Abstract
Background
GB virus C (GBV-C) is an enveloped positive-sense ssRNA virus belonging to the Flaviviridae family. Studies on the genetic variability of the GBV-C reveals the existence of six genotypes: genotype 1 predominates in West Africa, genotype 2 in Europe and America, genotype 3 in Asia, genotype 4 in Southwest Asia, genotype 5 in South Africa and genotype 6 in Indonesia. The aim of this study was to determine the frequency and genotypic distribution of GBV-C in the Colombian population.
Methods
Two groups were analyzed: i) 408 Colombian blood donors infected with HCV (n = 250) and HBV (n = 158) from Bogotá and ii) 99 indigenous people with HBV infection from Leticia, Amazonas. A fragment of 344 bp from the 5' untranslated region (5' UTR) was amplified by nested RT PCR. Viral sequences were genotyped by phylogenetic analysis using reference sequences from each genotype obtained from GenBank (n = 160). Bayesian phylogenetic analyses were conducted using Markov chain Monte Carlo (MCMC) approach to obtain the MCC tree using BEAST v.1.5.3.
Results
Among blood donors, from 158 HBsAg positive samples, eight 5.06% (n = 8) were positive for GBV-C and from 250 anti-HCV positive samples, 3.2%(n = 8) were positive for GBV-C. Also, 7.7% (n = 7) GBV-C positive samples were found among indigenous people from Leticia. A phylogenetic analysis revealed the presence of the following GBV-C genotypes among blood donors: 2a (41.6%), 1 (33.3%), 3 (16.6%) and 2b (8.3%). All genotype 1 sequences were found in co-infection with HBV and 4/5 sequences genotype 2a were found in co-infection with HCV. All sequences from indigenous people from Leticia were classified as genotype 3. The presence of GBV-C infection was not correlated with the sex (p = 0.43), age (p = 0.38) or origin (p = 0.17).
Conclusions
It was found a high frequency of GBV-C genotype 1 and 2 in blood donors. The presence of genotype 3 in indigenous population was previously reported from Santa Marta region in Colombia and in native people from Venezuela and Bolivia. This fact may be correlated to the ancient movements of Asian people to South America a long time ago.
Similar content being viewed by others
Background
GB virus C/Hepatitis G virus (GBV-C/HGV) is an enveloped, positive-sense, single-strand RNA virus belonging to the family Flaviviridae with a genomic size of about 9.3 Kb [1]. It is genomic organization mainly consists of a large open reading frame (ORF) that encodes a single polyprotein precursor in which the structural (E1 and E2) and nonstructural proteins (NS2 to NS5B) are positioned at the N-terminal and C-terminal end, respectively [2]. It was first identified in 1995 in serum from individuals with idiopathic hepatitis [3]. Although it was initially identified as a possible etiological agent of viral hepatitis in humans, and despite its similarity in genome structure with hepatitis C virus (HCV), in contrast to HCV, GBV-C does not appear to be a hepatotrophic virus neither replicates in hepatocytes nor causes acute or chronic hepatitis [4–6].
GBV-C can be transmitted parenterally through blood and derivates transfusion, intravenous drug use, hemodialysis and vertical transmission [7–10]. There is extensive evidence that GBV-C is transmitted by sexual and percutaneous routes and is frequently found in populations at risk for blood-borne or sexually transmitted viruses [5, 11]. Male to male sex has been reported as an effective way of transmission [12] and intrafamilial transmission has been determined based on the sequences analysis [13, 14]. GBV-C has not been associated with any particular disease despite numerous investigations. Alteration in the host's cellular immune response to HIV seems to be responsible for a protective effect of GBV-C but the exact mechanism to it still have to be defined. In contrast, GBV-C infection does not appear to have any effect on chronic liver disease due to HCV or HBV [12].
GBV-C infection is relatively common and has a worldwide distribution. At least 1 to 4% of healthy blood donors have GBV-C RNA [1, 5, 7]. Most people clear the virus and develop antibodies to the E2 envelope glycoprotein. Furthermore, infection is common in the normal population with up to 12.9% prevalence among paid blood donors in the United States of America [15], 11% to 14% in West Africa or South Africa [8], and as high as 37% among HIV-infected individuals [16]. GBV-C incidence amongst of patients with HCV infection varies from 11 to 24% [17–19]. Further, little work has been done on coinfection of GBV-C and HBV but no significant effect of this co-infection was reported [12].
The geographical distribution of GBV-C is related to the coevolution of the viruses with human during the migration along the history, suggesting that GBV-C is an ancient virus [20, 21]. Genotype 1 is found in West Africa [22], genotype 2 (sub-classified as both 2a and 2b) in the United States and Europe [23], genotype 3 in Asia [24–26], genotype 4 in Myanmar and Vietnam [27], genotype 5 in South Africa [28] and genotype 6 in Indonesia [29]. In South America, the genotypes 1, 2a, 2b and 3 have been reported [30–34]. In Colombia, a high prevalence of GBV-C RNA and presence of genotype 3 were found among Colombian native Indians from Wayuu, Kamsa and Inga ethic groups [34].
The aim of this study was determined the frequency of GBV-C RNA and the GBV-C genotypes circulating in Colombia. This is the first study that characterizes the presence of GBV-C among blood donors in coinfection with HCV and HBV in Colombia.
Materials and methods
Study Population
In order to identified the frequency of GBV-C in Colombia, i) 408 samples from Colombian blood donors positive for anti-HCV (n = 250) and positive for HBsAg (n = 158) using third generation ELISA in the blood bank of Cruz Roja Colombiana in Bogota city, Colombia, and ii) 99 samples from indigenous people with HBV infection from Leticia, Amazonas were obtained for this study. These samples were collected between 2003 and 2007 and the presence of HCV RNA and HBV DNA were previously reported by our group [35, 36].
The protocol of this study was approved from Ethical Committees from the Pontificia Universidad Javeriana, Bogotá, Colombia and University of São Paulo Medical School, São Paulo, Brazil. All patients have signed an informed consent before joining the study.
GBV-C RNA Extraction
To avoid false-positive results, rigorous procedures used for nucleic acid amplification diagnostic techniques were followed [37]. HCV-RNA extraction was carried out from 140 ml of serum using QIAamp Viral RNA Kit (QIAGEN, Valencia, CA), following the manufacturer's instructions. The synthesis of the complementary DNA (cDNA) was carried out immediately after RNA extraction.
Synthesis of the complementary DNA (cDNA)
Reverse transcriptase reaction was performed using the Moloney Murine Leukemia Virus Reverse Transcriptase (MMLV-RT) and random primers. The final volume of the reaction was 60 ml in the following concentrations: 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 0.5 mM of each dNTP, 450 ng random primers, 30 U RNAse enzyme inhibitor (RNaseOUT™), and 300 U MMLV-RT. Samples were submitted to the following temperature cycles: 70°C for 10 min, 25°C for 15 min, 37°C for 60 min, and 95°C for 15 min in a thermocycler (Eppendorf Mastercycler 1, Eppendorf, Hamburg, Germany).
Polymerase chain reaction (PCR)
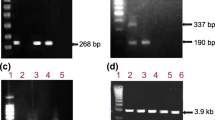
A fragment of 344 bp from 5' untranslated region (5'UTR) was amplified by nested RT PCR [28, 38]. Amplification consisted of 40 cycles for first and second round of PCR, with the following incubation times and temperatures: 94°C 30 s, 50°C 30 s and 72°C 30s for the first round and 94°C 30s, 60°C 30 s and 72°C 30s for the second round.
GBV-C Sequencing
Amplified cDNA was purified using the ChargeSwitch PCR Clean-Up Kit. Sequencing was done in an ABI Prism 3500 Automatic Sequencer (Applied Biosystems, Foster City, CA) using dideoxy nucleoside triphosphates (ddNTPs) containing fluorescent markers (Big Dye1 Terminator v3.1 Cycle Sequencing Ready Reaction Kit-Applied Biosystems).
The consensus sequences were obtained by alignment of both sequenced strands (sense and antisense) using the SEQUENCHER software (Gene Codes Corporation Ann Arbor, Michigan, United States of America).
Phylogenetic Analysis
The sequences obtained in this work were genotyped by phylogenetic reconstructions using reference sequences from each GBV-C genotype obtained from GenBank (n = 160). Sequences were aligned and edited using Clustal X [39] and Se-AL (available at: http://tree.bio.ed.ac.uk/software/seal/) softwares respectively. Bayesian phylogenetic analyses were conducted using the Markov Chain Monte Carlo (MCMC) simulation implemented in BEAST v.1.5.3 [40]. The dataset was analyzed under relaxed uncorrelated lognormal and relaxed uncorrelated exponential molecular clock using the best model of nucleotide substitution (GTR+G+I) chosen by MODELTEST [41] and 20 million generations were sufficient to obtain the convergence of parameters. The molecular clock that best fitted the data was chosen by Bayes factor (BF) comparison. The maximum clade credibility (MCC) tree was obtained from summarizing the 20,000 substitution trees after excluding 10% of burn-in using Tree Annotator v.1.5.3 [40]. Phylogenetic trees were visualized in FigTree v.1.2.2 (available at: http://tree.bio.ed.ac.uk/software/figtree/).
Statistical analyses
Statistical analyses were performed using Minitab Software v. 15. The χ2 test for linear trend (α = 0.05) was used to examine the variations in the presence of GBV-C RNA adjusted for age group, sex and co-infection group (Anti-HCV or HBsAg).
Results
Detection of GBV-C RNA
Among Colombian blood donors, from 158 HBsAg positive samples, 5.06% (n = 8) were positive for GBV-C RNA and from 250 anti-HCV positive samples, 3.2% (n = 8) were positive for GBV-C. Among 99 indigenous people from Leticia (n = 99), 7.7% (n = 7) GBV-C positive samples were found (Table 1).
Phylogenetic Analysis
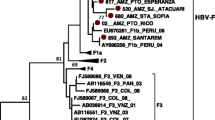
Eighteen out of 23 sequences with good quality were used for phylogenetic analysis. The phylogenetic tree constructed with the GBV-C sequences (n = 160) is shown in Figure 1. This analysis revealed the presence of the following GBV-C genotypes among blood donors: 2a (41.6%), 1 (33.3%), 3 (16.6%) and 2b (8.3%). All genotype 1 sequences were found in co-infection with HBV and 4/5 sequences genotype 2a were found in co-infection with HCV. All sequences from indigenous people from Leticia were classified as genotype 3. The presence of GBV-C infection among blood donors group and indigenous people was not correlated with sex (p = 0.43), age (p = 0.38) or origin of the samples (p = 0.17). The Colombian GBV-C sequences were deposited in the GenBank database under accession numbers JF832366 to JF832383.
The maximum clade credibility (MCC) tree was estimated by Bayesian analysis of 160 sequences with 344 bp of GB virus C strains. The posterior probabilities of the key nodes are depicted above the respective nodes. Samples obtained from Colombian blood donors (n = 12) and Amerindian population (n = 6) were analyzed together with other worldwide strains. Genotype 4 and genotype 5 branches were collapsed.
Discussion
In this study, we determined the frequency and genotypic characteristics of GBV-C virus in Colombia. Among Colombian blood donors, from 158 HBsAg positive samples, eight (5.06%) were positive for GBV-C and from 250 anti-HCV positive samples, eight (3.2%) were positive for GBV-C. This is the first report showing the frequency of GBV-C in HBV and HCV positive blood donors in Colombia.
There are few studies that reported GBV-C frequency in blood donors populations. In Salvador and in Rio de Janeiro, Brazil it was reported 10% of frequency of GBV-C among blood donors [42, 43]. Also, in São Paulo, Brazil it was reported a high prevalence among blood donors with normal and elevated ALT levels: 5.2% (5/95) and 6.5% (5/76), respectively [14]. Furthermore, the prevalence of GBV-C was 9.7% among 545 blood donors in São Paulo [44] and 8.3% in 1.039 healthy individuals [45].
In Iranian volunteer blood donors the prevalence of GBV-C was around 1% [46]. A study of prevalence of GBV-C among northeastern Thai blood donors carrying HBsAg and anti-HCV revealed a higher frequency of GBV-C RNA (10% and 11%, respectively) in the co-infected when compared with the controls [47]. In United States, GBV-C prevalence in blood donors was reported ranging from 0.8% to 12.9% [48, 49]. In Turkey the prevalence of GBV-C was 14% in hemodialysis patients and 5% in blood donors [50]. Also, in Thailand the GBV-C RNA positivity among blood donors was 4.8% [51]. In France, among 306 HCV RNA-positive donors, 19.3% were GBV-C RNA positive [52]. In Egypt, El-Zayadi et al., [53] found 12.2% of GBV-C prevalence among blood donors and GBV-C coinfection in HBV and HCV infected patients were 7.6 and 64.9%, respectively. GBV-C infection is generally more common in groups with risk factors for percutaneous and sexual transmission of infectious agents [54].
The presence of GBV-C infection in our study was not correlated with sex (p = 0.43), age (p = 0.38) or origin (p = 0.17). These results are correlated with the previous study performed among three different Indian groups from Colombia (Wayuu, Inga and Kamsa) where no significant differences were found [34].
A phylogenetic analysis revealed the presence of GBV-C genotypes 2a, 1, 3 and 2b among Colombian blood donors. There are few studies that reported GBV-C genotypes among blood donor populations in the world. In Bolivia, among blood donors, it was found that the major genotype was genotype 3 followed by genotype 2 [32]. In Shanghai, China, genotype 3 was the most prevalent [55]. In Martinique, Césaire et al., [56], reported genotypes 2a, 1 and 2b among blood donors.
Furthermore, all genotype 1 sequences from Colombian blood donors were found in co-infection with HBV and 4/5 sequences from blood donors genotype 2a were found in co-infection with HCV, it was not found a significant association between the presence of HBV or HCV co-infection and GBV-C genotype (p = 0.431). These results are similar to other studies where no significant differences of HBV, HCV and GBV-C infection rates were found [57].
Phylogenetic analysis showed that genotype 3 is the most common in Leticia, Amazon region of Colombia. The finding of an Asian GBV-C genotype in the Americas was first suggested by the analysis of 5'UTR hemophiliac patients from nine locations in Nicaragua [58], Colombian Amerindians [34] and Bolivia [32]. Similarity between Nicaraguan and Asian GBV-C genotype 3 strains indicates that these strains in the region presumably have an Amerindian origin [58].
In Colombia, from 163 native Indians, 6.1% (n = 10) were positive for GBV-C RNA and it was concluded that the incidence of GBV-C infection in native Indians tended to be high compared with the general population [34]. Furthermore, most Colombian native Indians harbored an Asian GBV-C genotype.
In an Amerindian population from Venezuela, a high prevalence of GBV-C genotype 3 was observed, ranging from 5% (9 out of 162) in the West to 25% (14 out of 56) in the south region of the country [33]. Whereas GBV-C genotypes 1, 2 and 3 were presented in Venezuela, genotype 3 (Asian genotype) was found infecting Amerindians and rural population [33].
Together with other studies, our results corroborate the hypothesis that GBV-C is an old virus [21], having been probably introduced in the America continent with the fist men coming thought the Bering Strait [32–34, 58]. It is generally believed that Colombian native Indians migrated from Asia to Colombia approximately 12,000 years ago and were isolated from other people for religious reasons [59]. On the other hand, since HTLV-1 or HTLV-2 may have been brought from Asia to Colombia together with the first human migrants [60], GBV-C/HGV apparently followed the same route [34]. These results are correlated with the evolutionary analysis of GBV-C performed by Suzuki et al., [61], that assumed that this virus diverged 100.000 years ago. However, further analysis needs to be performer to test this hypothesis. Actually, more sequences of GBV-C genotype 3 need to be reported with sampling date, allowing to estimate a specific substitution rate for this genotype that is needed for a more detailed phylogeographic analysis.
Conclusions
In conclusion, this is the first study that reported the GBV-C frequency and the distribution of genotypes among Colombian blood donors. The result presented indicated the circulation of the genotype 3 among Amerindian population in Colombia and blood donors.
References
Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P: The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol 2011, 92: 233-246. 10.1099/vir.0.027490-0
Leary TP, Muerhoff AS, Simons JN, Pilot-Matias TJ, Erker JC, Chalmers ML, Schlauder GG, Dawson GJ, Desai SM, Mushahwar IK: Sequence and genomic organization of GBV-C: a novel member of the flaviviridae associated with human non-A-E hepatitis. J Med Virol 1996, 48: 60-67. 10.1002/(SICI)1096-9071(199601)48:1<60::AID-JMV10>3.0.CO;2-A
Simons JN, Leary TP, Dawson GJ, Pilot-Matias TJ, Muerhoff AS, Schlauder GG, Desai SM, Mushahwar IK: Isolation of novel virus-like sequences associated with human hepatitis. Nat Med 1995, 1: 564-569. 10.1038/nm0695-564
Laskus T, Radkowski M, Wang LF, Vargas H, Rakela J: Lack of evidence for hepatitis G virus replication in the livers of patients coinfected with hepatitis C and G viruses. J Virol 1997, 71: 7804-7806.
Alter HJ: G-pers creepers, where'd you get those papers? A reassessment of the literature on the hepatitis G virus. Transfusion 1997, 37: 569-572. 10.1046/j.1537-2995.1997.37697335149.x
Zhu WF, Yin LM, Li P, Huang J, Zhuang H: Pathogenicity of GB virus C on virus hepatitis and hemodialysis patients. World J Gastroenterol 2003, 9: 1739-1742.
Moaven LD, Hyland CA, Young IF, Bowden DS, McCaw R, Mison L, Locarnini SA: Prevalence of hepatitis G virus in Queensland blood donors. Med J Aust 1996, 165: 369-371.
Casteling A, Song E, Sim J, Blaauw D, Heyns A, Schweizer R, Margolius L, Kuun E, Field S, Schoub B, Vardas E: GB virus C prevalence in blood donors and high risk groups for parenterally transmitted agents from Gauteng, South Africa. J Med Virol 1998, 55: 103-108. 10.1002/(SICI)1096-9071(199806)55:2<103::AID-JMV4>3.0.CO;2-6
Thomas DL, Nakatsuji Y, Shih JW, Alter HJ, Nelson KE, Astemborski JA, Lyles CM, Vlahov D: Persistence and clinical significance of hepatitis G virus infections in injecting drug users. J Infect Dis 1997, 176: 586-592. 10.1086/514078
Feucht HH, Zollner B, Polywka S, Laufs R: Vertical transmission of hepatitis G. Lancet 1996, 347: 615-616.
Bjorkman P, Naucler A, Winqvist N, Mushahwar I, Widell A: A case-control study of transmission routes for GB virus C/hepatitis G virus in Swedish blood donors lacking markers for hepatitis C virus infection. Vox Sang 2001, 81: 148-153. 10.1046/j.1423-0410.2001.00098.x
Berzsenyi MD, Bowden DS, Roberts SK: GB virus C: insights into co-infection. J Clin Virol 2005, 33: 257-266. 10.1016/j.jcv.2005.04.002
Seifried C, Weber M, Bialleck H, Seifried E, Schrezenmeier H, Roth WK: High prevalence of GBV-C/HGV among relatives of GBV-C/HGV-positive blood donors in blood recipients and in patients with aplastic anemia. Transfusion 2004, 44: 268-274. 10.1111/j.1537-2995.2004.00665.x
Pinho JR, Zanotto PM, Ferreira JL, Sumita LM, Carrilho FJ, da Silva LC, Capacci ML, Silva AO, Guz B, Goncales FL Jr, Goncales N, Buck G, Meyers G, Bernardini P: High prevalence of GB virus C in Brazil and molecular evidence for intrafamilial transmission. J Clin Microbiol 1999, 37: 1634-1637.
Dawson GJ, Schlauder GG, Pilot-Matias TJ, Thiele D, Leary TP, Murphy P, Rosenblatt JE, Simons JN, Martinson FE, Gutierrez RA, Lentino JR, Pachucki C, Muerhoff AS, Widell A, Tegtmeier G, Desai S, Mushahwar IK: Prevalence studies of GB virus-C infection using reverse transcriptase-polymerase chain reaction. J Med Virol 1996, 50: 97-103. 10.1002/(SICI)1096-9071(199609)50:1<97::AID-JMV16>3.0.CO;2-V
Tillmann HL, Manns MP: GB virus-C infection in patients infected with the human immunodeficiency virus. Antiviral Res 2001, 52: 83-90. 10.1016/S0166-3542(01)00172-3
Di Bisceglie AM: Hepatitis G virus infection: a work in progress. Ann Intern Med 1996, 125: 772-773.
Tanaka E, Alter HJ, Nakatsuji Y, Shih JW, Kim JP, Matsumoto A, Kobayashi M, Kiyosawa K: Effect of hepatitis G virus infection on chronic hepatitis C. Ann Intern Med 1996, 125: 740-743.
Feucht HH, Zollner B, Polywka S, Knodler B, Schroter M, Nolte H, Laufs R: Prevalence of hepatitis G viremia among healthy subjects, individuals with liver disease, and persons at risk for parenteral transmission. J Clin Microbiol 1997, 35: 767-768.
Naito H, Abe K: Genotyping system of GBV-C/HGV type 1 to type 4 by the polymerase chain reaction using type-specific primers and geographical distribution of viral genotypes. J Virol Methods 2001, 91: 3-9. 10.1016/S0166-0934(00)00207-X
Smith DB, Cuceanu N, Davidson F, Jarvis LM, Mokili JL, Hamid S, Ludlam CA, Simmonds P: Discrimination of hepatitis G virus/GBV-C geographical variants by analysis of the 5' non-coding region. J Gen Virol 1997,78(Pt 7):1533-1542.
Muerhoff AS, Simons JN, Leary TP, Erker JC, Chalmers ML, Pilot-Matias TJ, Dawson GJ, Desai SM, Mushahwar IK: Sequence heterogeneity within the 5'-terminal region of the hepatitis GB virus C genome and evidence for genotypes. J Hepatol 1996, 25: 379-384. 10.1016/S0168-8278(96)80125-5
Muerhoff AS, Smith DB, Leary TP, Erker JC, Desai SM, Mushahwar IK: Identification of GB virus C variants by phylogenetic analysis of 5'-untranslated and coding region sequences. J Virol 1997, 71: 6501-6508.
Okamoto H, Nakao H, Inoue T, Fukuda M, Kishimoto J, Iizuka H, Tsuda F, Miyakawa Y, Mayumi M: The entire nucleotide sequences of two GB virus C/hepatitis G virus isolates of distinct genotypes from Japan. J Gen Virol 1997,78(Pt 4):737-745.
Mukaide M, Mizokami M, Orito E, Ohba K, Nakano T, Ueda R, Hikiji K, Iino S, Shapiro S, Lahat N, Park YM, Kim BS, Oyunsuren T, Rezieg M, Al-Ahdal MN, Lau JY: Three different GB virus C/hepatitis G virus genotypes. Phylogenetic analysis and a genotyping assay based on restriction fragment length polymorphism. FEBS Lett 1997, 407: 51-58. 10.1016/S0014-5793(97)00136-1
Katayama K, Kageyama T, Fukushi S, Hoshino FB, Kurihara C, Ishiyama N, Okamura H, Oya A: Full-length GBV-C/HGV genomes from nine Japanese isolates: characterization by comparative analyses. Arch Virol 1998, 143: 1063-1075. 10.1007/s007050050356
Naito H, Win KM, Abe K: Identification of a novel genotype of hepatitis G virus in Southeast Asia. J Clin Microbiol 1999, 37: 1217-1220.
Tucker TJ, Smuts H, Eickhaus P, Robson SC, Kirsch RE: Molecular characterization of the 5' non-coding region of South African GBV-C/HGV isolates: major deletion and evidence for a fourth genotype. J Med Virol 1999, 59: 52-59. 10.1002/(SICI)1096-9071(199909)59:1<52::AID-JMV9>3.0.CO;2-D
Muerhoff AS, Dawson GJ, Desai SM: A previously unrecognized sixth genotype of GB virus C revealed by analysis of 5'-untranslated region sequences. J Med Virol 2006, 78: 105-111. 10.1002/jmv.20510
Oubina JR, Mathet V, Feld M, Della Latta MP, Ferrario D, Verdun R, Libonatti O, Fernandez J, Carballal G, Sanchez DO, Quarleri JF: Genetic diversity of GBV-C/HGV strains among HIV infected-IVDU and blood donors from Buenos Aires, Argentina. Virus Res 1999, 65: 121-129. 10.1016/S0168-1702(99)00109-4
Ramos Filho R, Carneiro MA, Teles SA, Dias MA, Cardoso DD, Lampe E, Yoshida CF, Martins RM: GB virus C/hepatitis G virus infection in dialysis patients and kidney transplant recipients in Central Brazil. Mem Inst Oswaldo Cruz 2004, 99: 639-643. 10.1590/S0074-02762004000600019
Konomi N, Miyoshi C, La Fuente Zerain C, Li TC, Arakawa Y, Abe K: Epidemiology of hepatitis B, C, E, and G virus infections and molecular analysis of hepatitis G virus isolates in Bolivia. J Clin Microbiol 1999, 37: 3291-3295.
Loureiro CL, Alonso R, Pacheco BA, Uzcategui MG, Villegas L, Leon G, De Saez A, Liprandi F, Lopez JL, Pujol FH: High prevalence of GB virus C/hepatitis G virus genotype 3 among autochthonous Venezuelan populations. J Med Virol 2002, 68: 357-362. 10.1002/jmv.10211
Tanaka Y, Mizokami M, Orito E, Ohba K, Nakano T, Kato T, Kondo Y, Ding X, Ueda R, Sonoda S, Tajima K, Miura T, Hayami M: GB virus C/hepatitis G virus infection among Colombian native Indians. Am J Trop Med Hyg 1998, 59: 462-467.
Mora MV, Romano CM, Gomes-Gouvea MS, Gutierrez MF, Carrilho FJ, Pinho JR: Molecular characterization, distribution, and dynamics of hepatitis C virus genotypes in blood donors in Colombia. J Med Virol 2010, 82: 1889-1898. 10.1002/jmv.21908
Alvarado Mora MV, Romano CM, Gomes-Gouvea MS, Gutierrez MF, Botelho L, Carrilho FJ, Pinho JR: Molecular characterization of the Hepatitis B virus genotypes in Colombia: a Bayesian inference on the genotype F. Infect Genet Evol 2011, 11: 103-108. 10.1016/j.meegid.2010.10.003
Kwok S, Higuchi R: Avoiding false positives with PCR. Nature 1989, 339: 237-238. 10.1038/339237a0
Jarvis LM, Davidson F, Hanley JP, Yap PL, Ludlam CA, Simmonds P: Infection with hepatitis G virus among recipients of plasma products. Lancet 1996, 348: 1352-1355. 10.1016/S0140-6736(96)04041-X
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG: The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997, 25: 4876-4882. 10.1093/nar/25.24.4876
Drummond AJ, Rambaut A: BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 2007, 7: 214. 10.1186/1471-2148-7-214
Posada D, Crandall KA: MODELTEST: testing the model of DNA substitution. Bioinformatics 1998, 14: 817-818. 10.1093/bioinformatics/14.9.817
Lyra AC, Pinho JR, Silva LK, Sousa L, Saraceni CP, Braga EL, Pereira JE, Zarife MA, Reis MG, Lyra LG, Silva LC, Carrilho FJ: HEV, TTV and GBV-C/HGV markers in patients with acute viral hepatitis. Braz J Med Biol Res 2005, 38: 767-775.
Lampe E, de Oliveira JM, Pereira JL, Saback FL, Yoshida CF, Niel C: Hepatitis G virus (GBV-C) infection among Brazilian patients with chronic liver disease and blood donors. Clin Diagn Virol 1998, 9: 1-7. 10.1016/S0928-0197(97)10017-4
Levi JE, Contri DG, Lima LP, Takaoka DT, Garrini RH, Santos W, Fachini R, Wendel S: High prevalence of GB virus C/hepatitis G virus RNA among Brazilian blood donors. Rev Inst Med Trop Sao Paulo 2003, 45: 75-78. 10.1590/S0036-46652003000200004
Ribeiro-dos-Santos G, Nishiya AS, Nascimento CM, Bassit L, Chamone DF, Focaccia R, Eluf-Neto J, Sabino EC: Prevalence of GB virus C (hepatitis G virus) and risk factors for infection in Sao Paulo, Brazil. Eur J Clin Microbiol Infect Dis 2002, 21: 438-443. 10.1007/s10096-002-0752-y
Ramezani A, Gachkar L, Eslamifar A, Khoshbaten M, Jalilvand S, Adibi L, Salimi V, Hamkar R: Detection of hepatitis G virus envelope protein E2 antibody in blood donors. Int J Infect Dis 2008, 12: 57-61. 10.1016/j.ijid.2007.04.010
Barusruk S, Urwijitaroon Y: High prevalence of HGV coinfection with HBV or HCV among northeastern Thai blood donors. Southeast Asian J Trop Med Public Health 2006, 37: 289-293.
Dawson GJ, Schlauder GG, Pilot-Matias TJ, Thiele D, Leary TP, Murphy P, Rosenblatt JE, Simons JN, Martinson FE, Gutierrez RA, Lentina JR, Pachucki C, Muerhoff AS, Widell A, Tegtmeier G, Desai S, Mushahwar IK: Prevalence studies of GB virus-C infection using reverse transcriptase-polymerase chain reaction. J Med Virol 1996, 50: 97-103. 10.1002/(SICI)1096-9071(199609)50:1<97::AID-JMV16>3.0.CO;2-V
Linnen J, Wages J Jr, Zhang-Keck ZY, Fry KE, Krawczynski KZ, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih WK, Young L, Piatak M Jr, Hoover C, Fernandez J, Chen S, Chao-Zou JC, Morris T, Hyams K, Ismay S, Lifson J, Hess G, Foung S, Thomas H, Bradley D, Margolis H, Kim J: Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science 1996, 271: 505-508. 10.1126/science.271.5248.505
Hanci SY, Cevahir N, Kaleli I, Hanci V: Investigation of hepatitis G virus prevalence in hemodialysis patients and blood donors in Denizli, Turkey. Mikrobiyol Bul 2008, 42: 617-625.
Wiwanitkit V: Hepatitis G, hepatitis SEN, hepatitis TT and hepatitis TT-like viruses: New emerging hepatitis viruses in pediatric patients. J Ped Infec Dis 2006, 1: 83-88.
Bouchardeau F, Laperche S, Pillonel J, Elghouzzi MH, Maisonneuve P, Tirtaine C, Boiret E, Razer A, Girault A, Beaulieu MJ, Courouce AM: GB virus type C/HGV markers in HCV RNA-positive French blood donors: correlation with HCV genotypes and risk factors. Transfusion 2000, 40: 875-878. 10.1046/j.1537-2995.2000.40070875.x
El-Zayadi AR, Abe K, Selim O, Naito H, Hess G, Ahdy A: Prevalence of GBV-C/hepatitis G virus viraemia among blood donors, health care personnel, chronic non-B non-C hepatitis, chronic hepatitis C and hemodialysis patients in Egypt. J Virol Methods 1999, 80: 53-58. 10.1016/S0166-0934(99)00036-1
Giret MT, Miraglia JL, Sucupira MC, Nishiya A, Levi JE, Diaz RS, Sabino EC, Kallas EG: Prevalence, Incidence Density, and Genotype Distribution of GB Virus C Infection in a Cohort of Recently HIV-1-Infected Subjects in Sao Paulo, Brazil. PLoS One 2011, 6: e18407. 10.1371/journal.pone.0018407
Ding X, Mizokami M, Kang LY, Cao K, Orito E, Tanaka Y, Ueda R, Sasaki M: Prevalence of TT virus and GBV-C infections among patients with liver diseases and the general population in Shanghai, China. Virus Genes 1999, 19: 51-58. 10.1023/A:1008188623062
Cesaire R, Martial J, Maier H, Kerob-Bauchet B, Bera O, Duchaud E, Brebion A, Pierre-Louis S: Infection with GB virus C/hepatitis G virus among blood donors and hemophiliacs in Martinique, a Caribbean island. J Med Virol 1999, 59: 160-163. 10.1002/(SICI)1096-9071(199910)59:2<160::AID-JMV6>3.0.CO;2-Y
Ling BH, Zhuang H, Cui YH, An WF, Li ZJ, Wang SP, Zhu WF: A cross-sectional study on HGV infection in a rural population. World J Gastroenterol 1998, 4: 489-492.
Gonzalez-Perez MA, Norder H, Bergstrom A, Lopez E, Visona KA, Magnius LO: High prevalence of GB virus C strains genetically related to strains with Asian origin in Nicaraguan hemophiliacs. J Med Virol 1997, 52: 149-155. 10.1002/(SICI)1096-9071(199706)52:2<149::AID-JMV5>3.0.CO;2-3
Cavalli-Sforza LL: Genes, people and languages. Sci Am 1991, 265: 72-78.
Zaninovic V, Sanzon F, Lopez F, Velandia G, Blank A, Blank M, Fujiyama C, Yashiki S, Matsumoto D, Katahira Y: Geographic independence of HTLV-I and HTLV-II foci in the Andes highland, the Atlantic coast, and the Orinoco of Colombia. AIDS Res Hum Retroviruses 1994, 10: 97-101. 10.1089/aid.1994.10.97
Suzuki Y, Katayama K, Fukushi S, Kageyama T, Oya A, Okamura H, Tanaka Y, Mizokami M, Gojobori T: Slow evolutionary rate of GB virus C/hepatitis G virus. J Mol Evol 1999, 48: 383-389. 10.1007/PL00006482
Acknowledgements
This work has been supported by CNPq, Fundação de Amparo à Pesquisa do Estado de São Paulo-FAPESP 2007/53457-7 and 2008/50461-6 and Pontificia Universidad Javeriana, Bogotá, Colombia. We thank Banco Nacional de Sangre de la Cruz Roja Colombiana for their kind provision of blood donor samples for this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MVAM conducted the phylogenetic and evolutionary analysis, drafted the manuscript and participated in its design and coordination. LB, AN and MSGG participated in the PCR amplification and sequencing process. RAN conducts the statistical analysis. FJC and MFG participated in the design of the study. JRRP participated in the elaboration of the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Alvarado-Mora, M.V., Botelho, L., Nishiya, A. et al. Frequency and genotypic distribution of GB virus C (GBV-C) among Colombian population with Hepatitis B (HBV) or Hepatitis C (HCV) infection. Virol J 8, 345 (2011). https://doi.org/10.1186/1743-422X-8-345
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-8-345