Abstract
Background
Disrupted-in-Schizophrenia 1 (DISC1) gene is one of the most promising candidate genes for major mental disorders. In a previous study, a Finnish group demonstrated that DISC1 polymorphisms were associated with autism and Asperger syndrome. However, the results were not replicated in Korean population. To determine whether DISC1 is associated with autism in Chinese Han population, we performed a family-based association study between DISC1 polymorphisms and autism.
Methods
We genotyped seven tag single nucleotide polymorphisms (SNPs) in DISC1, spanning 338 kb, in 367 autism trios (singleton and their biological parents) including 1,101 individuals. Single SNP association and haplotype association analysis were performed using the family-based association test (FBAT) and Haploview software.
Results
We found three SNPs showed significant associations with autism (rs4366301: G > C, Z = 2.872, p = 0.004; rs11585959: T > C, Z = 2.199, p = 0.028; rs6668845: A > G, Z = 2.326, p = 0.02). After the Bonferroni correction, SNP rs4366301, which located in the first intron of DISC1, remained significant. When haplotype were constructed with two-markers, three haplotypes displayed significant association with autism. These results were still significant after using the permutation method to obtain empirical p values.
Conclusions
Our study provided evidence that the DISC1 may be the susceptibility gene of autism. It suggested DISC1 might play a role in the pathogenesis of autism.
Similar content being viewed by others
Background
Autism is a severe neurodevelopmental disorder mainly characterized by impairment in social interaction, communicative deficits, and repetitive and stereotyped patterns of behaviors or interests [1]. Since autism was first described as a disorder by Dr. Leo Kanner in 1943 [2], the speculation about its etiology has become an intriguing field which attracts a large number of scientists. To date, compelling evidence from twin and family studies has indicated a strong genetic involvement in the etiology of autism and the estimated heritability is over 90% [3–7]. However, the genetic etiology remains elusive.
Several lines of evidence from postmortem [8–10] and structural magnetic resonance imaging (MRI) [11–16] supported the existence of brain abnormality in autism. It indicated that brain abnormalities in autism are not limited to a single brain area but involve different structures within a globally affected neuronal network. Furthermore, functional MRI studies in autism patients have indicated that alterations in task related connectivity, including enhanced activation and connectivity in posterior areas, enhanced reliance on visuospatial abilities for verbal and visual reasoning and reduced frontal systems connectivity [17–21]. All together, these previous studies suggested that abnormalities of neurodevelopment might be the etiology of autism. Considering the heritability of autism is relatively high, genes which play important roles in neurodevelopment might be candidate genes for autism.
Disrupted-in-Schizophrenia 1 (DISC1) is a candidate gene of autism, which has been demonstrated to involve in neuronal migration [22–24], neurite outgrowth [25, 26] and axon targeting [27] during brain development. The DISC1, on chromosome 1q42, was originally identified from the breakpoint of a hereditary chromosomal translocation in a large Scottish pedigree [28–30]. It has 13 exons spanning over 410 kb and encodes a cytosolic scaffold protein with coiled-coil-rich C-terminus which interacts with multiple proteins involved in various functions. As one of the most interesting candidate genes for major mental illness, DISC1 have been demonstrated to associate with schizophrenia, depression, bipolar disorder and schizoaffective disorder in several independent populations by multiple association studies [31–37]. Meanwhile, investigations into function of DISC1 have revealed that it may contribute to risk for psychiatric disorders through its effects on the processes of neurodevelopment [25, 38–43]. Recently, Niwa et al. reported that transient knockdown of Disc1 during embryo development of mice resulted in selective abnormalities in postnatal mesocortical dopaminergic maturation and adult behavioral deficits [44].
Autism and schizophrenia share neurocognitive defects such as impaired executive function and social functioning [45, 46]. Thus, the impairment of neurodevelopment may be the common underlying mechanism of these two disorders. Recent studies have showed a genetic overlap between these two disorders [47, 48]. Therefore, DISC1 was selected as a candidate gene for autism. Two reports have revealed some abnormalities of DISC1 in three individuals with autism spectrum disorders (ASD) [49, 50]. Moreover, a Finnish group has represented a significant association of DISC1 with ASD. They established association between autism and a DISC1 intragenic microsatellite D1S2709 and found that one intragenic SNP, rs1322784, was associated with Asperger syndrome [51]. Their study indicated that DISC1 might also play a role in the etiology of autism. However, the results of association studies were not consistent. In another association study, no significant association of DISC1 polymorphism with ASD was found in Korean population [52].
Considering the biological functions of DISC1 protein and the positive results of previous association studies between DISC1 and mental disorders, we hypothesized that DISC1 might be involved in the etiology of autism. To explore whether DISC1 is the susceptibility gene of autism in Chinese Han population, we performed this family-based association study to identify the association between DISC1 polymorphisms and autism.
Materials and methods
Subjects
Three hundred and sixty seven Chinese Han family trios (singleton autistic disorder patients and their unaffected biological parents) were recruited for the present study at the Institute of Mental Health, Peking University, China. Of the 367 autistic child probands, 336 were male and 31 were female. The mean age of the children at the time of testing was 7.5 years (ranged from 3 to 17 years). Diagnoses of autism were established by two senior psychiatrists. All patients fulfilled the DSM-IV (Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition) criteria for autistic disorder. To assess the cases, childhood autism rating scale (CARS) [53] and autism behavior checklist (ABC) [54] were used. Children with fragile X syndrome, tuberous sclerosis, a previously identified chromosomal abnormality, or any other neurological condition suspected to be associated with autism were excluded. All subjects provided written informed consent signed by their legal guardians (i.e., their parents) for participation in this study. This study was approved by the Ethics Committee of the Institute of Mental Health, Peking University.
Genotyping
Genomic DNA was extracted from the blood using a Qiagen QIAamp DNA Mini Kit. We selected seven single nucleotide polymorphisms (SNPs) in the DISC1 gene according to the dbSNP http://www.ncbi.nlm.nih.gov/SNP/ and the HapMap phase II Chinese Han in Beijing (CHB) genotype dataset http://hapmap.ncbi.nlm.nih.gov/, including rs4366301, rs11585959, rs1322784, rs6668845, rs10864698, rs872624 in introns and rs821616 in exon (Figure 1). SNPs with minor allele frequency (MAF) >0.05 were selected and pair-wise tagging in the Tagger module implemented in Haploview v4.1 program was used to select SNPs that could capture >80% of the markers with r2>0.8. Meanwhile, the physical position of SNPs and the previous positive results were considered too.
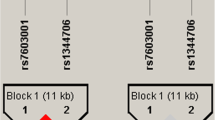
DISC1 gene locus and linkage disequilibrium (LD) structure. (A) A diagram showing the exonic structure of DISC1 gene (black). The single nucleotide polymorphisms (SNPs) used in this study are shown in relation to the exons. (B) The linkage disequilibrium (LD) structure of the DISC1 region in the total autism samples according to Haploview (solid spine of LD, D'> 0.7). Markers with LD (D' < 1 and LOD > 2) are shown in red through pink (color intensity decreases with decreasing D' value). Regions of low LD (D' < 1 and LOD < 2) are shown in white. Two blocks were generated by Haploview.
Direct DNA sequencing was used for analyzing rs11585959. The other six SNPs (rs4366301, rs1322784, rs6668845, rs10864698, rs872624 and rs821616) were analyzed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis. The information of primers and PCR-RFLP analysis is given in Table 1. The PCR amplification was performed in a 25 μl volume containing GC Buffer (TaKaRa), 200 μM of each dNTPs, 0.3 μM of each primer, 1 U of Taq DNA polymerase, and 40 ng of the genomic DNA. The conditions used for PCR amplification were an initial denaturation phase at 94°C for 5min, followed by 38 cycles at 94°C for 30 sec, annealing at 53-65°C for 30-45 sec, and extension at 72°C for 30 sec, followed by a final extension phase at 72°C for 7 min. A 15 μL aliquot of the PCR product mixtures was completely digested with 4 units of restriction enzyme overnight. Digestion products were visualized through ethidium bromide staining after electrophoresis in 1.5%-2% agarose gels. The DNA sequencing was performed after cleaning the PCR product using a BigDye Terminator Cycle Sequencing Ready Reaction Kit with Ampli Taq DNA polymerase (PE Biosystem). The inner primers were used for the cycle-sequencing reaction, and the fragments were separated by electrophoresis on an ABI PRISM 377-96 DNA Sequencer (Applied Biosystem, Foster City, U.S.A).
Statistical Analyses
Prior to analysis, Mendelian inconsistencies were checked using the PEDCHECK program, version 1.1 [55]. Deviations in the genotype counts from the Hardy-Weinberg equilibrium were tested using a chi-square goodness-of-fit test. The pairwise linkage disequilibrium (LD) analysis was applied to detect the inter-marker relationship with Haploview http://www.broad.mit.edu/mpg/haploview/, using D' values. The family-based association test (FBAT) was performed with FBAT program v1.5.1 [56]. The FBAT program uses generalized score statistics to perform a variety of transmission disequilibrium tests (TDT), including haplotype analyses. Moreover, the FBAT program provides estimates of haplotype frequencies and pairwise linkage disequilibrium (LD) between the specified markers http://www.biostat.harvard.edu/~fbat/default.html. The global haplotype tests of association were performed under "multiallelic" mode in haplotype FBAT. Meanwhile, the individual haplotype tests were conducted under "biallelic" mode in haplotype FBAT. Family-based association tests were performed under an additive model in the present study. The significance level for all statistical tests was two-tailed (p < 0.05). We also performed the association analysis by Haploview version 4.1 http://www.broad.mit.edu/mpg/haploview [57].
Results
We tested a total of seven SNPs over the 338 kb region of 1q42 with DISC1 in 367 Chinese Han autism trios. All seven SNPs were polymorphic with minor allele frequency (MAF) >5% and were then used as genetic markers for the association study. A number of sample genotypes could not be assigned due to repeated PCR failure or unclear genotype results, including 10 genotypes for rs4366301 and rs11585959; 5 for rs1322784; 9 for rs6668845; 7 for rs10864698; 30 for rs872624 and 3 for rs821616. None of the genotype distributions of these SNPs in patients significantly deviated from Hardy-Weinberg equilibrium (p > 0.05, data not shown). Allele frequencies and the results of FBAT for single SNP analysis are shown in Table 2. Univariate (single-marker) FBAT demonstrated that variant alleles at three SNPs showed a preferential transmission (rs4366301 G: Z = 2.872, p = 0.004; rs11585959 T: Z = 2.199, p = 0.028; rs6668845 A: Z = 2.326, p = 0.020) (Table 2). Moreover, rs4366301 remained significant with autism after the Bonferroni correction (significantly corrected p < 0.007, i.e. α = 0.05/n) which was considered as a conservative correction method. The LD patterns of the SNPs were measured with the solid spine of LD option of Haploview (D'>0.7). Two LD blocks were identified (Figure 1). To determine whether any specific haplotype would confer a higher risk for autism, the specific and global-haplotype FBAT tests of association were performed (Table 3). Three haplotypes displayed significant association. The haplotype G-T (rs4366301-rs11585959) revealed significant excess transmission from parents to affected offspring both in the specific and global haplotype FBAT (p = 0.0015 and 0.0062, respectively). In addition, haplotype constructed from the A allele of rs6668845 and the G allele of rs10864698 demonstrated an excess transmission (p = 0.024 and 0.0064, respectively) and haplotype constructed from the G allele of rs6668845 and the A allele of rs10864698 displayed an undertransmission (p = 0.0499 and 0.0064, respectively). These results were still significant, after using the permutation method to obtain empirical p values.
We also performed the single SNP association and haplotype analysis with Haploview, the results were similar to those using FBAT method. (Additional file 1, Table S1, Table S2).
Discussion
DISC1 is one of the most promising susceptibility genes for major mental disorders. Genetic studies had indicated that DISC1 was a susceptibility gene for schizophrenia, depression, bipolar disorder and schizoaffective disorder [31–37]. Many investigators have considered the possibility that adult brain function and behavior are influenced by neuronal network formation during early development [58–61]. Considering the established biological functions of the DISC1 protein and the structural and functional abnormalities in brains of autism patients, DISC1 was a candidate gene for autism. Up to know, besides two case reports, only one published research detected DISC1 was associated with autism and Asperger syndrome using family-based association analysis. However, the results did not replicated in a Korean population. To investigate DISC1 as potential autism susceptibility gene, we performed a family-based association study in Chinese Han population. In the present study, we analyzed seven tag SNPs in DISC1 for association with autism in 367 Chinese Han family trios. The results of our study demonstrated that three SNPs, rs4366301, rs11585959, and rs6668845 were associated with autism (Table 2). Among these three SNPs, rs4366301 had the most significance (p = 0.004). Even after Bonferroni correction, rs4366301 was still significant. When haplotypes were constructed with two markers, three haplotypes displayed positive association. The results remained significant when the global haplotype FBAT was performed (Table 3). Our research established the association between autism and the DISC1 gene.
In a previous study, a Finnish group reported a positive association of rs1322784 with Asperger syndrome besides a significant association of a DISC1 intragenic microsatellite with autism [51]. However, we didn't replicate this positive finding in 367 autism trios in Chinese Han population. No significant association was obtained for rs1322784 in our study (p = 0.695). One reason is that our research mainly focused on the genetic research of autism, while the Finnish study founded that rs1322784 was associated with Asperger syndrome. Although autism and Asperger syndrome are included in autism spectrum disorder (ASD), these two disorders have different clinical features. Therefore, the genetic etiology of autism and Asperger syndrome may be not consistent. Second, as the authors mentioned in their paper, the sample size of the Finnish study was relatively small [51]. The result should be replicated in an expanded sample. Ethnic genetic heterogeneity might be another reason for these discordant findings. In our study, major allele frequency for rs1322784 in autism cases was 0.617 compare to 0.83 in Finnish population (Additional file 1, Table S3)[51, 52]. The allele frequency for rs1322784 varies in different population as shown in the data from the International HapMap project. In CEU (Utah residents with Northern and Western European ancestry from the CEPH collection) population the major allele frequency (MAF) of rs1322784 is 0.786, as well as 0.589 in Chinese Han population in Beijing (CHB). Though our study did not replicate the association between rs1322784 and autism in Chinese Han population, we observed strong association of rs4366301 in DISC1 with autism.
In the Finnish study, Kilpinen et al. detected suggestive association with haplotype HEP3 (rs751229 and rs3738401) [62] in the Asperger syndrome study sample using TRANSMIT software. However, this haplotype was not associated with Asperger syndrome and autism when analysis was performed by FBAT software. Moreover, single SNP association analysis of rs751229 and rs3738401 obtained no significant association with Asperger syndrome and autism [51]. We constructed the LD patterns between rs751229 and rs3738401 in the Haploview by using the genotyping date (only SNPs with MAF > 0.05) from the HapMap project http://www.hapmap.org. In Chinese Han population, genotyping date of rs751229 was not available in HapMap project. However, SNP rs2082552 and rs823165 were genotyped. The positions of these two SNPs were very close to rs751229 (the distance is 649 bp and 907 bp, respectively). LD analysis demonstrated that rs3728401, rs2082552, and rs823165 were not located within one linkage disequilibrium block (Additional file 1, Figure S1). Pairwise D' between rs3728401 and rs2082552 is 0.64, while D' between rs3728401 and rs823165 is 0.66. These results suggested linkage disequilibrim of rs3738401 and rs751229 might not exist in Chinese Han population. Therefore, we did not perform the association study of these two SNPs with autism in Chinese Han population. In our study, we detected two other haplotypes which were associated with autism. The haplotype G-T (rs4366301-rs11585959) and A-G (rs6668845-rs10864698) demonstrate significant excess transmission from parents to affected offspring.
Many association studies have replicated a positive association of rs821616 which is a missense polymorphism leading to a serine-to-cysteine substitution at amino acid 704 (Ser704Cys)in DISC1 with schizophrenia [32, 36, 37]. Our group have also demonstrated the association between rs821616 and schizophrenia in Chinese Han population[32]. Function studies have reported that the substitution induced by rs821616 polymorphism may affect cognition [37, 63] and the underlying mechanism may be an altered interaction of DISC1 with its binding partners or disruption of its signaling pathways [64, 65]. Therefore, we genotyped this SNP in our study However, we obtained no significant support for the association of rs821616 with autism (p = 0.467). This result is consistent with the results of the previous studies in Korean population [52] and Finnish population [51]. It indicated that rs821616 may not involve in the etiology of autism. However, we found that other SNPs were significantly associated with autism in Chinese Han population.
DISC1 is a pleiotropic protein with many purported functions, subcellular locations, binding partners and plays roles in many aspects of neurodevelopment [22, 39, 66–69]. The middle part of DISC1 had been identified as a DISC1 self-association domain [23]. In contrast, the C-termianl part of DISC1 binds to important interacting molecules.. Interactions with LIS1, NDEL, FEZ1, integrin and other proteins indicated that many of the proteins of the "DISC1 network" are involved in pathways critical for neuronal polarization and migration, formation of synapses, and dendritic growth and branching. Up to now, researches have demonstrated that DISC1 is a susceptibility gene for mental disorders such as autism, schizophrenia and bipolar disorder. However, it is possible that the underlying pathogeneses of DISC1 for different psychiatric disorders might be different.
In conclusion, the present study demonstrated a positive association of DISC1 with autism in Chinese Han population. Our results provided evidence that DISC1 might involve in the etiology of autism. Further researches of association analysis across populations are needed.
References
American Psychiatric Association: The Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV). 1994, Washington, DC: American Psychiatric Publishing Inc
Kanner L: autistic disturbances of affective contact. Nervous Child. 1943, 2: 217-250.
Ronald A, Happe F, Bolton P, Butcher LM, Price TS, Wheelwright S, Baron-Cohen S, Plomin R: Genetic heterogeneity between the three components of the autism spectrum: a twin study. J Am Acad Child Adolesc Psychiatry. 2006, 45: 691-699. 10.1097/01.chi.0000215325.13058.9d.
Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M: Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995, 25: 63-77. 10.1017/S0033291700028099.
Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, Jakobsson G, Bohman M: A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry. 1989, 30: 405-416. 10.1111/j.1469-7610.1989.tb00254.x.
Folstein S, Rutter M: Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry. 1977, 18: 297-321. 10.1111/j.1469-7610.1977.tb00443.x.
Folstein SE, Rosen-Sheidley B: Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet. 2001, 2: 943-955. 10.1038/35103559.
Ritvo ER, Freeman BJ, Scheibel AB, Duong T, Robinson H, Guthrie D, Ritvo A: Lower Purkinje cell counts in the cerebella of four autistic subjects: initial findings of the UCLA-NSAC Autopsy Research Report. Am J Psychiatry. 1986, 143: 862-866.
Fatemi SH, Halt AR, Realmuto G, Earle J, Kist DA, Thuras P, Merz A: Purkinje cell size is reduced in cerebellum of patients with autism. Cell Mol Neurobiol. 2002, 22: 171-175. 10.1023/A:1019861721160.
Bailey AJ: Postmortem studies of autism. Autism Res. 2008, 1: 265-10.1002/aur.51.
Paya Gonzalez B, Fuentes Menchaca N: Neurobiology of autism: neuropathology and neuroimaging studies. Actas Esp Psiquiatr. 2007, 35: 271-276.
Courchesne E, Carper R, Akshoomoff N: Evidence of brain overgrowth in the first year of life in autism. JAMA. 2003, 290: 337-344. 10.1001/jama.290.3.337.
Saitoh O, Karns CM, Courchesne E: Development of the hippocampal formation from 2 to 42 years: MRI evidence of smaller area dentata in autism. Brain. 2001, 124: 1317-1324. 10.1093/brain/124.7.1317.
Verhoeven JS, De Cock P, Lagae L, Sunaert S: Neuroimaging of autism. Neuroradiology. 2010, 52: 3-14. 10.1007/s00234-009-0583-y.
Paakki JJ, Rahko J, Long X, Moilanen I, Tervonen O, Nikkinen J, Starck T, Remes J, Hurtig T, Haapsamo H: Alterations in regional homogeneity of resting-state brain activity in autism spectrum disorders. Brain Res. 2010, 1321: 169-179.
Griebling J, Minshew NJ, Bodner K, Libove R, Bansal R, Konasale P, Keshavan MS, Hardan A: Dorsolateral prefrontal cortex magnetic resonance imaging measurements and cognitive performance in autism. J Child Neurol. 2010, 25: 856-863. 10.1177/0883073809351313.
Koshino H, Kana RK, Keller TA, Cherkassky VL, Minshew NJ, Just MA: fMRI investigation of working memory for faces in autism: visual coding and underconnectivity with frontal areas. Cereb Cortex. 2008, 18: 289-300.
Allen G, Muller RA, Courchesne E: Cerebellar function in autism: functional magnetic resonance image activation during a simple motor task. Biol Psychiatry. 2004, 56: 269-278. 10.1016/j.biopsych.2004.06.005.
Allen G, Courchesne E: Differential effects of developmental cerebellar abnormality on cognitive and motor functions in the cerebellum: an fMRI study of autism. Am J Psychiatry. 2003, 160: 262-273. 10.1176/appi.ajp.160.2.262.
Sahyoun CP, Belliveau JW, Soulieres I, Schwartz S, Mody M: Neuroimaging of the functional and structural networks underlying visuospatial vs. linguistic reasoning in high-functioning autism. Neuropsychologia. 2010, 48: 86-95. 10.1016/j.neuropsychologia.2009.08.013.
Mostofsky SH, Powell SK, Simmonds DJ, Goldberg MC, Caffo B, Pekar JJ: Decreased connectivity and cerebellar activity in autism during motor task performance. Brain. 2009, 132: 2413-2425. 10.1093/brain/awp088.
Meyer KD, Morris JA: Disc1 regulates granule cell migration in the developing hippocampus. Hum Mol Genet. 2009, 18: 3286-3297. 10.1093/hmg/ddp266.
Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, Sawamura N, Park U, Kudo C, Okawa M: A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005, 7: 1167-1178. 10.1038/ncb1334.
Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, Liu XB, Yang CH, Jordan JD, Ma DK: Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007, 130: 1146-1158. 10.1016/j.cell.2007.07.010.
Miyoshi K, Honda A, Baba K, Taniguchi M, Oono K, Fujita T, Kuroda S, Katayama T, Tohyama M: Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry. 2003, 8: 685-694. 10.1038/sj.mp.4001352.
Hattori T, Shimizu S, Koyama Y, Yamada K, Kuwahara R, Kumamoto N, Matsuzaki S, Ito A, Katayama T, Tohyama M: DISC1 regulates cell-cell adhesion, cell-matrix adhesion and neurite outgrowth. Mol Psychiatry. 2010
Faulkner RL, Jang MH, Liu XB, Duan X, Sailor KA, Kim JY, Ge S, Jones EG, Ming GL, Song H, Cheng HJ: Development of hippocampal mossy fiber synaptic outputs by new neurons in the adult brain. Proc Natl Acad Sci USA. 2008, 105: 14157-14162. 10.1073/pnas.0806658105.
Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, St Clair DM, Muir WJ, Blackwood DH, Porteous DJ: Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000, 9: 1415-1423. 10.1093/hmg/9.9.1415.
St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, Gosden C, Evans HJ: Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990, 336: 13-16. 10.1016/0140-6736(90)91520-K.
Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ: Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001, 69: 428-433. 10.1086/321969.
Saetre P, Agartz I, De Franciscis A, Lundmark P, Djurovic S, Kahler A, Andreassen OA, Jakobsen KD, Rasmussen HB, Werge T: Association between a disrupted-in-schizophrenia 1 (DISC1) single nucleotide polymorphism and schizophrenia in a combined Scandinavian case-control sample. Schizophr Res. 2008, 106: 237-241. 10.1016/j.schres.2008.08.024.
Qu M, Tang F, Yue W, Ruan Y, Lu T, Liu Z, Zhang H, Han Y, Zhang D, Wang F: Positive association of the Disrupted-in-Schizophrenia-1 gene (DISC1) with schizophrenia in the Chinese Han population. Am J Med Genet B Neuropsychiatr Genet. 2007, 144B: 266-270. 10.1002/ajmg.b.30322.
Zhang F, Sarginson J, Crombie C, Walker N, St Clair D, Shaw D: Genetic association between schizophrenia and the DISC1 gene in the Scottish population. Am J Med Genet B Neuropsychiatr Genet. 2006, 141B: 155-159. 10.1002/ajmg.b.30274.
Hodgkinson CA, Goldman D, Jaeger J, Persaud S, Kane JM, Lipsky RH, Malhotra AK: Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am J Hum Genet. 2004, 75: 862-872. 10.1086/425586.
Zhang X, Tochigi M, Ohashi J, Maeda K, Kato T, Okazaki Y, Kato N, Tokunaga K, Sawa A, Sasaki T: Association study of the DISC1/TRAX locus with schizophrenia in a Japanese population. Schizophr Res. 2005, 79: 175-180. 10.1016/j.schres.2005.05.023.
Song W, Li W, Feng J, Heston LL, Scaringe WA, Sommer SS: Identification of high risk DISC1 structural variants with a 2% attributable risk for schizophrenia. Biochem Biophys Res Commun. 2008, 367: 700-706. 10.1016/j.bbrc.2007.12.117.
Callicott JH, Straub RE, Pezawas L, Egan MF, Mattay VS, Hariri AR, Verchinski BA, Meyer-Lindenberg A, Balkissoon R, Kolachana B: Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci USA. 2005, 102: 8627-8632. 10.1073/pnas.0500515102.
Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, Malloy MP, Chubb JE, Huston E, Baillie GS: DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005, 310: 1187-1191. 10.1126/science.1112915.
Kim JY, Duan X, Liu CY, Jang MH, Guo JU, Pow-anpongkul N, Kang E, Song H, Ming GL: DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009, 63: 761-773. 10.1016/j.neuron.2009.08.008.
Morris JA, Kandpal G, Ma L, Austin CP: DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum Mol Genet. 2003, 12: 1591-1608. 10.1093/hmg/ddg162.
Millar JK, Christie S, Porteous DJ: Yeast two-hybrid screens implicate DISC1 in brain development and function. Biochem Biophys Res Commun. 2003, 311: 1019-1025. 10.1016/j.bbrc.2003.10.101.
Brandon NJ, Handford EJ, Schurov I, Rain JC, Pelling M, Duran-Jimeniz B, Camargo LM, Oliver KR, Beher D, Shearman MS, Whiting PJ: Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Mol Cell Neurosci. 2004, 25: 42-55. 10.1016/j.mcn.2003.09.009.
Kamiya A, Tomoda T, Chang J, Takaki M, Zhan C, Morita M, Cascio MB, Elashvili S, Koizumi H, Takanezawa Y: DISC1-NDEL1/NUDEL protein interaction, an essential component for neurite outgrowth, is modulated by genetic variations of DISC1. Hum Mol Genet. 2006, 15: 3313-3323. 10.1093/hmg/ddl407.
Niwa M, Kamiya A, Murai R, Kubo K, Gruber AJ, Tomita K, Lu L, Tomisato S, Jaaro-Peled H, Seshadri S: Knockdown of DISC1 by in utero gene transfer disturbs postnatal dopaminergic maturation in the frontal cortex and leads to adult behavioral deficits. Neuron. 2010, 65: 480-489. 10.1016/j.neuron.2010.01.019.
Baron-Cohen S, Belmonte MK: Autism: a window onto the development of the social and the analytic brain. Annu Rev Neurosci. 2005, 28: 109-126. 10.1146/annurev.neuro.27.070203.144137.
Happe F, Booth R, Charlton R, Hughes C: Executive function deficits in autism spectrum disorders and attention-deficit/hyperactivity disorder: examining profiles across domains and ages. Brain Cogn. 2006, 61: 25-39. 10.1016/j.bandc.2006.03.004.
Carroll LS, Owen MJ: Genetic overlap between autism, schizophrenia and bipolar disorder. Genome Med. 2009, 1: 102-10.1186/gm102.
Burbach JP, van der Zwaag B: Contact in the genetics of autism and schizophrenia. Trends Neurosci. 2009, 32: 69-72. 10.1016/j.tins.2008.11.002.
Crepel A, Breckpot J, Fryns JP, De la Marche W, Steyaert J, Devriendt K, Peeters H: DISC1 duplication in two brothers with autism and mild mental retardation. Clin Genet. 2010, 77: 389-394. 10.1111/j.1399-0004.2009.01318.x.
Williams JM, Beck TF, Pearson DM, Proud MB, Cheung SW, Scott DA: A 1q42 deletion involving DISC1, DISC2, and TSNAX in an autism spectrum disorder. Am J Med Genet A. 2009, 149A: 1758-1762. 10.1002/ajmg.a.32941.
Kilpinen H, Ylisaukko-Oja T, Hennah W, Palo OM, Varilo T, Vanhala R, Nieminen-von Wendt T, von Wendt L, Paunio T, Peltonen L: Association of DISC1 with autism and Asperger syndrome. Mol Psychiatry. 2008, 13: 187-196. 10.1038/sj.mp.4002031.
Lim SM, Kim HJ, Nam M, Chung JH, Park YH: Association study of DISC1 in Korean population with autism spectrum disorders. Psychiatr Genet. 2009, 19: 160-10.1097/YPG.0b013e32832a9bd1.
Schopler E, Reichler RJ, DeVellis RF, Daly K: Toward objective classification of childhood autism: Childhood Autism Rating Scale (CARS). J Autism Dev Disord. 1980, 10: 91-103. 10.1007/BF02408436.
Krug DA, Arick J, Almond P: Behavior checklist for identifying severely handicapped individuals with high levels of autistic behavior. J Child Psychol Psychiatry. 1980, 21: 221-229. 10.1111/j.1469-7610.1980.tb01797.x.
O'Connell JR, Weeks DE: PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998, 63: 259-266. 10.1086/301904.
Rabinowitz D, Laird N: A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000, 50: 211-223. 10.1159/000022918.
Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005, 21: 263-265. 10.1093/bioinformatics/bth457.
Cannon TD, van Erp TG, Bearden CE, Loewy R, Thompson P, Toga AW, Huttunen MO, Keshavan MS, Seidman LJ, Tsuang MT: Early and late neurodevelopmental influences in the prodrome to schizophrenia: contributions of genes, environment, and their interactions. Schizophr Bull. 2003, 29: 653-669.
Harrison PJ, Weinberger DR: Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005, 10: 40-685. 10.1038/sj.mp.4001558. image 4
Lisman JE, Coyle JT, Green RW, Javitt DC, Benes FM, Heckers S, Grace AA: Circuit-based framework for understanding neurotransmitter and risk gene interactions in schizophrenia. Trends Neurosci. 2008, 31: 234-242. 10.1016/j.tins.2008.02.005.
Jaaro-Peled H, Hayashi-Takagi A, Seshadri S, Kamiya A, Brandon NJ, Sawa A: Neurodevelopmental mechanisms of schizophrenia: understanding disturbed postnatal brain maturation through neuregulin-1-ErbB4 and DISC1. Trends Neurosci. 2009, 32: 485-495. 10.1016/j.tins.2009.05.007.
Hennah W, Varilo T, Kestila M, Paunio T, Arajarvi R, Haukka J, Parker A, Martin R, Levitzky S, Partonen T: Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Hum Mol Genet. 2003, 12: 3151-3159. 10.1093/hmg/ddg341.
Prata DP, Mechelli A, Fu CH, Picchioni M, Kane F, Kalidindi S, McDonald C, Kravariti E, Toulopoulou T, Miorelli A: Effect of disrupted-in-schizophrenia-1 on pre-frontal cortical function. Mol Psychiatry. 2008, 13: 915-917. 10.1038/mp.2008.76. 909
Eastwood SL, Walker M, Hyde TM, Kleinman JE, Harrison PJ: The DISC1 Ser704Cys substitution affects centrosomal localization of its binding partner PCM1 in glia in human brain. Hum Mol Genet. 2010, 19: 2487-2496. 10.1093/hmg/ddq130.
Hashimoto R, Numakawa T, Ohnishi T, Kumamaru E, Yagasaki Y, Ishimoto T, Mori T, Nemoto K, Adachi N, Izumi A: Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Hum Mol Genet. 2006, 15: 3024-3033. 10.1093/hmg/ddl244.
Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, Kerrien S, Bonnert TP, Whiting PJ, Brandon NJ: Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry. 2007, 12: 74-86. 10.1038/sj.mp.4001880.
Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, Makino Y, Seshadri AJ, Ishizuka K, Srivastava DP: Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 2010, 13: 327-332. 10.1038/nn.2487.
Ming GL, Song H: DISC1 partners with GSK3beta in neurogenesis. Cell. 2009, 136: 990-992. 10.1016/j.cell.2009.03.005.
Shinoda T, Taya S, Tsuboi D, Hikita T, Matsuzawa R, Kuroda S, Iwamatsu A, Kaibuchi K: DISC1 regulates neurotrophin-induced axon elongation via interaction with Grb2. J Neurosci. 2007, 27: 4-14. 10.1523/JNEUROSCI.3825-06.2007.
Acknowledgements and Funding
We thank all the patients and their families for their support and participation.
This work was supported by grants from the National High Technology Research and Development Program of China (2010CB833905), the National Natural Science Foundation of China (30870897, 81071110), and the Beijing Natural Science Foundation (7081005).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
FZ and LW contributed equally to this work.
FZ and LW designed the study and drafted the manuscript. FZ performed the experiment, analyzed the data and interpreted the results. MJ, WY, YR, TL, JL and JL participated in data collection. WY and DZ supervised the study. All authors read and approved the final manuscript.
Electronic supplementary material
12993_2010_331_MOESM1_ESM.DOC
Additional file 1: 1-DISC1 and autism. Table S1. Results of single marker association for seven SNPs in DISC1 gene in 367 trios by Haploview. Table S2. Results of Haplotype association analysis for the SNPs with linkage disequilibrium in 367 trios by Haploview. Table S3. Frequencies of different alleles and genotypes in different association studies Figure S1. DISC1 gene linkage disequilibrium (LD) structure in Chinese Han in Beijing (CHB) using HapMap date by Haploview. Tables and figures (DOC 110 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Zheng, F., Wang, L., Jia, M. et al. Evidence for association between Disrupted-in-schizophrenia 1 (DISC1) gene polymorphisms and autism in Chinese Han population: a family-based association study. Behav Brain Funct 7, 14 (2011). https://doi.org/10.1186/1744-9081-7-14
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1744-9081-7-14