Abstract
Background
Avian Leukosis virus (ALV) of subgroup J (ALV-J) belong to retroviruses, which could induce tumors in domestic and wild birds. Myelocytomatosis was the most common neoplasma observed in infected flocks; however, few cases of hemangioma caused by ALV-J were reported in recent year.
Results
An ALV-J strain SCDY1 associated with hemangioma was isolated and its proviral genomic sequences were determined. The full proviral sequence of SCDY1 was 7489 nt long. Homology analysis of the env, pol and gag gene between SCDY1 and other strains in GenBank were 90.3-94.2%, 96.6-97.6%, and 94.3-96.5% at nucleotide level, respectively; while 85.1-90.7%, 97.4-98.7%, and 96.2-98.4% at amino acid level, respectively. Alignment analysis of the genomic sequence of ALV-J strains by using HPRS-103 as reference showed that a special 11 bp deletion was observed in U3 region of 3'UTR of SCDY1 and another ALV-J strain NHH isolated from case of hemangioma, and the non-functional TM and E element were absent in the genome of SCDY1, but the transcriptional regulatory elements including C/EBP, E2BP, NFAP-1, CArG box and Y box were highly conserved. Phylogenetic analysis revealed that all analyzed ALV-J strains could be separated into four groups, and SCDY1 as well as another strain NHH were included in the same cluster.
Conclusion
The variation in envelope glycoprotein was higher than other genes. The genome sequence of SCDY1 has a close relationship with that of another ALV-J strain NHH isolated from case of hemangioma. A 11 bp deletion observed in U3 region of 3'UTR of genome of ALV-J isolated from case of hemangioma is interesting, which may be associated with the occurrence of hemangioma.
Similar content being viewed by others
Background
Avian Leucosis virus subgroup J (ALV-J) was first reported in 1989 from commercial meat-type chickens with myelocytomatosis [1]. It is a novel subgroup different from classical ALV subgroup A, B, C, D and E, and a recombinant between exogenous avian leukosis viruses (ALVs) and the endogenous avian retrovirus (EAV) family of EAVs [2, 3]. ALV-J could be transmitted from broiler breeders to progeny more frequently than other ALV subgroups by Vertical transmission [4], and has caused severe economic losses in breeding farms all over the world. Recent studies showed that the host range of ALV-J had been largely widened, it could infect not only broilers but also commercial layers. Meanwhile, some new clinical pathotypes were observed in diseased flocks, such as hemangioma, which might be observed on the skin of the trunk, digitus, joint, wing and internal organs.
In China, the type of neoplasma induced by ALV-J was mainly myeloma leucosis (ML), and it occurred mainly in white meat-type chickens [5]. However, few cases of hemangioma caused by ALV subgroup J were reported in recent years [6, 7], and they were diagnosed mainly by histopathological examination and PCR detection. To updates, two complete proviral genomic sequences of ALV-J strain associated with hemangioma were reported, which were NHH (HM235668) and JL0901 (not published in NCBI).
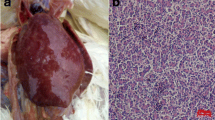
In 2009, an infectious disease occurred in a grandparent breeding farm in Sichuan province of China, characteristics mainly of weakness, inappetance and emaciation. Hemangioma was observed in the digiti of the sick. The gross pathology seen in organs of diseased chickens included hepatomegaly, splenomegaly and gray-white nodules of various sizes on the surface of the liver, but no myeloma leucosis was observed. In this study, an ALV-J strain SCDY1 was isolated from the blood samples of the diseased chickens, and its complete genome sequence was amplified and analyzed, which should be helpful for the research on the pathogenesis of hemangioma induced by ALV-J.
Methods
Chicken embryo fibroblast (CEF)
Specific-pathogen-free (SPF) eggs were obtained from the Beijing experimental animal center (Merial Inc., Beijing, China) and hatched at our lab. Ten-day-old chicken embryos were used to prepare the Chicken embryo fibroblast (CEF) cell cultures.
Virus isolation
The whole blood sample was inoculated onto the CEF monolayers prepared from 11-day-old embryonated eggs, and incubated at 37°C with 5% CO2 for five days for each passage. Uninfected CEF monolayers were used as negative control. After one blind passage, the existence of ALV-J in CEF was verified by PCR detection of 545 bp repeated sequence. Three blind passage were conducted until the result of PCR detection of cellular genome and RT-PCR detection of supernatant were both positive.
Primers
First, a pair of primers designed by smith [8] was synthesized and used to detect the proviral genomic DNA of ALV-J in infected CEF, or the viral RNA of ALV-J in the supernatant of infected CEF. Next, nine pairs of primers (Table 1) were designed according to the published sequences of HPRS-103 to amplify nine fragments which overlapped with each other and covered the whole genome of ALV-J. The ninth pair was used specifically to amplify the LTRs of circular DNA of ALV-J.
Genomic DNA extract and PCR amplification
The total DNA was extracted from chicken embryo fibroblast (CEF) infected with virus by using a sodium dodecyl sulfate (SDS) - proteinase K and phenol/chloroform/isoamylol (25:24:1) protocol. Genomic DNA PCR amplification was performed according to the manual of rTaq kit (Takara, Dalian, China), by using the proviral genomic DNA as template. Primers pairs 1-8 were used to amplify the internal region of SCDY1 genome; while primer pair 9 was used to amplify the LTRs of circular DNA by using total DNA of CEF infected by SCDY1 for 8-24 h as templates. The optimum conditions for PCR were as follows: 94°C for 4 min, ten cycles at 94°C for 35 s, 55°C for 35 s (a decrease of 0.5°C per cycle), 72°C for 40 s and followed by 20 cycles at 94°C for 35 s, 50°C for 35 s, 72°C for 40 s and a final elongation at 72°C for 10 min. The PCR product was analyzed in 1% agarose in Tris-borate-EDTA (TBE) buffer gel containing 0.5 mg/ml ethidium bromide.
Cloning and sequencing of proviral genomic DNA of SCDY1
The PCR products were isolated from agarose gels and purified using E.Z.N. ATM Gel Extraction Kit (TaKaRa, Japan), then cloned into pMD19-T vector (Takara, Dalian) and transformed into DH5α E. Coli competent cell. Confirmation of clones containing recombinant plasmid was achieved by PCR and restriction enzyme (RE) digestion. The recombinant plasmid was sequenced by Shanghai Sanggong Biologiucal Engineering Tachnology & Services Co., Ltd (shanghai, China).
Sequence analysis
The complete genome sequences of the isolate was compared with these of other ALV-J strains published in GenBank, including Chinese strains isolated from white broilers and commercial layers by using the Clustal W method in MegAlign program of the DNASTAR package. Transcriptional regulatory elements in U3 were analyzed by the online service system of NSITE (Recognition of Regulatory motifs) of Soft Berry (http://linux1.softberry.com/berry.phtml). Phylogenetic analysis of the complete proviral sequences of ALV-J was performed with the neighbor-joining method using MEGA version 4.0. The background of the reference strains used in this study and their accession numbers are listed in table 2.
Results
Virus isolation and identification
An ALV-J strain was isolated from the sampled blood, and was designated as SCDY1. The genomic DNA of infected CEF were positive in PCR detection, and the supernatant of infected CEF was positive in RT-PCR detection (data not shown), indicating the existence of proviral genomic DNA of ALV-J in infected CEF and ALV in the supernatant of infected CEF.
PCR amplification of proviral genome
Total DNA from SCDY1-infected CEF was used as PCR templates, and nine DNA fragments with the expected sizes of 0.614, 1.16, 1.098, 1.191, 1.376, 0.745, 1.921, 0.968, 0.466 kb long were amplified (data not shown).
Sequence analysis of the proviral genome of SCDY1
Sequences of the PCR products were aligned by using the EDITSEQ program in DNAstar software (DNASTAR Inc., Madison, WI 53715, USA). The complete proviral genome sequence of SCDY1, compiled from sequences of nine overlapping DNA fragments, was 7489 nt in length, and submitted to GenBank under accession No. HQ425636. The structure of SCDY1 proviral genome was presented in Figure 1.
Comparisons of the long terminal repeat (LTR) sequence showed that the LTR of SCDY1 shared a homology of 89.8-92.7% with these of other reference strains, and the LTR sequences of strains HPRS-103 and HN had the highest homology with these of SCDY1 (Table 3).
Comparisons of the three major structural genes revealed that the homology of nucleotide sequence of env, pol and gag gene between SCDY1 and other isolates in GenBank were 90.3-94.2%, 96.6-97.6% and 94.3-96.5%, respectively, and the homology of amino acid sequence of env, pol and gag gene between SCDY1 and other isolates in GenBank were 85.1-90.7%, 97.4-98.7% and 96.2-98.4%, respectively. The gag and pol genes of ALV-J were more conservative than the env gene relatively (Table 3, 4).
Homology analysis based on 3'UTR fragment of ALV-J associated with hemangioma was made by Clustal W method in MegAlign, and prototype strain HPRS-103 was used as a reference. The result showed that SCDY1 and NHH shared two almost identical deletions in 3'UTR, Which were the deletion of most part of the non-functional TM and a special 11 bp deletion in U3 region. Furthermore, most parts of E element in SCDY1 genome were absent; while, it was not observed in that of NHH (Figure 2). Online service analysis system of NSITE of SoftBerry showed that the transcriptional regulatory elements in U3 region of SCDY1 genome contained C/EBP, E2BP, CArG box and Y box, and they were highly conserved when comparing to these of the HPRS-103 strain. This special 11 bp deletion was found in the upstream of CArG box (Figure 3) and the sequence "TGATCAT" observed in this special 11 bp deletion region was recognized as a transcriptional regulatory element ABI REP1, which was the regulatory factor of human gene Apo-AI.
Phylogenetic relationships between SCDY1 and other ALV-J strains
Phylogenetic analysis of the complete genome sequences of SCDY1 and other eleven ALV-J strains demonstated that SCDY1 and other eleven reference strains could be grouped into four clusters, SCDY1 and another strain NHH isolated from case of hemangioma were in the same cluster (Figure 4).
Discussion
ALV-J has spread all over the world and caused severe economic losses in broiler breeder flocks since it was isolated [5, 9–11]. It can infect all species of chicken, but the types of neoplasma and the morbidity vary in different cases. The most common neoplasma observed in infected flocks was myeloid leucosis (ML) [12]; however, myelocytomas are not exclusive to ALV-J, other types of neoplasma such as histiocytic sarcomatosis, feather abnormalities, myelocyte infiltration in bones, hemangioma has also been reported [13–16]. Hemangioma has been even the most common neoplasm in broiler breeders in the UK [17], and an outbreak of hemangioma with 20% mortalities in young white Leghorn layers in Israel has also been reported [18]. In China, hemangioma was observed in several commercial layer flocks infected by ALV-J during 2007-2009 [19].
In this paper, a field ALV-J strain named as SCDY1 was isolated and its the complete proviral genome nucleotide sequence was determined. It contains 7489 nt with no EcoR I site, which could be found in the genome of NHH strain isolated from case of hemangioma. Homology analysis of the genes of SCDY1 and other reference strains showed that the envelope gene of ALV-J was prone to evolve compared to gag and pol genes, as the identity of the env gene between strain SCDY1 and other strains ranged from 90.3% to 94.2% at the nucleotides level and from 85.1% to 90.7% at the amino acid level respectively, lower than these of gag and pol genes.
The 3'UTR of avian retroviruses is important in viral pathogenesis and replication, the LTR enclosed within the 3' UTR contains powerful transcription regulatory elements that might differ among viruses and determine the virulence [20]. According to sequences analysis, the transcriptional regulatory elements of U3 region of SCDY1 genoem contained several transcriptional elements such as C/EBP, E2BP, CArG box and Y box, which played an important role in the efficient transcription of virus; while, NFAP-1 was not observed in SCDY1 genome, and it could be found in these of other analyzed strains. NFAP-1 could be recognized by the activator protein 1 (AP-1), and AP-1 is a heterodimeric protein composed of proteins belonging to the c-Fos, c-Jun, ATF and JDP families. It regulates gene expression in response to a variety of stimuli, including cytokines, growth factors, stress, and bacterial and viral infections [21]. The absent of NFAP-1 might affect the infection abilities of ALV-J virus.
Most of non-functional TM and E element of SCDY1 in 3'UTR were absent, which shared identical deletions with some ML strains isolated from meat-type chickens; while, the deletion of E element in SCDY1 genome was not observed in the genome of another strain NHH isolated from case of hemangioma. The E element has been found exclusively in some of the sarcoma viruses and ALV-J [22–24]. It may play a role in oncogenesis based on its close association with the src gene; however, it seemed not to be essential for oncogenesis, because other related oncogenic avian retroviruses lack the E element [25]. Therefore, the pathogenicity of SCDY1 may be unaffected by the deletion of TM and E element. The occurrence of hemangioma may be associated with multiple factors such as genetic backgroud, environment, concomitant infections, immunocompetence and the mode of transmission.
Interestingly, a special 11 bp deletion was observed in the U3 region of ALV-J strains associated with hemangioma. Online service analysis showed that a sequence "TGATCAT" within this special 11 bp region of U3 of ALV-J strains other than SCDY1 was a transcriptional regulatory element ABI REP1, which was the regulatory factor of human gene Apo-AI and function in the process of recognition of regulatory motifs. One binding factor of ABI REP1 named as HNF-4 was also found, and it served as an inhibitor modulator and played an important role in the regulation of eukaryotic genes. Although the special 11 bp deletion did not locate in CArG box, PRE box or Y box, its appearance may played a role in the transcriptional regulation of ALV-J genes, further study such as reverse genetics should be taken to test its function in oncogenesis.
So far, there are few proviral genomic sequence of ALV-J strains associated with hemangioma have been reported, the sequence comparison and phylogenetic analysis of SCDY1 and other ALV-J strains may help to reveal the evolution rhythm of ALV-J strains prevalent in fields, and benefit for the research on the pathogenicity and biological characteristics of ALV-J strain.
References
Payne LN, Brown SR, Bumstead N, Howes K, Frazier JA, Thouless ME: A novel subgroup of exogenous avian leukosis virus in chickens. J Gen Virol 1991,72(Pt 4):801-807. 10.1099/0022-1317-72-4-801
Smith LM, Toye AA, Howes K, Bumstead N, Payne LN, Venugopal K: Novel endogenous retroviral sequences in the chicken genome closely related to HPRS-103 (subgroup J) avian leukosis virus. J Gen Virol 1999,80(Pt 1):261-268.
Sacco MA, Flannery DM, Howes K, Venugopal K: Avian endogenous retrovirus EAV-HP shares regions of identity with avian leukosis virus subgroup J and the avian retrotransposon ART-CH. J Virol 2000, 74: 1296-1306. 10.1128/JVI.74.3.1296-1306.2000
Witter RL, Bacon LD, Hunt HD, Silva RE, Fadly AM: Avian leukosis virus subgroup J infection profiles in broiler breeder chickens: association with virus transmission to progeny. Avian Dis 2000, 44: 913-931. 10.2307/1593066
Cui Z, Du Y, Zhang Z, Silva RF: Comparison of Chinese field strains of avian leukosis subgroup J viruses with prototype strain HPRS-103 and United States strains. Avian Dis 2003, 47: 1321-1330. 10.1637/6085
Chao-nan L, Yu-long G, Hong-lei G, Xiao-le Q, Long J, Wei P, Xiao-mei W: Molecular characterization of a natural recombinant subgroup J avian leukosis virus with subgroup J avian leukosis virus with subgroup E partial env gene. Chinese journal of Preventive Veterinary Medicine 2009, 31: 978-981.
He-nan Z, Yan Q, Wei-wei S, Yi-yu L, Hong-bo L, Xiao-tao Z, Wei-sheng C, Ming L: Construction of an infectious clone of subgroup J Avian leukosis virus with E element deletion. Chinese journal of Preventive Veterinary Medicine 2010, 32: 94-97.
Smith LM, Brown SR, Howes K, McLeod S, Arshad SS, Barron GS, Venugopal K, McKay JC, Payne LN: Development and application of polymerase chain reaction (PCR) tests for the detection of subgroup J avian leukosis virus. Virus Res 1998, 54: 87-98. 10.1016/S0168-1702(98)00022-7
Fadly AM, Smith EJ: Isolation and some characteristics of a subgroup J-like avian leukosis virus associated with myeloid leukosis in meat-type chickens in the United States. Avian Dis 1999, 43: 391-400. 10.2307/1592636
Thu WL, Wang CH: Phylogenetic analysis of subgroup J avian leucosis virus from broiler and native chickens in Taiwan during 2000-2002. J Vet Med Sci 2003, 65: 325-328. 10.1292/jvms.65.325
Thapa BR, Omar AR, Arshad SS, Hair-Bejo M: Detection of avian leukosis virus subgroup J in chicken flocks from Malaysia and their molecular characterization. Avian Pathol 2004, 33: 359-363. 10.1080/0307945042000220435
Payne L: HPRS-103: A retro virus strikes back. The emergence of subgroup J avian leukosis virus. Avian pathology 1998, 27: 36-45. 10.1080/03079459808419291
Arshad SS, Bland AP, Hacker SM, Payne LN: A low incidence of histiocytic sarcomatosis associated with infection of chickens with the HPRS-103 strain of subgroup J avian leukosis virus. Avian Dis 1997, 41: 947-956. 10.2307/1592351
Nakamura K, Ogiso M, Tsukamoto K, Hamazaki N, Hihara H, Yuasa N: Lesions of bone and bone marrow in myeloid leukosis occurring naturally in adult broiler breeders. Avian Dis 2000, 44: 215-221. 10.2307/1592529
Landman WJ, Wilgen JL, Koch G, Dwars RM, Ultee A, Gruys E: Avian leukosis virus subtype J in ovo -infected specific pathogen free broilers harbour the virus in their feathers and show feather abnormalities. Avian Pathol 2001, 30: 675-684. 10.1080/03079450120092170
Williams SM, Reed WM, Bacon LD, Fadly AM: Response of white leghorn chickens of various genetic lines to infection with avian leukosis virus subgroup J. Avian Dis 2004, 48: 61-67. 10.1637/7052
Campbell JG, Appleby EC: Tumours in young chickens bred for rapid body growth (broiler chickens): a study of 351 cases. J Pathol Bacteriol 1966, 92: 77-90. 10.1002/path.1700920110
Burstein H, Gilead M, Bendheim U, Kotler M: Viral aetiology of haemangiosarcoma outbreaks among layer hens. Avian Pathol 1984, 13: 715-726. 10.1080/03079458408418568
Cheng Z, Liu J, Cui Z, Zhang L: Tumors associated with avian leukosis virus subgroup J in layer hens during 2007 to 2009 in China. J Vet Med Sci 72: 1027-1033. 10.1292/jvms.09-0564
Ruddell A: Transcription regulatory elements of the avian retroviral long terminal repeat. Virology 1995, 206: 1-7. 10.1016/S0042-6822(95)80013-1
Hess J, Angel P, Schorpp-Kistner M: AP-1 subunits: quarrel and harmony among siblings. J cell Sci 2004,117(Pt 25):5965-5973. 10.1242/jcs.01589
Laimins L, Tsichlis P, Khoury G: Multiple enhancer domains in the 3' terminus of the Prague strain of Rous sarcoma virus. Nucleic Acids Research 1984, 12: 6427. 10.1093/nar/12.16.6427
Hue D, Dambrine G, Denesvre C, Laurent S, Wyers M, Rasschaert D: Major rearrangements in the E element and minor variations in the U3 sequences of the avian leukosis subgroup J provirus isolated from field myelocytomatosis. Arch Virol 2006, 151: 2431-2446. 10.1007/s00705-006-0811-2
Chesters PM, Smith LP, Nair V: E (XSR) element contributes to the oncogenicity of Avian leukosis virus (subgroup J). J Gen Virol 2006, 87: 2685-2692. 10.1099/vir.0.81884-0
Bizub D, Katz RA, Skalka AM: Nucleotide sequence of noncoding regions in Rous-associated virus-2: comparisons delineate conserved regions important in replication and oncogenesis. J Virol 1984, 49: 557-565.
Acknowledgements
The work was financially supported by Program for Changjiang Scholars and Innovative Research Team in University "PCSIRT"(Grant No: IRTO848).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MS: carried out most of the experiments and wrote the manuscript, YH: carried out study design, and revised the manuscript. MXT, CL, YZ, YL, NLZ, PL, XTW, SJC helped in experiments. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Shi, M., Tian, M., Liu, C. et al. Sequence analysis for the complete proviral genome of subgroup J Avian Leukosis virus associated with hemangioma: a special 11 bp deletion was observed in U3 region of 3'UTR. Virol J 8, 158 (2011). https://doi.org/10.1186/1743-422X-8-158
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-8-158