Abstract
Background
There are only limited data on whether HIV infection occurs within the liver; therefore, we explored early and late stages of the HIV life cycle in two hepatocyte cell lines – Huh7.5 and Huh7.5JFH1 – as well as in primary human hepatocytes.
Results
Integrated HIV DNA was detected in Huh7.5 and Huh7.5JFH1 cells, as well as in primary hepatocytes, and was inhibited by the integrase inhibitor raltegravir in a dose-dependent manner. HIV p24 protein was also detected in cell culture supernatants at days 1, 3, 5, and 7 post-infection and was inhibited by AZT, although levels were modest compared to those in a lymphocyte cell line. Culture supernatants from HIV-infected hepatocytes were capable of infecting a non-hepatic HIV indicator cell line.
Conclusions
These results indicating low-level HIV replication in hepatoctyes in vitro complement evidence suggesting that HIV has deleterious effects on the liver in vivo.
Similar content being viewed by others
Background
HIV infection is associated with a number of hepatic and biliary tract disorders, hepatomegaly, and hepatic steatosis[1–5]. Moreover, liver enzyme elevations are frequent in persons with HIV infection even in the absence of viral hepatitis[6, 7]. Additionally, we have recently reported that HIV RNA levels are positively associated with FIB-4 score – a non-invasive serum index of hepatic fibrosis – in HIV mono-infected persons even after controlling for other causes of liver disease, thus supporting a potential association between HIV infection and hepatic fibrosis in vivo[8]. In vivo data further demonstrate the presence of HIV RNA, proviral DNA, and viral proteins in several hepatic cell types, including hepatocytes (reviewed in[9]). Thus, the effects of HIV itself on the liver deserve careful consideration.
Hepatitis C virus (HCV) – a major cause of chronic liver disease – is a significant public health threat worldwide with ~130 million infected persons[10]. Due to their shared routes of transmission, HIV/HCV co-infection is frequent, and liver disease is a major cause of morbidity and mortality in several HIV-positive cohorts[11–13]. It is well established that HIV co-infection is associated with accelerated liver disease and decreased HCV treatment response rates (reviewed in[14]). Until recently, in vitro systems to study the complete HCV life cycle were unavailable. Fortunately, a novel cell culture system – hereafter referred to as Huh7.5JFH1 – capable of producing infectious HCV particles now permits examination of the complete life cycle in the presence or absence of other pro-viral or antiviral factors[15–18]. Therefore, given the possibility that HIV infection of hepatocytes could have a profound effect on the liver, we investigated early and late stages of the HIV life cycle in the Huh7.5 and Huh7.5JFH1 hepatocyte cell lines, as well as in primary human hepatocytes. Such investigations represent an important first step in improving our understanding of HIV pathogenesis in the liver and characterizing pathways by which HIV interacts with co-morbid conditions such as viral hepatitis and/or alcoholic liver disease.
Methods
Cell lines and reagents
The Huh7.5 cell line – generated by curing an HCV replicon-containing cell line with interferon[19] – was provided by Apath LLC (St. Louis, MO). The Huh7.5JFH1 cell line – which produces infectious HCV genotype 2a virions – was provided by Dr. Guangxiang Luo[20]. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: zidovudine (AZT), TZM-bl cells[21] from Drs. John Kappes and Xiaoyun Wu and Tranzyme Inc., plasmids pNL4-3[22] from Dr. Malcolm Martin and pYK-JRCSF[23] from Drs. Irvin Chen and Yoshio Koyanagi, and raltegravir (catalog #11680) from Merck & Company, Inc. Jurkat and 293 T cells were obtained from ATCC. Primary human hepatocytes were purchased from Diagnostic Hybrids (Athens, OH) or BD Biosciences (San Jose, CA) after isolation from non-transplantable livers perfused with HEPES buffer followed by collagenase treatment. Hepatocytes were dissociated mechanically and filtered to obtain a cell suspension. After non-adherent cells were removed, trypan blue exclusion demonstrated >90% viability.
Virus preparation
HIVNL4-3 (CXCR4-utilizing) and HIVYK-JRCSF (CCR5-utilizing) were prepared by transfection of 1 × 106 293 T cells per well in a 24-well plate with 1 μg of the appropriate full-length infectious HIV plasmid using the FuGene6 transfection reagent (Roche). Transfected cells were incubated at 37°C for an additional 48–72 hours. Virus-containing supernatants were passed through a 0.20 μm filter to remove cellular debris and precipitated in polyethylene glycol at 4°C. Precipitated virus was then centrifuged at 14,000 g for 20 minutes, resuspended in PBS, and frozen at −80°C until use. The level of p24 protein in cell culture supernatants was determined by p24 ELISA (Perkin-Elmer, Boston,MA; lower limit of detection = 4.3 pg/mL) or by titering on TZM-bl cells. For experiments quantifying integrated HIV DNA, viruses were pre-treated with DNase I at 20 U/mL in 10 mM MgCl2 for 1 hour at room temperature to eliminate any cellular DNA carryover from virus production.
HIV infections
5 × 105 cells (Huh7.5, Huh7.5JFH1, or primary hepatocytes) were seeded per well of a 24-well, collagen-coated plate and incubated at 37°C in 5% CO2 with infectious HIV at a multiplicity of infection (MOI) of 0.25-1.0 in a volume of ~500 uL per well unless otherwise noted. After 4 hours, viruses were removed by washing cells five times with buffer, and fresh medium was added. Cells and cell culture supernatants were removed at various timepoints post-infection to measure HIV p24 protein levels by ELISA. For Western Blots, 100,000 cells were harvested at days 1, 3, 5, and 7 post-infection and lysed in 100 uL of buffer; 10 uL was loaded per well. HIV p24 was detected using a 1:1000 dilution of a rabbit monoclonal antibody (Ab) from Epitomics (Burlingame, CA; catalog # 1523–1) as the primary Ab and a 1:5000 dilution of a rabbit polyclonal Ab to mouse IgG from Abcam (Cambridge, MA; catalog # 6728) as the secondary Ab. As an additional loading control, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was detected using a rabbit polyclonal Ab from Santa Cruz Biotechnology (Santa Cruz, CA; catalog #SC-25778) at a 1:1000 dilution.
For preparation of non-infectious HIV with functionally intact envelope glycoproteins, HIVNL4-3 was first prepared by transfection of 293 T cells as described above. After filter sterilization, virus was inactivated by adding 250 uM of aldrithiol-2 (AT-2; Sigma-Aldrich) to the filtered supernatants as described elsewhere[24]. AT-2-treated viruses were then incubated with the TZM indicator cell line to confirm that they were not infectious (data not shown), and HIV-1 p24 ELISA was used to quantify virus.
To evaluate the production of infectious virions, supernatants from HIV-infected hepatocytes or Jurkats were collected at day 3 post-infection and titered using TZM-bl cells – a HeLa-derived indicator cell line which expresses the β-galactosidase and luciferase genes under the control of the HIV promoter[25]. 1 × 105 TZM-bl cells were infected with supernatants for two days, washed, and fixed with 1% (v/v) formaldehyde and 0.2% (v/v) glutaraldehyde in PBS. Following several washes with PBS, TZM-bl cells were tested for β-galactosidase activity with a solution of 4 mM potassium ferrocyanide/ferricyanide, 2 mM MgCl2, and 0.4 mg/mL 5-bromo-4-chloro-3-indolyl-D-galactopyranoside (X-gal). The number of positive cells per well was then counted.
Quantification of integrated HIV DNA
After 4 hours of HIV infection, 5 × 105 cells were washed and incubated for an additional 24 hours. For experiments with antiviral drugs, cells were incubated with 1–1000 uM of the integrase inhibitor raltegravir or 100uM of the reverse transcriptase inhibitor AZT before and during HIV infection. 7,500 cells were then suspended in lysis buffer containing 10 mM Tris HCl pH9, 0.1% Tween 20-NP40 and 400 μg/mL Proteinase K (Invitrogen). Cellular lysates containing ~7,500 cells were used to quantify integrated HIV DNA by nested real time PCR as described elsewhere with minor modifications[26, 27]. Briefly, the first round amplification used Alu sequence specific primers and the HIV-1 long terminal repeat (LTR). This reaction was followed by a second round of amplification with specific primers and a specific labeled probe against the LTR performed in a Light Cycler (Roche). Using the ACH-2 cell line, a line of human T-lymphocytic leukemia that contains a single copy of HIV-1 proviral DNA, the limit of detection was 3 copies of integrated HIV DNA per reaction.
Results
Detection of integrated HIV DNA in hepatocytes
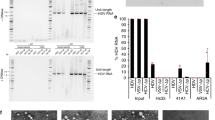
To assess HIV infection of hepatocytes, we first examined whether the Huh7.5 and Huh7.5JFH1 cell lines could support early steps in the HIV life cycle. As shown in Figure1A-B, both hepatocyte cell lines contained integrated HIV DNA after infection with the CXCR4-utilizing lab-adapted strain HIVNL4-3 that had been DNase-treated to remove residual input DNA. Addition of the integrase inhibitor raltegravir resulted in a dose-dependent decrease in integrated HIV DNA levels in both cell lines. Infection with the CCR5-utilizing lab-adapted strain HIVYK-JRCSF resulted in similar levels of integrated HIV DNA compared to HIVNL4-3 and was also inhibited by raltegravir in a dose-dependent manner (Figure1C). The Jurkat lymphocyte cell line was evaluated as a positive control, and demonstrated that HIV integration was approximately 10-fold higher in lymphocytes compared to hepatocytes (Figure1D). In this assay, integrated HIV DNA was regularly tested for in 1) DNase-treated stock virus preparations, 2) washes after removal of residual unbound virus, and 3) mock infected cell lines. In all cases, no integrated HIV DNA was detected, strongly suggesting that these findings were not due to residual input virus.
Detection of integrated HIV DNA in hepatocytes. Huh7.5 (A), Huh7.5JFH1(B), or Jurkat (D) cells were incubated in the presence of 0, 1, 10, 100, or 1000 uM of the integrase inhibitor raltegravir for one hour prior to and during infection with DNase-treated HIVNL4-3. Huh7.5 (C) cells were incubated in the presence of raltegravir for one hour prior to and during infection with DNase-treated HIVYK-JRCSF. After 24 hours, integrated HIV DNA was quantified in cell lysates by nested real-time PCR. Error bars represent the standard deviation between duplicates.
Expression of p24 and production of infectious HIV in hepatocytes
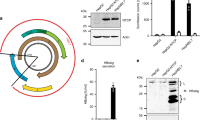
We next examined whether hepatocytes could also support late stages within the HIV life cycle. Because of the high autofluorescence associated with hepatocytes, flow cytometry was not an optimal choice for these experiments; rather, cumulative HIV p24 antigen expression was quantified in cell culture supernatants collected from Huh7.5 cells infected with HIVNL4-3 at day 1 (47.0 ± 4.9 pg/mL), day 3 (115.1 ± 26.8 pg/mL), day 5 (165.7 ± 3.7 pg/mL), and day 7 (203.9 ± 9.8 pg/mL) (Figure2A). Incubation of Huh7.5 cells before and during infection with 100 uM AZT resulted in a significant decrease in p24 expression (61.4 ± 0.6 pg/mL). These p24 levels in the supernatants of HIV-infected Huh7.5 cells were lower than those observed after HIVNL4-3 infection of the Jurkat lymphocyte cell line (data not shown). Additionally, p24 protein expression was detected at days 3, 5, and 7 post-infection via Western Blot in HIV-infected Huh7.5JFH1 cell lysates, although this approach was less sensitive than detection via ELISA (Figure2B).
HIV protein expression in hepatocytes. (A) HIV p24 protein levels were quantified by ELISA in culture supernatants from Huh7.5 cells infected with HIVNL4-3 at days 1, 3, 5, and 7 post-infection. Infection was also performed in the presence of 100 uM AZT for one hour prior to and during infection. Error bars represent the standard deviation between duplicates. (B) Huh7.5JFH1 cells were incubated with HIVNL4-3. At days 1, 3, 5, and 7 post-infection, cells were harvested, lysed, and subjected to Western Blot analysis using a rabbit monoclonal Ab (Epitomics) as the primary Ab and a rabbit polyclonal Ab to mouse IgG from (Abcam) as the secondary Ab. As a loading control, GAPDH was detected using a rabbit polyclonal Ab (Santa Cruz Biotechnology).
To determine if HIV released from hepatocytes could productively infect cells of non-hepatic origin, the TZM-bl indicator cell line was incubated with supernatants collected on day 3 from HIV-infected hepatocytes. Cell counts were 35.5 ± 9.2 or 49.0 ± 4.2 positive cells per well after incubation with supernatants from Huh7.5 or Huh7.5JFH1 cells infected with HIVNL4-3, while background values were 0.0 ± 0.0 positive cells for both cell lines treated with the 5th wash of the HIV input or mock infected cells (Figure3A). Similarly, no positive cells were detected in both cell lines after incubation with 100 ng AT-2-treated HIVNL4-3 or 100 uM AZT, suggesting that there was no non-specific activation of the HIV LTR and that TZM-bl positive cells were the result of bona fide virion production. Incubation of Huh7.5 or Huh7.5JFH1 cells with HIVYK-JRCSF-infected supernatants gave similar results, although overall positivity was lower (data not shown). When TZM-bl cells were incubated with supernatants from HIVNL4-3-infected Jurkat cells, the number of positive cells was 5125 ± 2651 per well (Figure3B) suggesting that production of virions is significantly higher in Jurkats than hepatocyte cell lines.
Infectious HIV production in hepatocytes. TZM-bl indicator cells were incubated with culture supernatants collected at day 3 post-infection from Huh7.5 or Huh7.5JHF1 cells (A) or Jurkat cells (B) infected with HIVNL4-3 or 100 ng AT-treated HIVNL4-3and tested for β-galactosidase activity. The mean number of positive TZM-bl cells per well is shown with the error bars representing the standard deviation between duplicates. β-gal expression was also evaluated in the presence of 100 uM AZT for one hour prior to and during infection.
HIV infection of primary hepatocytes
Because liver biopsies are not routinely performed in HIV mono-infected persons in the absence of viral hepatitis, there are limited data on HIV infection of hepatocytes in vivo. Moreover, mechanical dissociation of explant livers into their component cell types is challenging due to the infrequency of liver transplantation in HIV-positive indivixduals and the frequent cirrhosis that is found in these patients. Therefore, we further examined whether HIV could infect primary hepatocytes in vitro. Similar to the Huh7.5 and Huh7.5JFH1 cell lines, integrated HIV DNA was detected in primary hepatocytes after infection with HIVNL4-3 (Figure4A). Incubation of supernatants from HIV-infected primary hepatocytes with the TZM-bl indicator cell line also showed modest numbers of positive cells (21 ± 14.5; Figure4B). No TZM-bl positive cells were observed using supernatants from primary hepatocytes that were mock infected or exposed to AT-2-treated HIVNL4-3.
HIV infection of primary hepatocytes. (A) Primary human hepatocytes were incubated with DNase-treated HIVNL4-3 for two hours. After 24 hours, integrated HIV DNA was quantified in cell lysates by nested real-time PCR. (B)TZM-bl indicator cells were incubated with culture supernatants collected at day 3 post-infection from primary human hepatocytes infected with HIVNL4-3 or 100 ng AT-2 treated HIVNL4-3 and tested for β-galactosidase activity. The mean number of positive TZM-bl cells per well is shown with the error bars representing the standard deviation between duplicates. β-gal expression was also evaluated in the presence of 100 uM AZT for one hour prior to and during infection.
Conclusions
Several lines of evidence suggest the potential for HIV to interact directly with multiple liver cell populations in vivo (reviewed in[9]). For instance, HIV RNA and proviral DNA have been detected in liver biopsies from persons with HIV infection[28–30]. Subsequent immunohistochemistry and in situ hybridization studies using liver specimens from HIV-infected patients have demonstrated HIV p24 protein and HIV RNA in Kupffer cells, inflammatory mononuclear cells, sinusoidal cells, intrahepatic lymphocytes, and/or hepatocytes[28, 31–35]. Efficient activation of the HIV long terminal repeat also occurs in hepatocytes[36–38]. HIV compartmentalization in the liver has also been reported recently, suggesting that viral adaptation for replication within the liver may occur in vivo[30]. Soluble HIV proteins, such as gp120, also induce caspase expression and hepatic apoptosis[39–41], as well as interleukin 8 (IL-8), a pro-inflammatory chemokine that is an important mediator of hepatic inflammation and is known to antagonize the antiviral effects of IFN[42–45].
Despite multiple studies assessing the effects of HIV proteins on hepatocytes in vitro, contradictory reports of HIV entry receptor expression[39, 40, 46, 47] may have lessened enthusiasm for investigating the ability of HIV to infect and replicate within hepatocytes. Nonetheless, several recent reports provide evidence that HIV can productively infect hepatocytes at low levels. For example, Xiao et al. isolated a CD4-independent strain of HIV from a patient with advanced HIV disease that was capable of infecting hepatocyte cell lines, as well as primary hepatocytes[48]. Fromentin et al. demonstrated that Huh7.5 cells bind to and internalize HIV particles and that HIV infection of CD4+ T cells was enhanced after interacting with virus-loaded hepatocytes compared to cell-free virus[49]. Finally, Iser et al. observed increased HIV reverse transcriptase activity following HIV infection of hepatocyte cell lines[50]. While the precise receptors used by HIV to enter hepatocytes remain elusive, Iser et al. reported that, despite the inability to detect surface expression of CD4, CCR5, or CXCR4 by flow cytometry, infection with R5 or X4 HIV was inhibited by maraviroc or AMD3100, respectively, suggesting CCR5- or CXCR4-dependent entry[50]. We have also detected CCR1, CCR2, and CCR3 co-receptors on Huh7.5 cells (Cardona-Maya and Blackard, unpublished data). Therefore, these receptors may also contribute to HIV infection of hepatocytes, although this requires additional investigation.
The current investigation was considered a proof-of-principle study designed to assess whether HIV was capable of infecting hepatocytes or not. Thus, several cell lines (HCV infected and HCV uninfected), as well as primary hepatocytes, were utilized. This study complements previous investigations and provides several novel findings. First, the Huh7.5 and Huh7.5JFH1 cell lines, as well as primary hepatocytes, supported early steps in the HIV life cycle as demonstrated by detection of integrated HIV DNA. Second, hepatocyte cell lines and primary hepatocytes also support late steps in the HIV life cycle as demonstrated by p24 detection in the supernatants and/or lysates of infected cells and infection of an HIV indicator cell line with hepatocyte-derived HIV. This finding is particularly interesting in light of the recent study by Fromentin et al. demonstrating that circulating CD4+ T cells can be potentially infected with HIV through contact with hepatocytes[49]. Our data further suggest that replication of HIV in hepatocytes is occurring at relatively low levels as integrated DNA levels in hepatocytes were lower than levels in Jurkats. Similarly, supernatants from HIV-infected hepatocytes contained fewer HIV particles that were able to infect an indicator cell line than HIV-infected Jurkats. These findings are in agreement with other recent reports that hepatocytes permit low-level replication of HIV[49, 50].The levels of HIV infection achieved in primary hepatocytes versus hepatocyte cell lines were not equivalent. This may reflect differing levels of permissiveness to HIV in primary hepatocytes versus hepatocyte cell lines, possibly due to different levels of entry receptors or innate antiviral defense molecules. Interestingly, single-cell cloning has been utilized to examine permissiveness of Huh7 cells to HCV infection[51]. The finding of sub-populations of the parental cell line with distinct phenotypic characteristics implies that HIV infectivity could also be impacted, although a comprehensive analysis of HIV infection of primary hepatocytes derived from distinct patients has not been performed to date. Thus, future studies that address the expression of HIV entry receptors, as well as cellular factors that restrict HIV infection should be examined in primary hepatocytes from multiple donors and correlated with overall levels of HIV replication.
We were quite interested to find that HIV could indeed infect an HCV-infected hepatocyte cell line. In a preliminary investigation, we found that HIV infection of Huh7.5JFH1 cells resulted in increased positive- and negative-sense HCV RNA levels, as well as increased HCV protein expression, compared to HIV uninfected cells[52]. However, HIV-induced HCV replication was abolished in the presence of antiretroviral agents. These data complement in vivo reports that HCV RNA levels are elevated in individuals with HIV/HCV co-infection compared to those with HCV mono-infection[11, 53, 54] and suggest that the HCV-infected Huh7.5JFH1 cell line could be developed as an in vitro model to characterize mechanisms by which HIV and HCV interact at the cellular level and contribute to accelerated liver disease.
In conclusion, the present results demonstrate that the Huh7.5 and Huh7.5JFH1 cell lines, as well as primary hepatocytes, can be infected with HIV. These studies provide the necessary systems to further expand our understanding of virus-virus and virus-host interactions that are relevant for enhancing our understanding of how HIV impacts liver disease, as well as increased replication of hepatitis viruses. Moreover, these investigations could ultimately lead to optimization of current therapies for HIV and ameliorate the deleterious effects of HIV on liver disease.
References
Keaveny AP, Karasik M: Hepatobiliary and pancreatic infections in AIDS: Part one. AIDS Patient Care and STDs 1998,12(5):347-357. 10.1089/apc.1998.12.347
Kahn JO, Walker B: Acute human immunodeficiency virus type 1 infection. N Engl J Med 1998,339(1):33-39. 10.1056/NEJM199807023390107
Bonacini M: Hepatobiliary complications in patients with human immunodeficiency virus infection. Am J Med 1992, 92: 404-411. 10.1016/0002-9343(92)90271-C
Ingiliz P, et al.: Liver damage underlying unexplained transaminase elevation in human immunodeficiency virus-1 mono-infected patients on antiretroviral therapy. Hepatology 2009,49(2):436-442. 10.1002/hep.22665
Lefkowitch J: Pathology of AIDS-related liver disease. Dig Dis 1994,12(6):321-330. 10.1159/000171468
Sterling R, et al.: Impact of highly active antiretroviral therapy on the spectrum of liver disease in HCV-HIV coinfection. Clin Gastroenterol Hepatol 2004,2(5):432-439. 10.1016/S1542-3565(04)00129-6
Mata-Marin JA, et al.: Correlation between HIV viral and aminotransferases as liver damage markers in HIV infected naive patients: a concordance cross-sectional study. Virol J 2009, 6: 181. 10.1186/1743-422X-6-181
Blackard JT, et al.: HIV mono-infection is associated with FIB-4 - a noninvasive index of liver fibrosis - in women. Clin Infect Dis 2011,52(5):674-680. 10.1093/cid/ciq199
Blackard JT, Sherman K: HCV/HIV co-infection: time to re-evaluate the role of HIV in the liver? Journal of Viral Hepatitis 2008,15(5):323-330. 10.1111/j.1365-2893.2008.00970.x
Alter M: Epidemiology of hepatitis C virus infection. World J Gastroenterol 2007,13(17):2436-2441.
Tedaldi E, et al.: Influence of coinfection with hepatitis C virus on morbidity and mortality due to human immunodeficiency virus infection in the era of highly active antiretroviral therapy. Clin Infect Dis 2003, 36: 363-367. 10.1086/345953
Bica I, et al.: Increasing mortality due to end-stage liver disease in patients with human immunodeficiency virus infection. Clin Infect Dis 2001, 32: 492-497. 10.1086/318501
Salmon-Ceron D, et al.: Liver disease as a major cause of death among HIV infected patients: role of hepatitis C and B viruses and alcohol. Journal of Hepatolology 2005,42(6):799-805. 10.1016/j.jhep.2005.01.022
Operskalski EA, Kovacs A: HIV/HCV co-infection: pathogenesis, clinical complications, treatment, and new therapeutic technologies. Current HIV/AIDS Reports 2011,8(1):12-22. 10.1007/s11904-010-0071-3
Wakita T, et al.: Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nature Medicine 2005,11(7):791-796. 10.1038/nm1268
Zhong J, et al.: Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci 2005,102(26):9294-9299. 10.1073/pnas.0503596102
Heller T, et al.: An in vitro model of hepatitis C virion production. Proc Natl Acad Sci 2005,102(7):2579-2583. 10.1073/pnas.0409666102
Lindenbach B, et al.: Complete replication of hepatitis C virus in cell culture. Science 2005,309(5734):623-626. 10.1126/science.1114016
Blight KJ, McKeating JA, Rice C: Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 2002,76(24):13001-13014. 10.1128/JVI.76.24.13001-13014.2002
Cai Z, et al.: Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J Virol 2005,79(22):13963-13973. 10.1128/JVI.79.22.13963-13973.2005
Platt EJ, et al.: Effects of CCR5 and CD4 cell surface concentrations on infections by macrophage tropic isolates of human immunodeficiency virus type 1. J Virol 1998,72(4):2855-2864.
Adachi A, et al.: Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 1986,59(2):284-291.
Haltiner M, Kempe T, Tjian R: A novel strategy for constructing clustered point mutations. Nucleic Acids Res 1985,13(3):1015-1025. 10.1093/nar/13.3.1015
Rossio JL, et al.: Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J Virol 1998,72(10):7992-8001.
Kimpton J, Emerman M: Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol 1992,66(4):2232-2239.
Chomont N, et al.: HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nature Medicine 2009,15(8):893-900. 10.1038/nm.1972
Moreno-Fernandez ME, et al.: Human regulatory T cells are targets for human immunodeficiency virus (HIV) infection, and their susceptibility differs depending on the HIV type 1 strain. J Virol 2009,83(24):12925-12933. 10.1128/JVI.01352-09
Cao YZ, et al.: Identification and quantitation of HIV-1 in the liver of patients with AIDS. AIDS 1992,6(1):65-70. 10.1097/00002030-199201000-00008
Donaldson YK, et al.: Redistribution of HIV outside the lymphoid system with onset of AIDS. Lancet 1994,343(8894):383-385.
Blackard JT, et al.: HIV variability in the liver and evidence of possible compartmentalization. AIDS Research and Human Retroviruses 2011,27(10):1117-1126. 10.1089/aid.2010.0329
Housset C, Lamas E, Brechot C: Detection of HIV1 RNA and p24 antigen in HIV-1-infected human liver. Res Virol 1990,141(2):153-159. 10.1016/0923-2516(90)90017-D
Housset C, et al.: Presence of HIV-1 in human parenchymal and non-parenchymal liver cells in vivo. J Hepatol 1993,19(2):252-256. 10.1016/S0168-8278(05)80579-3
Hoda SA, White JE, Gerber M: Immunohistochemical studies of human immunodeficiency virus-1 in liver tissues of patients with AIDS. Mod Pathol 1991,4(5):578-581.
Lang ZW, et al.: A pathological study on liver tissues of patients with HIV infection. Zhonghua Gan Zang Bing Za Zhi 2005,13(12):930-932.
Jiang TJ, et al.: Immunohistochemical evidence for HIV-1 infection in the liver of HIV-infected patients. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 2005,19(2):152-154.
Zhu M, Duan L, Pomerantz R: TAR- and Tat-independent replication of human immunodeficiency virus type 1 in human hepatoma cells. AIDS Research and Human Retroviruses 1996,12(12):1093-1101. 10.1089/aid.1996.12.1093
Pizzella T, Banerjee R: Identification of a human immunodeficiency virus type 1 TAR binding protein in human hepatoblastoma HepG2 cells that trans-activates HIV-1 LTR-directed gene expression. DNA Cell Biology 1994,13(1):67-74. 10.1089/dna.1994.13.67
Hsu ML, et al.: Cytokine regulation of HIV-1 LTR transactivation in human hepatocellular carcinoma cell lines. Cancer Lett 1995,94(1):41-48. 10.1016/0304-3835(95)03820-M
Vlahakis S, et al.: Human immunodeficiency virus-induced apoptosis of human hepatocytes via CXCR4. J Infect Dis 2003, 188: 1455-1460. 10.1086/379738
Munshi N, et al.: Hepatitis C and human immunodeficiency virus envelope proteins cooperatively induce hepatocytic apoptosis via an innocent bystander mechanism. J Infect Dis 2003, 188: 1192-1204. 10.1086/378643
Balasubramanian A, et al.: Molecular mechanism of hepatic injury in coinfection with hepatitis C virus and HIV. Clin Infect Dis 2005,41(Suppl 1):S32-S37.
Balasubramanian A, Ganju R, Groopman J: HCV and HIV envelope proteins collaboratively mediate IL-8 secretion through activation of p38 MAP kinase and SHP2 in hepatocytes. J Biol Chem 2003,278(37):35755-35766. 10.1074/jbc.M302889200
Polyak S, et al.: Elevated levels of interleukin-8 in serum are associated with hepatitis C infection and resistance to interferon therapy. J Virol 2001,75(13):6209-6211. 10.1128/JVI.75.13.6209-6211.2001
Polyak S, et al.: Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J Virol 2001,75(13):6095-6106. 10.1128/JVI.75.13.6095-6106.2001
Khabar K, et al.: The alpha chemokine, interleukin 8, inhibits the antiviral action of interferon a. Journal of Experimental Medicine 1997,186(7):1077-1085. 10.1084/jem.186.7.1077
Cao YZ, et al.: CD4-independent, productive human immunodeficiency virus type 1 infection of hepatoma cell lines in vitro. J Virol 1990,64(6):2553-2559.
Banerjee R, et al.: Inhibition of HIV-1 productive infection in hepatoblastoma HepG2 cells by recombinant tumor necrosis factor-a. AIDS 1992,6(10):1127-1131. 10.1097/00002030-199210000-00010
Xiao P, et al.: Characterization of a CD4-independent clinical HIV-1 that can efficiently infect human hepatocytes through chemokine (C-X-C motif) receptor 4. AIDS 2008,22(14):1749-1757. 10.1097/QAD.0b013e328308937c
Fromentin R, MR T, Tremblay M: Human hepatoma cells transmit surface bound HIV-1 to CD4+ T cells through an ICAM-1/LFA-1-dependent mechanism. Virology 2010, 398: 168-175. 10.1016/j.virol.2009.12.008
Iser DM, et al.: Coinfection of hepatic cell lines with human immunodeficiency virus and hepatitis B virus leads to an increase in intracellular hepatitis B surface antigen. J Virol 2010,84(12):5860-5867. 10.1128/JVI.02594-09
Akazawa D, et al.: CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J Virol 2007,81(10):5036-5045. 10.1128/JVI.01573-06
Kong L, Welge JA, Blackard J: Infectious HIV increases hepatitis C virus (HCV) expression in hepatocytes. In 18th Conference on Retroviruses and Opportunistic Infections. MA: Boston; 2011.
Yokozaki S, et al.: Immunological dynamics in hemophiliac patients infected with hepatitis C and human immunodeficiency virus: influence of antiretroviral therapy. Blood 2000,96(13):4293-4299.
Beld M, et al.: Evidence that both HIV and HIV-induced immunodeficiency enhance HCV replication among HCV seroconverters. Virology 1998, 244: 504-512. 10.1006/viro.1998.9130
Acknowledgements
These data were presented at the XVII International AIDS Conference in Mexico City in August 2008 and the 13thInternational Symposium on Viral Hepatitis and Liver Disease in Washington, DC in March 2009. This work was supported in part by a Dean’s Scholar Award from the University of Cincinnati College of Medicine (to JTB), Merck Inc, (to JTB), NIH RO1 AI068524 (to CC), and a fellowship from COLCIENCIAS (to WCM).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LK participated in the study design and carried out virologic studies. WCM participated in the study design, carried out virologic studies, and helped draft the manuscript. MMF and GM assisted with virologic studies. MTS, KES, and CC participated in study design and coordination and assisted with manuscript editing. JTB designed and coordinated the study and drafted and edited the manuscript. All authors have read and approved the final manuscript.
Ling Kong, Walter Cardona Maya, Maria E Moreno-Fernandez contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kong, L., Maya, W.C., Moreno-Fernandez, M.E. et al. Low-level HIV infection of hepatocytes. Virol J 9, 157 (2012). https://doi.org/10.1186/1743-422X-9-157
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-9-157