Abstract
Background
DNA deletion and duplication were determined as the major mutation underlying Duchenne muscular dystrophy (DMD) and Becker muscular dystrophy (BMD).
Method
Applying multiplex ligation-dependent probe amplification (MLPA), we have analyzed 179 unrelated DMD/BMD subjects from northern China.
Results
Seventy-three percent of the subjects were found having a deletion (66.25%) or duplication (6.25%). Exons 51–52 were detected as the most common fragment deleted in single-exon deletion, and the region of exons 45–50 was the most common exons deleted in multi-exon deletions. About 90% of DMD/BMD cases carry a small size deletion that involves 10 exons or less, 26.67% of which carry a single-exon deletion. Most of the smaller deletions resulted in an out-of-frame mutation. The most common exons deleted were determined to be between exon 48 and exon 52, with exon 50 was the model allele. Verifying single-exon deletion, one sample with a deletion of exon 53 that was initially observed from MLPA showed that there was a single base deletion that abolished the ligation site in MLPA. Confirmation of single-exon deletion is recommended to exclude single base deletion or mutation at the MLPA ligation site.
Conclusion
The frequency of deletion and duplication in northern China is similar to global ethnic populations.
Similar content being viewed by others
Background
Duchenne muscular dystrophy (DMD) and its less severe allelic form Becker muscular dystrophy (BMD) are common X-linked recessive neuromuscular degeneration diseases [1, 2]. The incidence of DMD is 1/3500 and BMD is 1/18,000 in male living births, respectively [3, 4]. They both are caused by genetic defects of the dystrophin gene, which is located at Xq21.2, spanning 2.4 Mb [5, 6]. The dystrophin gene consists of 79 exons and encodes a 14.6 kb mRNA, which is expressed in skeleton, muscles and brain [7]. Deletions, duplications and point mutations have been reported in most exons of this gene. There were two deletion hot-spot regions, one is located at the 5' exons 2–20 and the other, located between exons 44–53 [8, 9]. Mutations affecting open-reading frame (ORF), due to a frameshift, may result in an aberrantly spliced mRNA and generate a truncated, nonfunctional dystrophin protein that usually gives rise to the DMD/BMD phenotypes. However, mutations not affecting the ORF may produce a semi-functional dystrophin protein and usually correlate with mild phenotypes [10]. Levels of dystrophin protein in DMD are very low (<3% of normal) or absent, however, BMD patients have 10–40% of the normal amount of dystrophin with an abnormal molecular weight [11, 12]. About 65% of cases are caused by deletion [8, 9, 13], approximately 5% by duplication [9, 14] and the remaining is point mutation of the dystrophin gene [15]. Due to no effective treatment available for DMD/BMD, presently an accurate genetic diagnosis may offer prenatal diagnosis for the familial DMD/BMD although about one-third of mutations are de novo [16]. Although the frequency of complex rearrangement was reported as high as 4% [17], usually complex rearrangement is considered as a rare condition [18–20] that includes double deletion [17] and non-contiguous duplications [21–23].
In this study, we have analyzed a subset of DMD/BMD patients from Northern China.
Methods
Subjects
A total number of 179 unrelated patients in northern China were studied. There were 160 affected male probands and 19 females who had an affected son clinically diagnosed as DMD/BMD. This study was approved by the Institutional Ethic Committee and informed consent was obtained from all participants.
Multiplex ligation-dependent probe amplification (MLPA)
MLPA is a reliable technology for quantitative analysis of the copy number in clinical diagnosis of genetic diseases [24]. It has the capability of detecting either large or small genomic deletions or duplications [20, 25]. An MLPA kit with probes of P034 and P035 for detecting deletion and/or duplication of the DMD gene was purchased commercially from MRC Holland (Amsterdam, Netherlands). Procedures were following the manufacturer's instruction. Ligation and amplification were carried out with an ABI 9800 Thermal Cycler. The PCR conditions were 35 cycles at 95°C for 30s, 60°C for 30s, and 72°C for 60s; followed by a final incubation at 72°C for 20 min.
The PCR products were separated by capillary electrophoresis in an ABI 310 Genetic Analyzer with Genescan 3.7 (Applied Biosystems, Foster City, California) software. The raw data was analyzed by Genescan Analysis 3.7 software. The peaks obtained after the analysis of the row data (Fig. 1A) could be distinguished and assigned to specific exons on the basis of their different lengths representing the variability of their stuffer sequences. Peak area of raw data can be also exported into a Microsoft excel spreadsheet program (Fig. 1B), which normalizes each peak to that of a known normal control (Fig. 1, 1). Peaks derived from DMD/BMD patients (Fig. 1, 2–5) that vary more than 20% from the normal should be flagged for review. If a deletion of a single exon was observed, conventional PCR and DNA sequencing analysis of the specific region embedding the deleted exon was performed to verify the deletion [20, 21].
MLPA results were selected from a normal control (1), a multi-exon deletion (2), a multi-exon duplication (3), a single-exon duplication (4), and a single-exon deletion (5). Raw data are shown on the left panels (A). Each peak represents one exon of the dystrophin gene. Arrow(s) indicate missing signals representing deleted exon(s). The corresponding relative peak area (RPA) ratios, after statistical analysis, are shown on the right panels (B), in which each bar represents the RPA ratio compared to the mean of two male control samples.
Results
Detection of DMD gene deletion with MLPA
Among 160 male probands, 106 were found to have deletions, accounting for 66.25% of male patients analyzed. Among 19 females who had an affected boy but without previous genetic confirmation, 14 with exonic deletion were detected, which brought the deletion rate to 67.04% (120/179). Duplication mutation was identified from 10 (16.95%) of the remaining 59 people (54 male patients and 5 female carriers) who had no deletion identified. Totally, 130 of 179 (72.63%) were found to have mutations (Table 1).
Detection of single-exon deletion may result from 1-bp deletion
When a single-exon deletion was observed, DNA sequencing would be applied for verification. Among 32 DNA samples with single-exon deletion, DNA sequencing observed a 1-bp deletion (NM_004006.1, c.8027delG) in one case that had a deletion of exon 53 detected by initial MLPA. This 1-bp deletion (Fig. 2) was located precisely at the ligation site and destroyed the ligation that resulted in failure of amplification of the probes in MLPA.
Deletion of small fragment is predominant in DMD gene
Analyses of 106 males and 14 females (Table 1) showed that 26.67% (29 males and 3 females) of deletions involved a single-exon; 14.17% (15 males and 2 females) exhibited 3-exon deletion; 10.83% (12 males and 1 female) had 5- or 6-exon deletion, 8.33% (10 males) had 2-exon deletion, 7.50% (5 males and 4 females) showed 4-exon deletion, 5.0% (6 males) had 10-exon deletion, 4.17% (5 males) had 8-exon deletion, and 1.67% (2 males) had 7-exon deletion. Our results showed that deletion of six exons and smaller accounts for 77.5% (93/120). To combine all deletions smaller than 10 exons, this group deletion may account for 88.33% (106/120) of patients and carriers.
Duplication detected in this study involved a large fragment
Ten patients with duplication were identified. Most of the duplications involved large fragments, among which six spun 11 to 41 exons. Eight duplications involved previously identified 3' region. A large 21-exon duplication was beyond the 3' hot-spot region that has reached exon 74. Only three duplications in this study were found to involve small fragment.
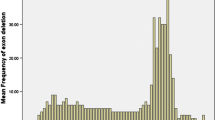
Exons 48–52 were the most common region deleted
Analysis of the single-exon deletion among the 32 (29 males, three females) patients showed that both exons 51 and 52 were the most common exons deleted, accounting for 18.75% (6/32) respectively, followed by exon 45 (15.63%). Single-exon deletion was not found in exons 1–6, 8–16, 20–34, 36–42, 46–49, and 55–79. Among the 88 cases with multiple exons (multi-exon), deletion of six exons in 45–50 were the most common (7.95%), followed by deletion of three exons in 48–50 (6.82%), and then two exons in 48–49 and eight exons in 45–52. When the frequency of deletion for individual exon of dystrophin gene was considered, the most common deleted was exon 50, followed by exon 49 (6.54%), then exon 48 (6.01%), and exons 51 (5.10%). Deletion of exons from 45 to 54 accounts for 50.16%. No deletion was found in either 5' end exons 1–2 or 3' end exons 56–79 (Fig. 3).
Out-of-frame mutation is predominant in DMD/BMD
It has been considered that clinical severity is correlated to in frame or out-of-frame deletion of the dystrophine gene [26]. An interesting result we noticed from this study was that for smaller deletions, they usually resulted in out-of-frame mutations. Among 106 small deletion that involved ten or less exons, there were only 11 cases that carried in-frame but 89.62% had out-of-frame. However, among 14 larger deletions, there were 11 (78.57%) cases had in-frame mutations. This data suggested that out-of-frame mutations in smaller deletion cases account for the majority of DMD mutations, which likely resulted in a severe phenotype. Among five larger duplications, there was only one that was an in-frame mutation while the others were out-of-frame mutations, and there was no in-frame mutation in two small duplications. Overall, 81.53% of duplications were out-of-frame. Correlating each DNA sample with its clinical phenotypes would be idea for drawing a clearer picture. A regret of this study, unfortunately, was that the clinical information had been incomplete which made us unable to perform an analysis on genotype-phenotype correlation. We encourage to perform a genotype-phenotype study with a larger cohort in order to draw a better clue.
Discussion
Several studies have shown that MLPA were useful in detecting mutation quantitatively, not only for deletions but also duplications and female carriers in the DMD gene [20, 25, 27–29]. We applied MLPA in a subset of DMD/BMD patients in northern China. Our results indicated that 66.25% (106/160) of our patients carry deletion and 6.25% (10/160) patients had a duplication, which brought the sensitivity of detecting DNA rearrangement up to 72.5%. In addition, MLPA may present higher sensitivity. In this study, three patients who had negative results in their previous DNA testing by multiplex PCR (data not shown) were identified. Furthermore, an advantage of employing MLPA in clinical genetic testing is that it is capable of identifying female carriers, as we have demonstrated among 14 out of 19 females who came to our prenatal clinics. Certainly, MLPA should be considered as the initial test for the clinically suspected DMD/BMD patients as well as for women who have a DMD/BMD family history, as suggested previously [29], to provide both confirmation of genetic defects on DMD gene and better genetic counseling.
Exons 2–20 and exons 44–53 had been previously reported as the two-hotspot regions [8, 9]. In this study, we have analyzed the distribution of individual exons regarding their frequency of deletion or duplication (Fig. 4). Our data indicates that the deletion occurs at exons 45 to 55 accounting for 47.67%, followed by exons 21–44 (26.67%) and exons 1–20 (23.66%). If both deletions and duplications are counted together, the most frequent mutant region occurs within exons 45–55 (41.06%) in the DMD gene, followed by exons 21–44 (30.43%), exons 1–20 (25.32%) and exons 56–79 (3.19%). We found the most common region in our study is the exons 45–55, and the least common region is the exons 56–79. These data are in agreement with other global populations [26]. We have noticed that the frequency of duplication is more than 20% and it appears there is no model peak of deletion in Taiwan (Fig. 4) [21, 22, 27, 30–32]. As well, the deletion rate in Hong Kong [31] showed difference from the northern Chinese. We hypothesize that the frequency of deletion is different in variant regions of China, and we propose that further investigation with molecular screening of deletion (and duplication) should be pursued among newborns in China. The pattern of duplication did not show significant difference among the global variant populations (Fig. 4).
Comparison of exon deletion (A) and duplication (B) among global populations. All these data showed that exons 45–55 are the most frequent deletion region of DMD gene. Deletion and duplication were not common in region of exons 56–79, although the data may derive from variant methodology (MLPA for Hong Kong, Taiwan, Denmark, Australia, and the current study; quantitative multiplex PCR for Canada; and multiplex PCR and Southern blotting for Japan and USA).
The structure of introns of dystrophin gene determines the breakpoint that is responsible for the deletion [33]. There were many repeated sequences in introns of dystrophin gene which resulted in many breakpoints. For example, in intron 44 there were 13 breakpoints resulted in the hot-spot deletion region [34]. The repeated sequences included direct repeated sequence, inverted repeat sequence and homology sequence. It was reported that 22 repeated sequences had been found in intron 51 and intron 52, which was a possible reason for the high frequency of single-exon deletion at exon 51 [35]. We believe this might be the same case for the most common deletion at exon 50.
Recently, White et al [25] reported that as high as 87% of duplications can be detected among three subsets of selected DMD/BMD cases, among which no deletion or point mutation had been found. In this study, duplication was only detected in 18.51% of the cases where no deletion could be detected. This lower rate possibly resulted from either our small size of samples or a high rate of point mutation, which should be further investigated. To combine the rates of duplication with deletion, we estimate that a point mutation might account for 20–28% of our cases. In addition, the duplications in our cases mainly involved a larger fragment; however, among 118 cases of the reported duplications [25], there were only 12 cases where the duplication was more than 20 exons.
In summary, we have applied MLPA to detect DNA rearrangements of deletions and duplications among DMD/BMD male patients and female potential carriers. We have determined that deletions and duplications were 72.5% of our samples. We observed that the majority of deletions was small in size that the number of deleted exons was less than 10 but duplications usually involved larger sizes. Finally, we determined that the most common region of DMD gene rearrangement is between exons 45 and 55 and the 5' hotspot region was not a hotspot region in our study.
Conclusion
MLPA is an effective and cost effective technique to detect a large number of samples. In our study, we found the frequency of deletion/duplication of DMD in the northern China is similar to other population. We recommend that MLPA should be applied as the first line for clinical molecular testing for DMD/BMD.
Abbreviations
- DMD:
-
Duchenne muscular dystrophy
- BMD:
-
Becker muscular dystrophy
- MLPA:
-
multiplex ligation-dependent probes amplification
- PCR:
-
ploymerase chain reaction.
References
Davies KE, Pearson PL, Harper PS, Murray JM, O'Brien T, Sarfarazi M, Williamson R: Linkage analysis of two cloned DNA sequences flanking the Duchenne muscular dystrophy locus on the short arm of the human X chromosome. Nucl Acids Res. 1983, 11 (8): 2303-2312. 10.1093/nar/11.8.2303.
Kingston HM, Thomas NS, Pearson PL, Sarfarazi M, Harper PS: Genetic linkage between Becker muscular dystrophy and a polymorphic DNA sequence on the short arm of the X chromosome. J Med Genet. 1983, 20 (4): 255-258.
Emery AE: Population frequencies of inherited neuromuscular diseases: a world survey. Neuromuscul Disord. 1991, 1 (1): 19-29. 10.1016/0960-8966(91)90039-U.
Bushby KM, Thambyayah M, Gardner-Medwin D: Prevalence and incidence of Becker muscular dystrophy. Lancet. 1991, 337 (8748): 1022-1024. 10.1016/0140-6736(91)92671-N.
Hoffman EP, Brown RH, Kunkel LM: Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987, 51 (6): 919-928. 10.1016/0092-8674(87)90579-4.
Den Dunnen JT, Grootscholten PM, Dauwerse JG, Walker AP, Monaco AP, Butler R, Anand R, Coffey AJ, Bentley DR, Steensma HY, van Ommen G: Reconstruction of the 2.4 Mb human DMD-gene by homologous YAC recombination. Hum Mol Genet. 1992, 1 (1): 19-28. 10.1093/hmg/1.1.19.
Mandel JL: Dystrophin, the gene and its product. Nature. 1989, 339: 584-586. 10.1038/339584a0.
Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM: Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987, 50 (3): 509-517. 10.1016/0092-8674(87)90504-6.
Den Dunnen JT, Grootscholten PM, Bakker E, Blonden LA, Ginjaar HB, Wapenaar MC, van Paassen HM, van Broeckhoven C, Pearson PL, van Ommen GJ: Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am J Hum Genet. 1989, 45 (6): 835-847.
Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM: An explantion for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988, 2 (1): 90-95. 10.1016/0888-7543(88)90113-9.
Hoffman EP, Fischbeck KH, Brown RH, Johnson M, Medori R, Loike JD, Harris JB, Waterston R, Brooke M, Specht L, Kupsky W, Chamberlain J, Caskey CT, Shapiro F, Kunkel LM: Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne's or Becker's muscular dystrophy. N Engl J Med. 1988, 318 (21): 1363-1368.
Nicholson LV, Johnson MA, Gardner-Medwin D, Bhattacharya S, Harris JB: Heterogeneity of dystrophin expression in patients with Duchenne and Becker muscular dystrophy. Acta Neuropathol (Berl). 1990, 80 (3): 239-250. 10.1007/BF00294640.
Forrest SM, Cross GS, Flint T, Speer A, Robson KJ, Davies KE: Further studies of gene deletions that cause Duchenne and Becker muscular dystrophies. Genomics. 1988, 2 (2): 109-114. 10.1016/0888-7543(88)90091-2.
Hu XY, Ray PN, Murphy EG, Thompson MVV, Worton RG: Duplicational mutation at the Duchenne muscular dystrophy locus: its frequency, distribution, origin, and phenotype-genotype correlation. Am J Hum Genet. 1990, 46: 682-695.
Roberts RG, Bobrow M, Bentley DR: Point mutations in the dystrophin gene. Proc Natl Acad Sci USA. 1992, 89 (6): 2331-2335. 10.1073/pnas.89.6.2331.
Mukherjee M, Chaturvedi LS, Srivastava S, Mittal RD, Mittal B: De novo mutations in sporadic deletional Duchenne muscular dystrophy (DMD) cases. Exp Mol Med. 2003, 35 (2): 113-117.
Bartlett RJ, Walker AP, Laing NG, Koh J, Secore SL, Speer MC, Pericak-Vance MA, Hung WY, Yamaoka LH, Siddique T, Kandt R, Roses AD: Inherited deletion at the Duchenne dystrophy locus in normal males. Lancet. 1989, 1: 496-497. 10.1016/S0140-6736(89)91394-9.
Laing NG, Layton MG, Johnsen RD, Chandler DC, Mears ME, Goldblatt J, Kakulas BA: Two distinct mutations in a single dystrophin gene: chance occurrence or premutation?. Am J Med Genet. 1992, 42 (5): 688-692. 10.1002/ajmg.1320420512.
Hoop RC, Russo LS, Riconda DL, Schwartz LS, Hoffman EP: Restoration of half of the normal dystrophin sequence in a double-deletion Duchenne muscular dystrophy family. Am J Med Genet. 1994, 49 (3): 323-327. 10.1002/ajmg.1320490316.
Janssen B, Hartmann C, Scholz V, Jauch A, Zschocke J: MLPA analysis for the detection of deletion, duplication and complex arrangements in the dystrophin gene: potential and pitfalls. Neurogenetics. 2005, 6 (1): 29-35. 10.1007/s10048-004-0204-1.
Lalic T, Vossen RH, Coffa J, Schouten JP, Guc-Scekic M, Radivojevic D, Djurisic M, Breuning MH, White SJ, den Dunnen JT: Deletion and duplication screening in the DMD gene using MLPA. Eur J Hum Genet. 2005, 13 (11): 1231-1234. 10.1038/sj.ejhg.5201465.
Hsiao-Lin Hwa, Yih-Yuan Chang, Chung-Hsiung Chen, Yen-Shi Kao, Yuh-Jyh Jong, Mei-Chyn Chao, Tsang-Ming Ko: Multiplex Ligation-dependent Probe Amplification Identification of Deletions and Duplications of the Duchenne Muscular Dystrophy Gene in Taiwanese Subjects. J Formos Med Assoc. 2007, 106 (5): 339-46.
El-Harouni AA, Amr KS, Effat LK, Eassawi ML, Ismail S, Gad YZ, El-Awady MK: The milder phenotype of the dystrophin gene double deletions. Acta Neurol Scand. 2003, 107 (6): 400-404. 10.1034/j.1600-0404.2003.00072.x.
Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G: Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucl Acids Res. 2002, 30 (12): e57-10.1093/nar/gnf056.
White SJ, Aartsma-Rus A, Flanigan KM, Weiss RB, Kneppers AL, Lalic T, Janson AA, Ginjaar HB, Breuning MH, den Dunnen JT: Duplications in the DMD gene. Hum Mutat. 2006, 27 (9): 938-945. 10.1002/humu.20367.
Leiden Muscular Dystrophy pages. http://www.dmd.nl
Schwartz M, Duno M: Improved molecular diagnosis of dystrophin gene mutations using the multiplex ligation dependent probe amplification method. Genet Test. 2004, 8 (4): 361-367. 10.1089/gte.2004.8.361.
Carsana A, Frisso G, Tremolaterra MR, Lanzillo R, Vitale DF, Santoro L, Salvatore F: Analysis of Dystrophin gene deletions indicates that the Hinge β III region of the protein correlates with disease severity. Ann Hum Genet. 2005, 69 (pt): 253-259. 10.1046/J.1469-1809.2005.00160.x.
Gatta V, Scarciolla O, Gaspari AR, Palka C, De Angelis MV, Di Muzio A, Guanciali-Franchi P, Calabrese G, Uncini A, Stuppia L: Identification of deletions and duplications of the DMD gene in affected males and carrier females by multiple ligation probe amplification (MLPA). Hum Genet. 2005, 117 (1): 92-98. 10.1007/s00439-005-1270-7.
Stockley TL, Akber S, Bulgin N, Ray PN: Strategy for comprehensive molecular testing for Duchenne and Becker Muscular dystrophies. Genet Test. 2006, 10 (4): 229-243. 10.1089/gte.2006.10.229.
Ivan Lo, Kent Lai, Tony Tong, Stephen Lam: A different spectrum of DMD gene mutations in local Chinese patients with Duchenne/Becker muscular dystrophy. Chin Med J. 2006, 119 (13): 1079-1087.
Thomas Prior, Scott Bridgeman: Experience and Strategy for the Molecular Testing of Duchenne Muscular Dystrophy. J Mol Diag. 2005, 7 (3): 317-326.
Pozzoli U, Sironi M, Cagliani R, Comi GP, Bardoni A, Bresolin N: Comparative analysis of teh human dystrophin and utrophin gene structures. Genetics. 2002, 160 (2): 793-798.
Blonden LA, Grootscholten PM, Den Dunnen JT, Bakker E, Abbs S, Bobrow M, Boehm C, Van Broeckhoven C, Baumbach L, Chamberlain J, Caskey CT, Denton M, Felicetti L, Galluzi G, Fischbeck KH, Francke U, Darras B, Gilgenkrantz H, k aplan JC, Herrmann FH, Junien C, Boileau C, Liechti-Gallati S, Lindlöf M, Matsumoto T, Niikawa N, Mûller CR, Poncin J, Malcolm S, Robertson E, Romeo G, Covone AE, Scheffer H, Schröder E, Schwartz M, Verellen C, Walker A, Worton R, Gillard E, Van Ommen GJB: 242 breakpoints in the 200-kb deletion prone p20 region of the DMD gene are widely spread. Genomics. 1991, 10 (3): 631-639. 10.1016/0888-7543(91)90445-K.
Sheng WL, Chai JH, Liu LZ: A study of the relationship between multiple repeats sequences in introns and the deletion of exons of DMD gene. Chin J Neur. 1997, 30: 214-217.
Acknowledgements
This project was partially funded by the Chinese Ministry of Education (985-2-035-39), the National Nature Science Fund of China (NSFC2004BA720A03 and 30671157), "973" Project (2007CB511902) and the New York State Office of Mental Retardation and Developmental Disabilities (NYS OMRDD). We would like to thank all participants in this study.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
XZW conducted experiments, analyzed and interpreted results, and drafted the manuscript. ZW assisted in data analysis and manuscript drafting. SZH participated in DNA sample collection. MY assisted in operating fragment analysis with sequencer ABI310. TJC directed raw data analysis. NZ conceived of the project, designed, supervised, and coordinated the entire studies, and is responsible for the final edition of the manuscript and approval of the submission. All authors read and approved the final manuscript.
Xiaozhu Wang, Zheng Wang contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Wang, X., Wang, Z., Yan, M. et al. Similarity of DMD gene deletion and duplication in the Chinese patients compared to global populations. Behav Brain Funct 4, 20 (2008). https://doi.org/10.1186/1744-9081-4-20
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1744-9081-4-20