Abstract
Background
Three* human polyomaviruses have been discovered recently, KIPyV, WUPyV and MCPyV. These viruses appear to circulate ubiquitously; however, their clinical significance beyond Merkel cell carcinoma is almost completely unknown. In particular, nothing is known about their preponderance in vertical transmission. The aim of this study was to investigate the frequency of fetal infections by these viruses. We sought the three by PCR, and MCPyV also by real-time quantitative PCR (qPCR), from 535 fetal autopsy samples (heart, liver, placenta) from intrauterine fetal deaths (IUFDs) (N = 169), miscarriages (120) or induced abortions (246). We also measured the MCPyV IgG antibodies in the corresponding maternal sera (N = 462) mostly from the first trimester.
Results
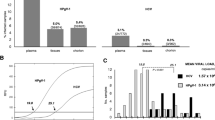
No sample showed KIPyV or WUPyV DNA. Interestingly, one placenta was reproducibly PCR positive for MCPyV. Among the 462 corresponding pregnant women, 212 (45.9%) were MCPyV IgG seropositive.
Conclusions
Our data suggest that none of the three emerging polyomaviruses often cause miscarriages or IUFDs, nor are they transmitted to fetuses. Yet, more than half the expectant mothers were susceptible to infection by the MCPyV.
Similar content being viewed by others
Background
Among the five* human polymaviruses known, aside from the BK virus (BKV) and JC virus (JCV) [1, 2], three* new ones, KI polyomavirus (KIPyV), WU polyomavirus (WUPyV), and Merkel cell polyomavirus (MCPyV) have been discovered during the past few years by use of advanced molecular techniques [3–5]. In their DNA sequences, KIPyV and WUPyV are interrelated more than MCPyV, which differs from all the human polyomaviruses known [6]. The KIPyV and WUPyV were discovered in nasopharyngeal aspirates (NPA) from children with respiratory tract infections [3, 4]. Although many reports have confirmed their presence in the upper airways of patients with respiratory illness, evidence is lacking of their pathogenicity in this context [7–10, 12]. For these viruses, their tropism and clinical significance are unknown. Likewise, for the tumorigenic MCPyV [5], also found in the nasopharynx [13–15], the mode of transmission and, host cells, as well as latency characteristics are yet to be established. MCPyV DNA has been detected in a variety of specimen types including skin, saliva, gut, and respiratory secretion samples [5, 16, 17]. Recovery of complete MCPyV genomes from the skin of 40% of healthy adults and PCR detection of MCPyV in the skin of almost all adults by cutaneous swabbing suggests that most adults are persistently infected with this polyomavirus [18]. Another recent study revealed the viral DNA in environmental samples (sewage and river water) [19], confirming that MCPyV indeed is a ubiquitous virus.
Serological studies have shown that initial exposure to KIPyV and WUPyV, as well as MCPyV occurs often in childhood, similar to that for BKV and JCV, and that MCPyV circulates widely in the human population [20–24]. Although most adults have been exposed to MCPyV, the exact site(s) of MCPyV infection remain unclear. Vertical transmission of many human DNA and RNA viruses is well established. However, for Polyomaviridae this mode of transmission is far from clear. Transplacental transmission of BKV was first suggested by detection of the virus DNA in fetal tissues [25], while others obtained no evidence of vertical transmission [26]. IgM studies of BKV and JCV in cord blood samples showed no apparent association with congenital infection [27, 28].
These findings prompted us to investigate a sizeable material of fetal autopsy samples (placenta, heart, liver) for the presence of the three polyomaviruses in order to determine whether these viruses give rise to fetal infections. We also studied the corresponding maternal sera for MCPyV IgG antibodies, by a newly established virus like particle (VLP) - based IgG assay (Chen et al, in revision).
Materials and methods
Clinical samples
The DNA studies were carried out using formalin-fixed, paraffin-embedded (FFPE) tissues - placenta, heart, and liver - after intrauterine fetal deaths (IUFDs, N = 169), miscarriages (N = 120), or using as controls, specimens from induced abortions (N = 246) performed exclusively due to medical indications. From each fetus, 3 organs (when available; placenta, heart, and liver) were initially studied in pools, by PCR for the three polyomaviruses (KI, WU, and MC). In the positive pool its constituent tissues (placenta and heart) were then re-examined separately. Thus, a total of 535 fetuses were included in the overall cohort. The sampling in the Helsinki region occurred from July 1992 to December 1995 and January 2003 to December 2005 [29]. The gestational weeks of the fetal deaths ranged from 11 to 42. In this study, IUFD corresponds to fetal loss having occurred during or after the 22nd gestational week, and miscarriage to fetal loss having occurred earlier. Our number of IUFDs represents 58% of all occurring in Helsinki during the study period.
We furthermore examined for the presence of MCPyV IgG antibodies all serum samples available from the corresponding mothers (n = 462). These samples had been collected at the municipal maternity centers during antenatal screening around the 9th gestational week (mean 9; median 9; range 2 to 36) and were stored frozen at the Finnish Maternity Cohort, National Institute for Health and Welfare, Oulu, Finland. The mothers' ages ranged between 18 and 45 years (mean 31, median 31).
The study was approved by the Coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa. Permission for use of fetal tissues was obtained from the National Supervisory Authority for Welfare and Health (Valvira).
DNA preparation and PCRs
The paraffin blocks were punch-biopsied, proteinase K-digested, and the DNA was prepared by salting out, as described [29]. Briefly, tissue lysates were heated at 95°C for 10 min. The paraffin then appeared floating on the surface, after centrifugation at 13,200 rpm for 5 minutes (Eppendorf; 4°C). Sodium chloride was added to achieve a final concentration of 1.2 mol/L, and the sample was mixed for 20 s and recentrifuged. The supernatant was transferred to a new tube, carefully avoiding particles. The DNA in the supernatant was precipitated with absolute ethanol and was redissolved in 60 mL of water. The DNA solution diluted 1 to 10 was stored at - 20°C until use. Water, as a negative control, was inserted between every 20 samples and was prepared along with the tissue pools.
All these DNA preparations were β -globin-PCR-positive, pointing to DNA stability and lack of appreciable PCR inhibition. The WUPyV and KIPyV nested PCRs employed primer set A (table 1) [12]. For MCPyV DNA detection by qualitative PCRs, two primer sets were used (table 1); all samples were first studied by the LT3 nested PCR [15], and were reanalyzed by the LT1/M1 nested PCR [30], and samples with positive results by agarose gel electrophoresis were DNA sequenced. As short PCR products ought to be used when working with FFPE tissues [31, 32] we reanalyzed all pools with a real-time quantitative PCR with an amplicon length of 59 bp, as described [13].
PCR sensitivity
For detection of KIPyV/WUPyV and MCPyV we performed a highly sensitive PCR assay [15]. As positive controls and to determine assay sensitivities by limiting dilution analysis, plasmids containing the VP2 gene of WUPyV (EU693907) and KIPyV (EU358767) and the LT3 amplification product of MCPyV (EU375803) were constructed; the amplification products of the VP2 genes were cloned into pCR8/GW/TOPO (Invitrogen; Carlsbad, CA, USA) whereas the MCPyV LT3 region was synthesized and cloned into pGOv4 by Gene Oracle, Inc. (San Leandro, USA). In the MCPyV and KIPyV/WUPyV assays, plasmid controls with 30 and 5 copies/reaction were reproducibly positive, respectively. Of note, the sensitivities were unaffected by the inclusion of genomic human DNA from cultured 293T cells at 100 ng per reaction (4 ng/μ l). In non-nested format with 40 PCR cycles the LT3 primers had a sensitivity 1 log lower than that of the nested assay both in the presence and absence of human genomic DNA. In addition, we also used a previously established real time quantitative PCR for MCPyV, where a plasmid control with 2 copies/reaction was reproducibly positive [13].
Sequence analyses
The MCPyV PCR products were purified for automated sequencing using the High Pure PCR product purification kit (Roche). The resulting DNA sequences using BLAST were aligned against the reference sequences in GenBank including accession numbers of [gb|EU375803.1]; [gb|EU375804.1]; [gb|FJ173815.1]; [gb|FJ464337.1]; [gb|HM011538] and [gb|HM011557].
MCPyV antibody EIA
MCPyV IgG antibodies were measured by EIA based on virus protein 1 (VP1) VLPs (Chen et al, in revision). Briefly, the VP1-VLPs expressed in insect cells and purified by CsCl density gradient centrifugation were biotinylated and attached (at 60 ng/well) to streptavidin-coated microtiter plates (Thermo Scientific) and saturated with a sample diluent (Ani Labsystems). The sera (1:200) were applied in duplicate, the bound IgG were quantified with peroxidase-conjugated anti-human IgG (1:2000; DakoCytomation) using H2O2 and OPD substrates, and the absorbances at 492 nm were read after blank subtraction. The EIA cut-off for IgG positivity is 0.120 OD units.
Results and Discussion
Among the 535 fetal autopsy samples studied in pools, none was PCR positive for KIPyV or WUPyV DNA. On the other hand, one pool was positive for MCPyV by the LT3 PCR. Tissue samples from 2 sites were available for further study of this fetus. On retesting of the placenta and fetal heart separately, the heart was PCR negative and the placenta was positive for MCPyV. However, it was negative by the LT1/M1 PCR for MCPyV. DNA preparations re-extracted from the other half of the same punch biopsy, as well as those obtained via another punch from the same paraffin block, showed exactly the same MCPyV DNA results. Furthermore, we studied all samples with a real-time qPCR for MCPyV of a different genomic region, with an identical result; the placenta was positive with a Ct value of 38.4, while the biopsy from the fetal heart was negative. To verify the specificity of the amplified products and to detect possible genomic variants, the MCPyV LT3 PCR products were sequenced. They showed 100% homology with all the existing database sequences. N.B., the miscarried fetus, with gastroschisis and umbilical cord complications, was deceased in 1994, in gestational week 17.
In addition, we examined for circulating MCPyV IgG antibodies the corresponding pregnant women. Of the 462 maternal sera, 45.9% (212) showed positive results.
We searched formalin-fixed, paraffin-embedded tissues - placenta, heart, and liver - of 535 fetal autopsy samples for the KI, WU, and MC polyomaviruses. We found no genomic DNAs of KIPyV or WUPyV in any of the stillborn or deceased fetuses. This suggests that during the study period neither of these two newly found viruses (i) often caused miscarriages or IUFDs, (ii) nor were incidentally (as bystanders) transmitted to remain in the fetuses succumbing for other reasons. Whether the exclusive mechanism in our mid-size series was the serendipitous absence of maternal infections (primary; secondary) caused by these viruses, or pathogenetic resistance by other mechanisms, remains to be shown. On the other hand, it was interesting to observe LT3 PCR and VP1 qPCR positivity for MCPyV (reproducibly, and of correct DNA sequence) in the placenta of one miscarriage in the 17th gestational week. The negativity of this placenta with the other MCPyV PCR (LT1/M1) may be due to the known sensitivity difference of the PCR assays [15, 30].
As for the previously known human polyomaviruses BKV and JCV, no fetal autopsy materials have been found positive for JCV DNA, whereas one study [25] reported a high genoprevalence of BKV DNA; BKV vertical transmission has been denied by others [11, 33, 34], however.
Ours is to our knowledge the first study in which fetal tissues have been searched for the newly discovered human polyomaviruses. Serology has shown that a high proportion of adults have been exposed to MCPyV, and that the infection can be acquired early in life [20–24]. We detected an IgG seroprevalence for MCPyV of 45.9% among the pregnant women. This value is in line with previous reports and shows that more than half our women were susceptible to MCPyV infection. Tolstov et al. studying serologically 6 children 1 year or younger found no evidence of MCPyV vertical transmission [24].
Conclusions
While the three* emerging polyomaviruses occur frequently in tissues of many different types, and MCPyV also in environmental samples [3, 4, 13, 15, 18, 35], our PCR data from 535 pregnancies suggest that none of these viruses are frequently transmitted vertically. Further studies with larger populations may, however, be warranted to determine which role, if any, MCPyV plays in pregnant women and their offspring.
References
Gardner SD, Field AM, Coleman DV, Hulme B: New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet 1971,1(7712):1253-1257. 10.1016/S0140-6736(71)91776-4
Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH: Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet 1971,1(7712):1257-1260. 10.1016/S0140-6736(71)91777-6
Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, Dalianis T, Ramqvist T, Andersson B: Identification of a third human polyomavirus. J Virol 2007,81(8):4130-4136. 10.1128/JVI.00028-07
Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D: Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog 2007,3(5):e64. 10.1371/journal.ppat.0030064
Feng H, Shuda M, Chang Y, Moore PS: Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 2008,319(5866):1096-1100. 10.1126/science.1152586
Dalianis T, Ramqvist T, Andreasson K, Kean JM, Garcea RL: KI, WU and Merkel cell polyomaviruses: a new era for human polyomavirus research. Semin Cancer Biol 2009,19(4):270-275. 10.1016/j.semcancer.2009.04.001
Le BM, Demertzis LM, Wu G, Tibbets RJ, Buller R, Arens MQ, Gaynor AM, Storch GA, Wang D: Clinical and epidemiologic characterization of WU polyomavirus infection, St. Louis, Missouri. Emerg Infect Dis 2007,13(12):1936-1938.
Abedi Kiasari B, Vallely PJ, Corless CE, Al-Hammadi M, Klapper PE: Age-related pattern of KI and WU polyomavirus infection. J Clin Virol 2008,43(1):123-125. 10.1016/j.jcv.2008.05.003
Ren L, Gonzalez R, Xie Z, Zhang J, Liu C, Li J, Li Y, Wang Z, Kong X, Yao Y, Hu Y, Qian S, Geng R, Yang Y, Vernet G, Paranhos-Baccala G, Jin Q, Shen K, Wang J: WU and KI polyomavirus present in the respiratory tract of children, but not in immunocompetent adults. J Clin Virol 2008,43(3):330-333. 10.1016/j.jcv.2008.08.003
Wattier RL, Vazquez M, Weibel C, Shapiro ED, Ferguson D, Landry ML, Kahn JS: Role of human polyomaviruses in respiratory tract disease in young children. Emerg Infect Dis 2008,14(11):1766-1768. 10.3201/eid1411.080394
Coleman DV, Wolfendale MR, Daniel RA, Dhanjal NK, Gardner SD, Gibson PE, Field AM: A prospective study of human polyomavirus infection in pregnancy. J Infect Dis 1980,142(1):1-8.
Norja P, Ubillos I, Templeton K, Simmonds P: No evidence for an association between infections with WU and KI polyomaviruses and respiratory disease. J Clin Virol 2007,40(4):307-311. 10.1016/j.jcv.2007.09.008
Goh S, Lindau C, Tiveljung-Lindell A, Allander T: Merkel cell polyomavirus in respiratory tract secretions. Emerg Infect Dis 2009,15(3):489-491. 10.3201/eid1503.081206
Bialasiewicz S, Lambert SB, Whiley DM, Nissen MD, Sloots TP: Merkel cell polyomavirus DNA in respiratory specimens from children and adults. Emerg Infect Dis 2009,15(3):492-494. 10.3201/eid1503.081067
Kantola K, Sadeghi M, Lahtinen A, Koskenvuo M, Aaltonen LM, Mottonen M, Rahiala J, Saarinen-Pihkala U, Riikonen P, Jartti T, Ruuskanen O, Soderlund-Venermo M, Hedman K: Merkel cell polyomavirus DNA in tumor-free tonsillar tissues and upper respiratory tract samples: implications for respiratory transmission and latency. J Clin Virol 2009,45(4):292-295. 10.1016/j.jcv.2009.04.008
Loyo M, Guerrero-Preston R, Brait M, Hoque M, Chuang A, Kim M, Sharma R, Liegeois N, Koch W, Califano J, Westra W, Sidransky D: Quantitative detection of merkel cell virus in human tissues and possible mode of transmission. Int J Cancer 2009.
Wieland U, Mauch C, Kreuter A, Krieg T, Pfister H: Merkel cell polyomavirus DNA in persons without merkel cell carcinoma. Emerg Infect Dis 2009,15(9):1496-1498. 10.3201/eid1509.081575
Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB: Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 2010,7(6):509-515. 10.1016/j.chom.2010.05.006
Bofill-Mas S, Rodriguez-Manzano J, Calgua B, Carratala A, Girones R: Newly described human polyomaviruses Merkel cell, KI and WU are present in urban sewage and may represent potential environmental contaminants. Virol J 2010,7(1):141. 10.1186/1743-422X-7-141
Kean JM, Rao S, Wang M, Garcea RL: Seroepidemiology of human polyomaviruses. PLoS Pathog 2009,5(3):e1000363. 10.1371/journal.ppat.1000363
Nguyen NL, Le BM, Wang D: Serologic evidence of frequent human infection with WU and KI polyomaviruses. Emerg Infect Dis 2009,15(8):1199-1205. 10.3201/eid1508.090270
Carter JJ, Paulson KG, Wipf GC, Miranda D, Madeleine MM, Johnson LG, Lemos BD, Lee S, Warcola AH, Iyer JG, Nghiem P, Galloway DA: Association of Merkel cell polyomavirus-specific antibodies with Merkel cell carcinoma. J Natl Cancer Inst 2009,101(21):1510-1522. 10.1093/jnci/djp332
Pastrana DV, Tolstov YL, Becker JC, Moore PS, Chang Y, Buck CB: Quantitation of human seroresponsiveness to Merkel cell polyomavirus. PLoS Pathog 2009,5(9):e1000578. 10.1371/journal.ppat.1000578
Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, Chang Y, Buck CB, Moore PS: Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int J Cancer 2009,125(6):1250-1256. 10.1002/ijc.24509
Pietropaolo V, Di Taranto C, Degener AM, Jin L, Sinibaldi L, Baiocchini A, Melis M, Orsi N: Transplacental transmission of human polyomavirus BK. J Med Virol 1998,56(4):372-376. 10.1002/(SICI)1096-9071(199812)56:4<372::AID-JMV14>3.0.CO;2-4
Boldorini R, Veggiani C, Barco D, Monga G: Kidney and urinary tract polyomavirus infection and distribution: molecular biology investigation of 10 consecutive autopsies. Arch Pathol Lab Med 2005,129(1):69-73.
Andrews CA, Daniel RW, Shah KV: Serologic studies of papovavirus infections in pregnant women and renal transplant recipients. Prog Clin Biol Res 1983, 105: 133-141.
Brown DW, Gardner SD, Gibson PE, Field AM: BK virus specific IgM responses in cord sera, young children and healthy adults detected by RIA. Arch Virol 1984,82(3-4):149-160. 10.1007/BF01311159
Riipinen A, Vaisanen E, Nuutila M, Sallmen M, Karikoski R, Lindbohm ML, Hedman K, Taskinen H, Soderlund-Venermo M: Parvovirus b19 infection in fetal deaths. Clin Infect Dis 2008,47(12):1519-1525. 10.1086/593190
Kassem A, Schopflin A, Diaz C, Weyers W, Stickeler E, Werner M, Zur Hausen A: Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res 2008,68(13):5009-5013. 10.1158/0008-5472.CAN-08-0949
Goelz SE, Hamilton SR, Vogelstein B: Purification of DNA from formaldehyde fixed and paraffin embedded human tissue. Biochem Biophys Res Commun 1985,130(1):118-126. 10.1016/0006-291X(85)90390-0
Dubeau L, Chandler LA, Gralow JR, Nichols PW, Jones PA: Southern blot analysis of DNA extracted from formalinfixed pathology specimens. Cancer Res 1986,46(6):2964-2969.
Taguchi F, Nagaki D, Saito M, Haruyama C, Iwasaki K: Transplacental transmission of BK virus in human. Jpn J Microbiol 1975,19(5):395-398.
Shah K, Daniel R, Madden D, Stagno S: Serological investigation of BK papovavirus infection in pregnant women and their offspring. Infect Immun 1980,30(1):29-35.
Foulongne V, Kluger N, Dereure O, Mercier G, Moles JP, Guillot B, Segondy M: Merkel cell polyomavirus in cutaneous swabs. Emerg Infect Dis 2010,16(4):685-687.
Acknowledgements
This study was supported by the Helsinki University Central Hospital Research & Education and Research & Development funds, the Sigrid Jusèlius Foundation, the Medical Society of Finland (FLS), and the Academy of Finland (project code 1122539). M.S. expresses his gratitude to the Ministry of Science, Research and Technology of Iran for a research scholarship as well as to Bu-Ali Sina University, Hamedan for the opportunity to advanced studies. For friendly help with language revision we are much indebted to Carolyn Brimley Norris from language services of Helsinki University.
*Note added in proof: Since the work described in this paper was completed and submitted for publication, another previously unknown human polyomavirus, TSPyV, was identified by Van der Meijden et al, (PLoS Pathog 2010;6, e1001024), bringing to six the number human polyomaviruses known.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MS carried out the PCR and serological assays, analyzed the data, and participated in writing. AR and EV organized the clinical materials and carried out the DNA extractions. TC accounted for the serology part. KK participated in the methods design and setup. HMS, RK, and HT collected the specimens and contributed to the data analysis. MSV and KH conceived the study, participated in its design and coordination and accounted for the manuscript writing. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Sadeghi, M., Riipinen, A., Väisänen, E. et al. Newly discovered KI, WU, and Merkel cell polyomaviruses: No evidence of mother-to-fetus transmission. Virol J 7, 251 (2010). https://doi.org/10.1186/1743-422X-7-251
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-7-251