. Firstly, obesity is always accompanied by other complications such as coronary artery diseases and hypertension. These confounding factors needed to be adjusted in the original case–control studies. We were unable to obtain the related information. Therefore we can’t exclude a chance of the positive findings confounded by these obesity-related factors. Secondly, the significant result of FAIM2 rs7138803 needs to be validated in the future. However, after Bonferroni’s correction by the number of testing, the association of FAIM2 rs7138803 was unable to retain significant. Thirdly, power analysis suggested moderate power in the meta-analyses of MTHFR rs1801133 (power = 78.2%) and SERPINE1 rs1799768 (power = 69.4%) The negative results of them might be caused by a lack of power in our meta-analyses. Future studies with larger samples may help clarify the contribution of these biomarkers to the risk of overweight/obesity.
Our results identified significant associations between 2 polymorphisms (SH2B1 rs7498665 and FAIM2 rs7138803) and overweight/obesity. Moreover, overweight/obesity is a complicated disease influenced by both genetic and environmental factors. The potential mechanism of interaction between gene and environment could be taken into consideration in the future study. Well-designed studies with large samples could help elucidate the contribution of above polymorphisms to overweight/obesity.
Authors’ information
Linlin Tang and Huadan Ye: co-first authors of this work.
References
Ogunbode AM, Ladipo M, Ajayi IO, Fatiregun AA: Obesity: an emerging disease. Niger J Clin Pract. 2011, 14 (4): 390-394. 10.4103/1119-3077.91741.
Haslam DW, James WP: Obesity. Lancet. 2005, 366 (9492): 1197-1209. 10.1016/S0140-6736(05)67483-1.
The situation and trends of obesity and overweight.http://www.who.int/gho/ncd/risk_factors/overweight/en/index.html,
Keaver L, Webber L, Dee A, Shiely F, Marsh T, Balanda K, Perry I: Application of the UK foresight obesity model in ireland: the health and economic consequences of projected obesity trends in ireland. PLoS One. 2013, 8 (11): e79827-10.1371/journal.pone.0079827.
Schwenk RW, Vogel H, Schurmann A: Genetic and epigenetic control of metabolic health. Mol Metab. 2013, 2 (4): 337-347. 10.1016/j.molmet.2013.09.002.
Latham KE, Sapienza C, Engel N: The epigenetic lorax: gene-environment interactions in human health. Epigenomics. 2012, 4 (4): 383-402. 10.2217/epi.12.31.
Wei D, Zhang X, Zou H, Wang L, Fu B, Wu X, Luo Z, Li X, Ge J, Li Y, Zhu H, Wang K, Wang T, Yang P, Hou Z, Wang W: WW domain containing oxidoreductase induces apoptosis in gallbladder-derived malignant cell by upregulating expression of P73 and PUMA. Tumour Biol. 2013, 35 (2): 1539-1550.
Fukuoka H, Mukai S, Taniguchi T: [Nutritional environment in utero and development of obesity]. Nihon Rinsho. 2013, 71 (2): 237-243.
Jiang D, Zheng D, Wang L, Huang Y, Liu H, Xu L, Liao Q, Liu P, Shi X, Wang Z, Sun L, Zhou Q, Li N, Le Y, Ye M, Shao G, Duan S: Elevated PLA2G7 gene promoter methylation as a gender-specific marker of aging increases the risk of coronary heart disease in females. PLoS One. 2013, 8 (3): e59752-10.1371/journal.pone.0059752.
Tisherman SA: Salvage techniques in traumatic cardiac arrest: thoracotomy, extracorporeal life support, and therapeutic hypothermia. Curr Opin Crit Care. 2013, 19 (6): 594-598.
Monda KL, Chen GK, Taylor KC, Palmer C, Edwards TL, Lange LA, Ng MC, Adeyemo AA, Allison MA, Bielak LF, Chen G, Graff M, Irvin MR, Rhie SK, Li G, Liu Y, Lu Y, Nalls MA, Sun YV, Wojczynski MK, Yanek LR, Aldrich MC, Ademola A, Amos CI, Bandera EV, Bock CH, Britton A, Broeckel U, Cai Q, Caporaso NE, et al.: A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet. 2013, 45 (6): 690-696. 10.1038/ng.2608.
Klenke S, Kussmann M, Siffert W: The GNB3 C825T polymorphism as a pharmacogenetic marker in the treatment of hypertension, obesity, and depression. Pharmacogenet Genomics. 2011, 21 (9): 594-606. 10.1097/FPC.0b013e3283491153.
Piedade MC, Galhardo MS, Battlehner CN, Ferreira MA, Caldini EG, de Toledo OM: Effect of ultrasound therapy on the repair of gastrocnemius muscle injury in rats. Ultrasonics. 2008, 48 (5): 403-411. 10.1016/j.ultras.2008.01.009.
Chmurzynska A, Malinowska AM, Twardowska-Rajewska J, Gawecki J: Elderly women: homocysteine reduction by short-term folic acid supplementation resulting in increased glucose concentrations and affecting lipid metabolism (C677T MTHFR polymorphism). Nutrition. 2013, 29 (6): 841-844. 10.1016/j.nut.2012.09.015.
Engeli S, Bohnke J, Feldpausch M, Gorzelniak K, Janke J, Batkai S, Pacher P, Harvey-White J, Luft FC, Sharma AM, Jordan J: Activation of the peripheral endocannabinoid system in human obesity. Diabetes. 2005, 54 (10): 2838-2843. 10.2337/diabetes.54.10.2838.
Di Marzo V, Matias I: Endocannabinoid control of food intake and energy balance. Nat Neurosci. 2005, 8 (5): 585-589. 10.1038/nn1457.
Feng Q, Jiang L, Berg RL, Antonik M, MacKinney E, Gunnell-Santoro J, McCarty CA, Wilke RA: A common CNR1 (cannabinoid receptor 1) haplotype attenuates the decrease in HDL cholesterol that typically accompanies weight gain. PLoS One. 2010, 5 (12): e15779-10.1371/journal.pone.0015779.
Kernie SG, Liebl DJ, Parada LF: BDNF regulates eating behavior and locomotor activity in mice. EMBO J. 2000, 19 (6): 1290-1300. 10.1093/emboj/19.6.1290.
Xu B, Goulding EH, Zang K, Cepoi D, Cone RD, Jones KR, Tecott LH, Reichardt LF: Brain-derived neurotrophic factor regulates energy balance downstream of melanocortin-4 receptor. Nat Neurosci. 2003, 6 (7): 736-742. 10.1038/nn1073.
Tsuchida A, Nonomura T, Nakagawa T, Itakura Y, Ono-Kishino M, Yamanaka M, Sugaru E, Taiji M, Noguchi H: Brain-derived neurotrophic factor ameliorates lipid metabolism in diabetic mice. Diabetes Obes Metab. 2002, 4 (4): 262-269. 10.1046/j.1463-1326.2002.00206.x.
Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB: Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996, 384 (6604): 83-87. 10.1038/384083a0.
Lieb W, Manning AK, Florez JC, Dupuis J, Cupples LA, McAteer JB, Vasan RS, Hoffmann U, O’Donnell CJ, Meigs JB, Fox CS: Variants in the CNR1 and the FAAH genes and adiposity traits in the community. Obesity (Silver Spring). 2009, 17 (4): 755-760. 10.1038/oby.2008.608.
Louis SN, Jackman GP, Nero TL, Iakovidis D, Louis WJ: Role of beta-adrenergic receptor subtypes in lipolysis. Cardiovasc Drugs Ther. 2000, 14 (6): 565-577. 10.1023/A:1007838125152.
Ueta CB, Fernandes GW, Capelo LP, Fonseca TL, Maculan FD, Gouveia CH, Brum PC, Christoffolete MA, Aoki MS, Lancellotti CL, Kim B, Bianco AC, Ribeiro MO: beta(1) Adrenergic receptor is key to cold- and diet-induced thermogenesis in mice. J Endocrinol. 2012, 214 (3): 359-365. 10.1530/JOE-12-0155.
Penas-Steinhardt A, Tellechea ML, Gomez-Rosso L, Brites F, Frechtel GD, Poskus E: Association of common variants in JAK2 gene with reduced risk of metabolic syndrome and related disorders. BMC Med Genet. 2011, 12: 166-10.1186/1471-2350-12-166.
Duan C, Li M, Rui L: SH2-B promotes insulin receptor substrate 1 (IRS1)- and IRS2-mediated activation of the phosphatidylinositol 3-kinase pathway in response to leptin. J Biol Chem. 2004, 279 (42): 43684-43691. 10.1074/jbc.M408495200.
Helwig M, Khorooshi RM, Tups A, Barrett P, Archer ZA, Exner C, Rozman J, Braulke LJ, Mercer JG, Klingenspor M: PC1/3 and PC2 gene expression and post-translational endoproteolytic pro-opiomelanocortin processing is regulated by photoperiod in the seasonal Siberian hamster (Phodopus sungorus). J Neuroendocrinol. 2006, 18 (6): 413-425. 10.1111/j.1365-2826.2006.01431.x.
Farooqi IS, Volders K, Stanhope R, Heuschkel R, White A, Lank E, Keogh J, O’Rahilly S, Creemers JW: Hyperphagia and early-onset obesity due to a novel homozygous missense mutation in prohormone convertase 1/3. J Clin Endocrinol Metab. 2007, 92 (9): 3369-3373. 10.1210/jc.2007-0687.
Dionne IJ, Garant MJ, Nolan AA, Pollin TI, Lewis DG, Shuldiner AR, Poehlman ET: Association between obesity and a polymorphism in the beta(1)-adrenoceptor gene (Gly389Arg ADRB1) in Caucasian women. Int J Obes Relat Metab Disord. 2002, 26 (5): 633-639. 10.1038/sj.ijo.0801971.
Naveilhan P, Hassani H, Canals JM, Ekstrand AJ, Larefalk A, Chhajlani V, Arenas E, Gedda K, Svensson L, Thoren P, Ernfors P: Normal feeding behavior, body weight and leptin response require the neuropeptide Y Y2 receptor. Nat Med. 1999, 5 (10): 1188-1193. 10.1038/13514.
Siddiq A, Gueorguiev M, Samson C, Hercberg S, Heude B, Levy-Marchal C, Jouret B, Weill J, Meyre D, Walley A, Froguel P: Single nucleotide polymorphisms in the neuropeptide Y2 receptor (NPY2R) gene and association with severe obesity in French white subjects. Diabetologia. 2007, 50 (3): 574-584. 10.1007/s00125-006-0555-2.
Somia NV, Schmitt MJ, Vetter DE, Van Antwerp D, Heinemann SF, Verma IM: LFG: an anti-apoptotic gene that provides protection from Fas-mediated cell death. Proc Natl Acad Sci U S A. 1999, 96 (22): 12667-12672. 10.1073/pnas.96.22.12667.
Leon-Mimila P, Villamil-Ramirez H, Villalobos-Comparan M, Villarreal-Molina T, Romero-Hidalgo S, Lopez-Contreras B, Gutierrez-Vidal R, Vega-Badillo J, Jacobo-Albavera L, Posadas-Romeros C, Canizalez-Roman A, Rio-Navarro BD, Campos-Perez F, Acuna-Alonzo V, Aguilar-Salinas C, Canizales-Quinteros S: Contribution of common genetic variants to obesity and obesity-related traits in mexican children and adults. PLoS One. 2013, 8 (8): e70640-10.1371/journal.pone.0070640.
Eriksson P, Reynisdottir S, Lonnqvist F, Stemme V, Hamsten A, Arner P: Adipose tissue secretion of plasminogen activator inhibitor-1 in non-obese and obese individuals. Diabetologia. 1998, 41 (1): 65-71. 10.1007/s001250050868.
Ferretti G, Bacchetti T, Masciangelo S, Bicchiega V: HDL-paraoxonase and membrane lipid peroxidation: a comparison between healthy and obese subjects. Obesity (Silver Spring). 2010, 18 (6): 1079-1084. 10.1038/oby.2009.338.
Pachocka LM, Wlodarczyk M, Nowicka G, Klosiewicz-Latoszek L, Wolanska D, Stolarska I: [CETP gene TaqIB polymorphism and plasma lipids in patients with overweight and obesity]. Rocz Panstw Zakl Hig. 2012, 63 (2): 149-154.
Chapman MJ, Le Goff W, Guerin M, Kontush A: Cholesteryl ester transfer protein: at the heart of the action of lipid-modulating therapy with statins, fibrates, niacin, and cholesteryl ester transfer protein inhibitors. Eur Heart J. 2010, 31 (2): 149-164. 10.1093/eurheartj/ehp399.
Cannon B, Nedergaard J: Brown adipose tissue: function and physiological significance. Physiol Rev. 2004, 84 (1): 277-359. 10.1152/physrev.00015.2003.
Miyaki K, Sutani S, Kikuchi H, Takei I, Murata M, Watanabe K, Omae K: Increased risk of obesity resulting from the interaction between high energy intake and the Trp64Arg polymorphism of the beta3-adrenergic receptor gene in healthy Japanese men. J Epidemiol. 2005, 15 (6): 203-210. 10.2188/jea.15.203.
Dalgaard LT, Pedersen O: Uncoupling proteins: functional characteristics and role in the pathogenesis of obesity and Type II diabetes. Diabetologia. 2001, 44 (8): 946-965. 10.1007/s001250100596.
Costford S, Gowing A, Harper ME: Mitochondrial uncoupling as a target in the treatment of obesity. Curr Opin Clin Nutr Metab Care. 2007, 10 (6): 671-678. 10.1097/MCO.0b013e3282f0dbe4.
Oram JF, Heinecke JW: ATP-binding cassette transporter A1: a cell cholesterol exporter that protects against cardiovascular disease. Physiol Rev. 2005, 85 (4): 1343-1372. 10.1152/physrev.00005.2005.
Ghorbanian B, Ravassi A, Kordi MR, Hedayati M: The Effects of Rope Training on Lymphocyte ABCA1 Expression, Plasma ApoA-I and HDL-c in Boy Adolescents. Int J Endocrinol Metab. 2013, 11 (2): 76-81.
Mahley RW: Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988, 240 (4852): 622-630. 10.1126/science.3283935.
Wang J, Perrard XD, Perrard JL, Mukherjee A, Rosales C, Chen Y, Smith CW, Pownall HJ, Ballantyne CM, Wu H: ApoE and the role of very low density lipoproteins in adipose tissue inflammation. Atherosclerosis. 2012, 223 (2): 342-349. 10.1016/j.atherosclerosis.2012.06.003.
Xu Z, Yu L, Zhang X: Association between the hOGG1 Ser326Cys polymorphism and lung cancer susceptibility: a meta-analysis based on 22,475 subjects. Diagn Pathol. 2013, 8: 144-10.1186/1746-1596-8-144.
de Matos LL, Del Giglio AB, Matsubayashi CO, de Lima Farah M, Del Giglio A, da Silva Pinhal MA: Expression of CK-19, galectin-3 and HBME-1 in the differentiation of thyroid lesions: systematic review and diagnostic meta-analysis. Diagn Pathol. 2012, 7: 97-10.1186/1746-1596-7-97.
Meta-analysis.http://en.wikipedia.org/wiki/Meta-analysis,
The criteria for overweight or obesity in children and adolescents.http://www.who.int/dietphysicalactivity/childhood_what/en/index.html,
The criteria for overweight or obesity in children.http://www.who.int/growthref/who2007_bmi_for_age/en/index.html,
Jiang H, Sun MW, Hefright B, Chen W, Lu CD, Zeng J: Efficacy of hypocaloric parenteral nutrition for surgical patients: a systematic review and meta-analysis. Clin Nutr. 2011, 30 (6): 730-737. 10.1016/j.clnu.2011.05.006.
Dupont WD, Plummer WD: Power and sample size calculations. A review and computer program. Control Clin Trials. 1990, 11 (2): 116-128. 10.1016/0197-2456(90)90005-M.
Srivastava N, Achyut BR, Prakash J, Agarwal CG, Pant DC, Mittal B: Association of cholesteryl ester transfer protein (TaqIB) and apolipoprotein E (HhaI) gene variants with obesity. Mol Cell Biochem. 2008, 314 (1–2): 171-177.
Maures TJ, Kurzer JH, Carter-Su C: SH2B1 (SH2-B) and JAK2: a multifunctional adaptor protein and kinase made for each other. Trends Endocrinol Metab. 2007, 18 (1): 38-45. 10.1016/j.tem.2006.11.007.
Speakman JR: Functional analysis of seven genes linked to body mass index and adiposity by genome-wide association studies: a review. Hum Hered. 2013, 75 (2–4): 57-79.
Richard AJ, Stephens JM: Emerging roles of JAK-STAT signaling pathways in adipocytes. Trends Endocrinol Metab. 2011, 22 (8): 325-332. 10.1016/j.tem.2011.03.007.
Ren D, Li M, Rui L: Identification of SH2-B as a key regulator of leptin sensitivity, energy balance, and body weight in mice. Cell Metab. 2005, 2 (2): 95-104. 10.1016/j.cmet.2005.07.004.
Paternoster L, Evans DM, Nohr EA, Holst C, Gaborieau V, Brennan P, Gjesing AP, Grarup N, Witte DR, Jorgensen T, Linneberg A, Lauritzen T, Sandbaek A, Hansen T, Pedersen O, Elliott KS, Kemp JP, St Pourcain B, McMahon G, Zelenika D, Hager J, Lathrop M, Timpson NJ, Smith GD, Sorensen TI: Genome-wide population-based association study of extremely overweight young adults–the GOYA study. PLoS One. 2011, 6 (9): e24303-10.1371/journal.pone.0024303.
Hotta K, Nakamura M, Nakamura T, Matsuo T, Nakata Y, Kamohara S, Miyatake N, Kotani K, Komatsu R, Itoh N, Mineo I, Wada J, Masuzaki H, Yoneda M, Nakajima A, Funahashi T, Miyazaki S, Tokunaga K, Kawamoto M, Ueno T, Hamaguchi K, Tanaka K, Yamada K, Hanafusa T, Oikawa S, Yoshimatsu H, Nakao K, Sakata T, Matsuzawa Y, Kamatani N, et al.: Association between obesity and polymorphisms in SEC16B, TMEM18, GNPDA2, BDNF, FAIM2 and MC4R in a Japanese population. J Hum Genet. 2009, 54 (12): 727-731. 10.1038/jhg.2009.106.
Li C, Qiu X, Yang N, Gao J, Rong Y, Xiong C, Zheng F: Common rs7138803 variant of FAIM2 and obesity in Han Chinese. BMC Cardiovasc Disord. 2013, 13: 56-10.1186/1471-2261-13-56.
Liu Y, Tang W, Wang J, Xie L, Li T, He Y, Qin X, Li S: Clinicopathological and prognostic significance of S100A4 overexpression in colorectal cancer: a meta-analysis. Diagn Pathol. 2013, 8 (1): 181-10.1186/1746-1596-8-181.
Wang Z, Zhang Y, Kong X, Li S, Hu Y, Wang R, Li Y, Lu C, Lin N, Chen W: Association of a polymorphism in PON-1 gene with steroid-induced osteonecrosis of femoral head in Chinese Han population. Diagn Pathol. 2013, 8 (1): 186-10.1186/1746-1596-8-186.
Zhang Y, Wang R, Li S, Kong X, Wang Z, Chen W, Lin N: Genetic polymorphisms in plasminogen activator inhibitor-1 predict susceptibility to steroid-induced osteonecrosis of the femoral head in Chinese population. Diagn Pathol. 2013, 8 (1): 169-10.1186/1746-1596-8-169.
Acknowledgments
The research was supported by the grants from: National Natural Science Foundation of China (31100919 and 81371469), Natural Science Foundation of Zhejiang Province (LR13H020003), K. C. Wong Magna Fund in Ningbo University, and Ningbo Social Development Research Projects (2012C50032).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
QH, LX and SB conceived the study idea and designed the study. FC, QL and QH reviewed the literature and performed statistical analyses. LT and HY extracted data and drafted the manuscript. SD, YM DW and MY reviewed and edited the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
13000_2014_1106_MOESM2_ESM.tiff
Additional file 2: Figure S1: Forest plots of the association studies of UCP1 rs1800592 in our subgroup meta-analysis. (TIFF 26 KB)
Additional file 3: Figure S2: Forest plots of the association studies of APOE ϵ2/ϵ3/ϵ4. (TIFF 23 KB)
13000_2014_1106_MOESM4_ESM.tiff
Additional file 4: Figure S3: Funnel plots of the studies related to UCP1 rs1800592 by subgroup meta-analysis and APOE ϵ2/ϵ3/ϵ4. (TIFF 26 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Tang, L., Ye, H., Hong, Q. et al. Meta-analyses between 18 candidate genetic markers and overweight/obesity. Diagn Pathol 9, 56 (2014). https://doi.org/10.1186/1746-1596-9-56
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1746-1596-9-56