Abstract
Background
Some UL45 gene function of Herpesvirus was reported. While there was no any report of the duck enteritis virus (DEV) UL45 protein as yet.
Results
The UL45 gene and des-transmembrane domain of UL45 (named UL45Δ gene, 295-675bp of UL45) of DEV were amplified by PCR and subcloned into the prokaryotic expression vector pET-32a(+). The constructed recombinant plasmids were transformed into the host strain BL21(DE3) PLysS and induced by IPTG. SDS-PAGE analysis showed the UL45 gene couldn't express while UL45Δ gene was highly expressed. His Purify Kit or salting-out could purify the protein effectively. Using the purified protein to immunize New-Zealand rabbits and produce polyclonal antibody. The agar diffusion reaction showed the titer of antibody was 1:32. Western blot analysis indicated the purified rabbit anti-UL45Δ IgG had a high level of specificity and the UL45 gene was a part of DEV genome. The transcription phase study of UL45 gene showed that expression of UL45 mRNA was at a low level from 0 to 18 h post-infection (pi), then accumulated quickly at 24 h pi and peaked at 42 h pi. It can be detected till 72 h pi. Besides, western blot analysis of purified virion and different viral ingredients showed that the UL45 protein resided in the purified virion and the viral envelope.
Conclusions
The rabbit anti-UL45Δ IgG was produced successfully and it can serve as a good tool for penetrating studies of the function of DEV UL45 protein. The transcription phase and protein characteristics analysis indicated that DEV UL45 gene was a late gene and UL45 protein may be a viral envelope protein.
Similar content being viewed by others
Background
Duck virus enteritis (DVE) or duck plague (DP), was an acute, febrile, contagious and septic disease of waterfowl (duck, goose, and swan) caused by Duck Enteritis Virus (DEV). It caused considerable economic losses to the duck-producing areas of the world due to its high mortality and decreased egg production [1–5]. DEV was currently classified to the Alphaherpesvirinae subfamily of the Herpesviridae, but had not been grouped into any genus yet [6]. For a long time, studies of the molecular biology of DEV had larged behind other members of the herpesviridae family. Luckily, during the past several years, some DEV genes had been reported successfully [7–27]. However, the function of potential proteins encoded by many of the DEV genes was still unclear, including UL45.
The conservatism of UL45 gene was low in different herpervirus subfamily, while in different strains of the same herpervirus it was highly conserved [28–31]. The UL45 protein was a true late protein and a component of the virion from other herpesvirinae [32–34]. The main function of UL45 protein which had reported included promoting the cell-cell fusion, anti-apoptosis, viral correct propagation, egress and keeping virulence of virus [35–37].
Using a series of software to analyze the bioinformatics of DEV UL45 gene, the results indicated that UL45 protein had 224 residues with a molecular mass of 24kDa, 73 to 95 amino acids was a potential membrane-spanning segment and no cleavage site of signal peptide. When the threshold was defined to 0.5, it had 13 potential phosphorylation sites and no glycosylation site.
In this article, the construction of cloning and expression plasmids, expression of UL45Δ fusion protein, production of polyclonal antibody, time course of transcription and protein characteristics analysis were detailedly introduced.
Results
Gene amplification, construct expression plasmids
Using genome of DEV CHv-strain to amplify the UL45 and UL45Δ gene. Electrophoresis analysis results of amplified products showed that the size of UL45 and UL45Δ gene was the same as expected.
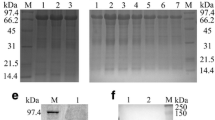
The UL45 and UL45Δ gene digested from pMD18-T/UL45 and PMD18-T/UL45Δ plasmids (constructed by TaKaRa) were respectively directionally inserted to pET-32a(+) plasmid to construct the expression plasmids (fig. 1, 2). PCR and restriction digestion analysis showed the UL45 and UL45Δ expression plasmids were successfully constructed (fig. 3, 4).
Protein expression, purification, polyclone antibody production and western blot analysis
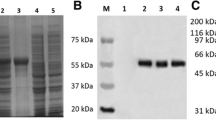
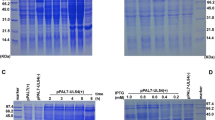
The protein expression condition was analyzed by SDS-PAGE. It showed that the UL45 gene couldn't express while UL45Δ gene was highly expressed in the supernatant. The optimized condition of expression was inducing 4 h at 30°C after adding 0.2 mmol/L IPTG. The UL45Δ protein could be purified effectively by IMAC on Ni2+-NTA agarose or salting-out (fig. 5). Using the protein to immune rabbits, after four injections the rabbit anti-UL45Δ serum was collected. Agar diffusion reaction showed the titer of antibody was 1:32. With the methods of ammonium sulfate precipitation and DEAE-Sepharose column, we got the homogeneous rabbit anti-UL45Δ IgG (fig. 6). Importantly, western blot analysis showed that the UL45Δ protein could be recognized by the rabbit anti-UL45Δ IgG and rabbit anti-DEV IgG but it couldn't be recognized by the negative control serum (fig. 7, 8). These showed that the rabbit anti-UL45Δ IgG had a good specificity and UL45 gene was a member of DEV genome.
Expression and purification of recombinant protein. M1: Protein Marker; 1: Total protein from pET-32a (+) after induction; 2: Total protein from recombinant plasmid PET32a(+)-UL45Δ after induction; 3: Uninduced control; 4: The clear supernatant after ultrasonic disruption; 5: Purified recombinant UL45Δ protein using a single chromatographic step of IMAC on Ni 2+ -NTA agarose.
Transcription characteristics analysis of UL45 gene
The standard curve of PMD18-T/UL45 and PMD18-T/β-actin showed the FQ-PCR was excellent at performance (fig. 9, 10, 11 and 12). The integrality and purity detecting of total RNA showed that OD260/OD280 was from 1.8 to 2.0, and the 28 S, 18 S and 5 S could be clearly seen by agarose gel electrophoresis. This indicated that the RNA could be used for further study. The condition of UL45 mRNA expression showed that the situation of transcription was changed during the whole cycle. The expression of DEV UL45 mRNA was at a low level from 0 to 18 h post-infection (pi), then accumulated quickly at 24 h pi and peaked at 42 h pi. It can be detected till 72 h pi (Fig. 13). This inferred that the UL45 gene of DEV-CHv was a late gene.
The fluorescent quantitative real-time PCR standard curve of β-actin. The x-axis represented ten-fold dilutions of PMD18-T/β-actin plasmids, and the y-axis represented corresponding cycle threshold (Ct) values. Each dot represents the result of triplicate amplification of each dilution. The standard curve equation is Y = -3.481 × + 5.836, the correlation coefficient and the slope value of the regression curve were indicated in the figure.
The fluorescent quantitative real-time PCR standard curve of UL45. The x-axis represented ten-fold dilutions of PMD18-T/UL45 plasmids, and the y-axis represented corresponding cycle threshold (Ct) values. Each dot represents the result of triplicate amplification of each dilution. The standard curve equation is Y = -3.366 × +8.995. The correlation coefficient and the slope value of the regression curve were indicated in the figure.
Characteristics analysis of UL45 protein
When analyzing the purified virion with the rabbit anti-UL45Δ IgG, the UL45 protein could be detected and this suggested that the UL45 protein was a component of virion. We extracted the virion with NP-40 detergent, and obtained a supernatant fraction (envelope and minor amounts of tegument proteins) and a pellet (nucleocapsids and tegument proteins). Equivalent amounts of the supernatant and pellet proteins were immunoblotted with the UL45Δ IgG. The results showed that the 24kDa UL45 protein was found almost exclusively in the NP-40 soluble fract, suggested that the UL45 protein was associated with the envelope of the virion (Fig. 14).
Discussion
Choosing the pET-32a(+), the E.coli strain DH5α and E.coli strain BL21(DE3) PlysS as the expression vector, cloning and expression host because of their unexampled advantages. Here, E. coli strain DH5α has very high transformation efficiency. The E. coli strain BL21(DE3) PlysS has the advantage of being deficient in both the lon and ompT proteases and harboring the T7 bacteriophage RNA polymerase gene which permits the specific expression of heterologous genes driven by the T7 promoter [38–40]. Prokaryotic expression vector pET-32a(+), which features a high stringency T7 lac promoter, His6 tag and T7 terminator, has been recognized as one of the most powerful tools for producing recombinant proteins in E. coli[41]. This fusion tag permits purification of the produced protein by metal chelate chromatography on a nitrilo-triacetic acid agarose matrix charged with nickel ions. Slight inorganic salt (ammonium sulfate, sodium sulfate etc) can promote the dissolution of proteins but fortis saline solution will induce proteins separated from the solution because of agglomeration, and this effect is called salting-out. The solubility of proteins in aqueous solution is determinated by the extent of hydration between the hydrophilic grouping of proteins and water, the situation of the protein electric charge. Adding the neutral salt to the protein solution, the hydration shell around the proteins becomes weaken or vanished, and the surface charge of protein is neutralized greatly. These lead to the depression of the proteins solubility and separation from the solution. Salting-out is a good method to purify the dissolvable proteins.
Here, the UL45 protein couldn't express regardless of the expression vector, expression host strain and the condition of expression. The possible reasons were as follows. First, the codon could influence the expression of protein. AGA, AGG, AUA, CCG, CCT, CTC, CGA, GTC etc codon was the rare codon of E.coli[42, 43]. If the exogenous gene had a high level of rare codon then the efficiency of expression was usually low. The statistics of the UL45 gene codon usage condition showed that rare codon had a high usage frequency in UL45 gene, and this may be a reason of the UL45 gene expression failure. Besides, the bioinformatics analysis showed that the 73 to 95 amino acids of UL45 protein was a membrane-spanning segment. As we know, the membrane-spanning segment was high hydrophobic and not good for protein expression. We designed a couple of primers to amplify the UL45Δ gene (deletion of the membrane-spanning segment) and it was expressed with high performance. So we inferred that the existence of membrane-spanning segment was the main reason of the UL45 gene expression failure.
There had been some function studies of UL45 protein from other herpesvirus but there wasn't any report about the function of DEV UL45 protein till now. We used the rabbit anti-UL45Δ IgG to study the DEV UL45 gene transcription phase and the UL45 protein characteristics. These may supply effective evidence to explain the function of UL45 protein in the DEV infection, replication of life cycle.
According to the different transcription sequence, gene can be classified as immediate early gene, early gene and late gene [44]. After the virus infected the target cell, linear double strands DNA which locates in intra-nuclear becomes cyclization, and the virus gene starts to transcript according to some sequence. The late gene encodes about thirty viral proteins, primarily including capsid protein, tegument protein and envelope protein. They usually express at last [45]. Up to now, there wasn't any report of transcription characterization about DEV UL45 gene, while the situation of transcription could reflect the viral genetic structure and the basic situation of gene expression. So the transcription course of UL45 gene was studied, and the results showed that DEV UL45 gene may be a late gene. It was consistent with the report of other herpesvirus and it could provide guidance for future study [28, 33]. While only using the FQ-PCR to affirm the UL45 gene is a late gene isn't sufficient, more penetrating studies need to be done. The UL45 gene temporal regulation condition should be analyzed when the infected cell is dealt with some canonical medicine. Such as γ gene can't be detected in the cell dealt with phosphonoacetate; α gene can be detected in the cell dealt with protein synthesis inhibitor; β gene can be inhibited by cycloheximide (protein synthesis inhibitor) while can't be inhibited by phosphonoacetate (DNA synthesis inhibition factor) [46].
The protein characteristics analysis showed that UL45 protein was related with the virion and may be an envelope protein. The UL45 protein may not be a tegument protein, because most tegument protein resided in the sediment and there was no positive signal can be detected in the sediment. However, we couldn't exclude the possibility of UL45 protein was noncohesive binding with the tegument. The result of protein characteristics analysis was coincidence with the transcription characteristics. They all showed that the DEV UL45 gene was a late gene. While the affirmation of UL45 protein characteristics needs more penetrating studies, such as using the protein which had known the characteristics do the parallel experiment or using the 35S mark the methionine to confirm the effect of virion purification and so on.
Conclusions
In conclusions, the UL45Δ protein and rabbit anti-UL45Δ IgG were produced successfully. Western blot analysis showed that the purified UL45Δ protein could be recognized by the rabbit anti-UL45Δ IgG and rabbit anti-DEV IgG, indicating that this specific antibody could serve as a good tool for penetrating studies of DEV UL45 protein function. The transcription phase and protein characteristics analysis indicated that UL45 gene of DEV was a late gene and UL45 protein may be associated with viral envelope. But we still need to do more penetrating studies to ascertain these conclusions.
Materials and methods
Virus, strains, vector and main reagents
DEV CHv-strain (a high-virulence field strain of DEV. Separated, identified and preserved by the author's laboratory); E. coli strain DH5α, E. coli BL21(DE3) PlysS and expression vector pET-32a(+) were preserved in the author's laboratory; Restriction enzymes and ligase mixture were purchased from TaKaRa.
PrimeSTAR HS (Premix) DNA polymerase, DNA and protein molecular weight markers, TIANpure Mini plasmid Kit (catalogue No: DP104-02), TIANgel Midi purification Kit (catalogue No: DP209-03), RNAprep Pure Cell/Bacteria Kit (catalogue No: DP430), Quant Reverse Transcriptase (catalogue No: ER103-04) were purchased from Tiangen Biotech Company; Horseradish peroxidase (HRP)-labeled goat anti-rabbit IgG was purchased from Zhongshan goldenbridge Biotechnology Company; Rabbit anti-DEV IgG was provided by the author's laboratory.
PCR amplification of the DEV UL45 and UL45Δ gene
Using 11 daytime duck-embryo (purchased from nonimmune region in Ya'an) to prepare the duck embryo fibroblast (DEF) [47]. 24 h later, inculated the DEV-CHv. When the cytopathic effect (CPE) reached 80% harvested the DEF. After three freeze-thaw cycles, using the method of phenol-chloroform to extracted DEV DNA [48].
Considering the order of UL45 (GenBank accession no: EU195107, not release yet), the primers for amplification of UL45 and UL45Δ gene were designed using biological software Oligo6.0 and synthesized by TaKaRa. The forward primers of UL45 and UL45Δ (F1/F2) were 5'-GGATCC CGGATCACCCTAACAATG-3' and 5'-GGATCC ACTACAGCGTGGGATACGA-3', the same reverse primer (R1) was 5'-CTCGAG AAACACGCATACAAATAACAAGTC -3', containing the BamHI and XHo I restriction sites (underlined), respectively. Using the genome of DEV CHv strain as the template, the PCR reactions (50 μl/tube) containing 25 μl PrimeSTAR HS (Premix) DNA polymerase, 1.4 μl of each primer (10pmol each), 2.6 μl DNA template and 19.6 μl ultrapure water was performed. The condition of PCR amplification was initial denaturation at 94°C for 5 min followed by 30 consecutive cycles of denaturation at 94°C for 60 s, annealing at 60.5°C for 60 s, and extension at 72°C for 60 s, and then a final extension at 72°C for 10 min. The amplified products were analyzed by electrophoresis on a 1.2% (w/v) agarose gel.
Construct the expression plasmids PET32a(+)/UL45 and PET32a(+)/UL45Δ
The PCR products of UL45 and UL45Δ gene were sent to TaKaRa for constructing cloning plasmids. The pMD18-T/UL45 and pMD18-T/UL45Δ plasmids were confirmed by sequencing. The UL45 gene digested from pMD18-T/UL45 (retrieving by TIANgel Midi purification Kit) was directionally ligated into the previously BamHI /XHcI -digested expression vector pET-32a(+). The ligation mixture was transformed into competent E. coli DH5α cells for storing. The positive colony was identified by PCR and restriction analysis. Extracted positive plasmids and transformed into competent E. coli BL21 (DE3) PlysS strain. Do the same operation to the cloning plasmids pMD18-T/UL45Δ.
Protein expression, Purification and polyclonal antibody production
Inoculating UL45 and UL45Δ positive cloning strain into 5 ml LB/AMP liquid medium respectively and cultivating overnight. 50 μl cultures were inoculated to 5 ml LB/Amp to activation. When the bacterium reached logarithmic phase (at OD600 of 0.5-0.6), adding IPTG (final concentration 0.2 mM) to induce the expression of UL45 and UL45Δ protein. The situation of protein expression was analyzed by SDS-PAGE. The un-induced and vector control culture were analyzed in parallel. To increase the production of the recombinant protein, expression conditions including the temperatures, concentrations of IPTG and durations of induction were optimized.
The UL45Δ protein was purified by IMAC on Ni2+-NTA affinity resin and salting-out. The samples from Ni-column and sediments from salting-out were assessed by SDS-PAGE. The purified protein was used to immune New Zealand white rabbits to raise antibody. The antiserum was harvested from the jugular vein and stored at -70°C.
Purify the antiserum and Western blot analysis
First, the rabbit anti-UL45ΔIgG was precipitated from the polyclonal antiserum by ammonium sulfate precipitation [49]. Then, using a DEAE-Sepharose column, the IgG fraction was purified by ion-exchange column chromatography following the manufacturer's instructions [50]. To characterize the antigenicity of the UL45Δ fusion protein, western blot analysis was performed according to the standard procedure using the purified rabbit anti-UL45Δ IgG and rabbit anti-DEV IgG [51].
Transcription characteristics analysis of DEV UL45 gene
The DEF was produced by the usual method [47]. When the DEF grew monostratum, inoculated the DEV-CHv. Total cellular RNA was extracted at 0, 0.5, 1, 2, 4, 6, 8, 10, 12, 18, 24, 30, 36, 42, 48, 54, 60, 66 and 72 h post-infection (pi) using the RNAprep Pure Cell/Bacteria Kit. Then the RNA was immediately inversed transcribed to the cDNA by Quant Reverse Transcriptase and stored at -70°C. At the same time detecting the integrality and purity of RNA by electrophoresis on a 3.0% agarose gel and nucleic acid-protein detecting instrument. The fluorescent quantitative real-time PCR (FQ-PCR) primers for UL45 and β-actin (used as the internal parameters, its expression level is relative constancy in cells) were UL45 F (5'- CATGGAGTTGGGTGTGCT -3') and UL45 R (5'-ACGCTGTAGTCGGTATCG -3'), β-actin F (5'-CCGGGCATCGCTGACA-3') and β-actin R (5'- GGATTCATCATACTCCTGCTTGCT-3'). Standard curve of PMD18-T/UL45 and PMD18-T/β-actin (constructed and preserved in the author's laboratory) was established. The transcription kinetics of the DEV UL45 gene during the viral infection was detected by the method of FQ-PCR. The real-time FQ-PCR was performed in an 20 μl reaction mixture containing cDNA 2 μl, SYBR Green I Mix 9 μl, each of the primer 25 pmo1, adding ultrapure water to total volume. Each run consisted of initial denaturation at 95°C for 1 min followed by 40 consecutive cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s. Then the fluorescence was measured by 94°C for 60 s and 60°C for 60 s, and then followed by 70 consecutive cycles of 60°C for 10 s, with each cycle increased 0.5°C. β-actin served as the internal parameters done the parallel experiment. Samples and internal parameters were tested in triplicate. The method of 2-ΔΔCt was convenient to measure the relative amount of the UL45 mRNA expression [52].
Characteristics analysis of UL45 protein
The purified DEV was got from ultra-high speed centrifugation [53, 54]. Extracted the purified virion with NP-40 detergent and centrifuged at 70,000 rpm for 60 min at 4°C (Hitachi), and obtaining a supernatant fraction containing the detergent soluble proteins (envelope and minor amounts of some tegument proteins) and a pellet (the nucleocapsids and tegument proteins) [28]. The purified virion and viral ingredients were then analyzed by western blot using the rabbit anti-UL45Δ IgG [51].
References
Baudet A: Mortality in ducks in the Netherlands caused by a filtrable virus; fowl plague. Tijdschr Diergeneeskd 1923, 50: 455-459.
Campagnolo E, Banerjee M, Panigrahy B, Jones R: An outbreak of duck viral enteritis (duck plague) in domestic Muscovy ducks (Cairina moschata domesticus) in Illinois. Avian Diseases 2001, 522-528. 10.2307/1592999
Proctor S: Pathogenesis of duck plague in the bursa of Fabricius, thymus, and spleen. American journal of veterinary research 1976, 37: 427-431.
Shawky S, Sandhu T, Shivaprasad H: Pathogenicity of a low-virulence duck virus enteritis isolate with apparent immunosuppressive ability. Avian Diseases 2000, 590-599. 10.2307/1593098
Sandhu T, Leibovitz L: Duck virus enteritis (Duck plague). Diseases of Poultry 2003, 11: 354-363.
Fauquet C, Mayo M, Maniloff J, Desselberger U, Ball L: Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. elsevier academic press London; 2005.
Pan H, Cao R, Liu L, Niu M, Zhou B, Chen P, Hu J: Molecular cloning and sequence analysis of the duck enteritis virus UL5 gene. Virus Research 2008, 136: 152-156. 10.1016/j.virusres.2008.05.003
Plummer P, Alefantis T, Kaplan S, O'Connell P, Shawky S, Schat K: Detection of duck enteritis virus by polymerase chain reaction. Avian Diseases 1998, 554-564. 10.2307/1592682
Han X, Wang J, Ma B: Cloning and sequence of glycoprotein H gene of duck plague virus. Agricultural Sciences in China 2006, 5: 397-402. 10.1016/S1671-2927(06)60067-9
Jia R, Cheng A, Wang M, Xin H, Guo Y, Zhu D, Qi X, Zhao L, Ge H, Chen X: Analysis of synonymous codon usage in the UL24 gene of duck enteritis virus. Virus Genes 2009, 38: 96-103. 10.1007/s11262-008-0295-0
Liu S, Chen S, Li H, Kong X: Molecular characterization of the herpes simplex virus 1 (HSV-1) homologues, UL25 to UL30, in duck enteritis virus (DEV). Gene 2007, 401: 88-96. 10.1016/j.gene.2007.06.022
An R, Li H, Han Z, Shao Y, Liu S, Kong X: The UL31 to UL35 gene sequences of Duck enteritis virus correspond to their homologs in Herpes simplex virus 1. Acta virologica 2008, 52: 23-30.
Wei X, Anchun C, Mingshu W, Hua C, Dekang Z, Qihui L, Renyong J, Xiaoyue C: Expression and characterization of the UL31 protein from duck enteritis virus. Virology Journal 2009, 6: 19-28. 10.1186/1743-422X-6-19
Cai M, Cheng A, Wang M, Zhao L, Zhu D, Luo Q, Liu F, Chen X: His6-Tagged UL35 Protein of Duck Plague Virus: Expression, Purification, and Production of Polyclonal Antibody. Intervirology 2009, 52: 141-151. 10.1159/000221833
Zhao L, Cheng A, Wang M, Yuan G, Jia R, Zhou D, Qi X, Ge H, Sun T: Identification and characterization of duck enteritis virus dUTPase gene. Avian Diseases 2008, 52: 324-331. 10.1637/8169-110607-ResNote.1
Shen C, Guo Y, Cheng A, Wang M, Zhou Y, Lin D, Xin H, Zhang N: Characterization of subcellular localization of duck enteritis virus UL 51 protein. Virology Journal 2009, 6: 92. 10.1186/1743-422X-6-92
Liu F, Ma B, Zhao Y, Zhang Y, Wu Y, Liu X, Wang J: Characterization of the gene encoding glycoprotein C of duck enteritis virus. Virus Genes 2008, 37: 328-332. 10.1007/s11262-008-0266-5
Chang H, Cheng A, Wang M, Guo Y, Xie W, Lou K: Complete nucleotide sequence of the duck plague virus gE gene. Archives of Virology 2009, 154: 163-165. 10.1007/s00705-008-0284-6
Zhao Y, Wang J, Liu F, Ma B: Molecular analysis of US10, S3, and US2 in duck enteritis virus. Virus Genes 2009, 38: 243-248. 10.1007/s11262-008-0315-0
Liting Lu, Anchun Cheng, Mingshu Wang, Jinfeng Jiang, Dekang Zhu, Renyong Jia, Qihui Luo, Fei Liu, Zhengli Chen, Xiaoyue Chen, Yang J: Polyclonal antibody against DPV UL46 M protein can be a diagnostic candidate. Virology Journal 2010, 7: 83. 10.1186/1743-422X-7-83
Fu xiao Shen, An chun Cheng, Ming shu Wang, Hai bin Huang, Chuan feng Li, Jin feng Jiang, Ren yong Jia, De kang Zhu, Chen Xy: Factors affecting synonymous codon usage bias in the gC gene of DPV CHv strain. African Journal of Microbiology Research 2010, 4: 343-353.
YongPing Wen, Anchun Cheng, Mingshu Wang, Han Ge, ChanJuan Shen, Jun Xiang, RenYong Jia, Dekang Zhu, Xiaoyue Chen, Bei Lian, et al.: A TK recombinant protein-based ELISA for detecting antibodies to Duck Plague Virus. Virology Journal 2010, 7: 77. 10.1186/1743-422X-7-77
Qing Zou, Kunfeng Sun, Anchun Cheng, Mingshu Wang, Chao Xu, Dekang Zhu, Renyong Jia, Qihui Luo, Yi Zhou, Zhengli Chen, Chen X: Detection of anatid herpesvirus 1 gC gene by TaqManTM fluorescent quantitative real-time PCR with specific primers and probe. Virology Journal 2010, 7: 37. 10.1186/1743-422X-7-37
Xin H, Cheng A, Wang M, Jia R, Shen C, Chang H: Identification and Characterization of a Duck Enteritis Virus US3-Like Gene. Avian Diseases 2009, 53: 363-369. 10.1637/8643-020409-Reg.1
Yao Zhang, Anchun Cheng, Mingshu Wang, Dekang Zhu, Renyong Jia, Fei Liu, Qihui Luo, Chen. X: Analysis of Synonymous Codon Usage in the UL26. 5 Gene of Duck Enteritis Virus. IEEE, the 2nd International Conference on BioMedical Engineering and Informatics, Tianjing, China 2009, 4: 1829-1835.
Jun Xiang, Anchun Cheng, Mingshu Wang, Hua Chang, Chen. W: Molecular Cloning and Sequence Analysis of the Duck Enteritis Virus Nucleocapsid Gene(UL38). IEEE, the 2nd International Conference on BioMedical Engineering and Informatics, Tianjing, China 2009, 4: 1874-1880.
Zhong XR, Cheng AC, Wang MS, Gong YQ, Zhu DK, Jia RY, Luo QH: Molecular cloning and sequence analysis of UL34 gene from duck plague virus CHv strain. IEEE, the 2nd International Conference on BioMedical Engineering and Informatics, Tianjing, China 2009, 3: 1478-1485.
Visalli RJ, Brandt CR: The HSV-1 UL45 18 kDa gene product is a true late protein and a component of the virion. Virus Res 1993, 29: 167-178. 10.1016/0168-1702(93)90057-T
Yanagida N, Yoshida S, Nazerian K, Lee L: Nucleotide and predicted amino acid sequences of Marek's disease virus homologues of herpes simplex virus major tegument proteins. Journal of General Virology 1993, 74: 1837-1845. 10.1099/0022-1317-74-9-1837
Kato A, Sato I, Ihara T, Ueda S, Ishihama A, Hirai K: Homologies between herpesvirus of turkey and Marek's disease virus type-1 DNAs within two co-linearly arranged open reading frames, one encoding glycoprotein A. Gene 1989, 84: 399-405. 10.1016/0378-1119(89)90136-4
Ushijima Y, Luo C, Goshima F, Yamauchi Y, Kimura H, Nishiyama Y: Determination and analysis of the DNA sequence of highly attenuated herpes simplex virus type 1 mutant HF10, a potential oncolytic virus. Microbes and infection 2007, 9: 142-149. 10.1016/j.micinf.2006.10.019
Lembo D, Donalisio M, Hofer A, Cornaglia M, Brune W, Koszinowski U, Thelander L, Landolfo S: The ribonucleotide reductase R1 homolog of murine cytomegalovirus is not a functional enzyme subunit but is required for pathogenesis. The Journal of Virology 2004, 78: 4278-4288. 10.1128/JVI.78.8.4278-4288.2004
Patrone M, Percivalle E, Secchi M, Fiorina L, Pedrali-Noy G, Zoppe M, Baldanti F, Hahn G, Koszinowski UH, Milanesi G, Gallina A: The human cytomegalovirus UL45 gene product is a late, virion-associated protein and influences virus growth at low multiplicities of infection. J Gen Virol 2003, 84: 3359-3370. 10.1099/vir.0.19452-0
Visalli RJ, Brandt CR: Mutation of the herpes simplex virus 1 KOS UL45 gene reveals dose dependent effects on central nervous system growth. Arch Virol 2002, 147: 519-532. 10.1007/s007050200004
Haanes EJ, Nelson CM, Soule CL, Goodman JL: The UL45 gene product is required for herpes simplex virus type 1 glycoprotein B-induced fusion. J Virol 1994, 68: 5825-5834.
Oettler D, Kaaden OR, Neubauer A: The equine herpesvirus 1 UL45 homolog encodes a glycosylated type II transmembrane protein and is involved in virus egress. Virology 2001, 279: 302-312. 10.1006/viro.2000.0690
Visalli RJ, Brandt CR: The HSV-1 UL45 gene product is not required for growth in Vero cells. Virology 1991, 185: 419-423. 10.1016/0042-6822(91)90790-I
Mierendorf R, Yeager K, Novy R: The pET system: your choice for expression. Innovations 1994, 1: 1-3.
Studier F, Rosenberg A, Dunn J, Dubendorff J: Use of T7 RNA polymerase to direct expression of cloned genes. Methods in enzymology 1990, 185: 60-89. full_text
Studier F, Moffatt B: Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. Journal of Molecular Biology 1986, 189: 113-130. 10.1016/0022-2836(86)90385-2
Brown C, Stockwell P, Trotman C, Tate W: The signal for the termination of protein synthesis in procaryotes. Nucleic acids research 1990, 18: 2079-2086. 10.1093/nar/18.8.2079
Chen H, Xu Z, P C: High-level expression of human beta-defensin-2 gene with rare codons in E. coil cell-free system. Protein Pept Lett 2006, 13: 155-162. 10.2174/092986606775101724
Chumpolkulwong N, Aakamoto K, Hayashi A: Translation of 'rare'codons in a cell-free protein synthesis system from Esche-richia coli. J Struct Funct Genomics 2006, 7: 31-36. 10.1007/s10969-006-9007-y
Weir J: Regulation of herpes simplex virus gene expression. Gene 2001, 271: 117-130. 10.1016/S0378-1119(01)00512-1
Gale M Jr, Tan S, Katze M: Translational control of viral gene expression in eukaryotes. Microbiology and molecular biology reviews 2000, 64: 239-280. 10.1128/MMBR.64.2.239-280.2000
Gale MJ, Tan SL, Katze MG: Translational control of viral gene expression in eukaryotes. Microbiol Mol Biol Rev 2000, 64: 239-280. 10.1128/MMBR.64.2.239-280.2000
Chen SH, Han ZX, Shao YH, Liu SW, G KX: Molecular characterization of UL6 gene of duck enteritis virus. Chinese Journal of Virology 2006, 22: 391-396.
J LZ, F LY, S C, X SM, L LY: Comparison of extraction method of genomic DNA of duck plague virus. Journal of Shandong Normal University(Natural Science) 2008, 23: 134-136.
Temponi M, Kageshita T, Perosa F: Purification of murine IgG monoclonal antibodies by precipitation with caprylic acid: comparison with other methods of purification. Hybridoma 1989, 8: 85-95. 10.1089/hyb.1989.8.85
McGuire JM, Douglas M, D SK: The resolution of the neutral N-linked oligosaccharides of IgG by high pH anion-exchange chromatography. Carbohydral Research 1996, 292: 1-9.
Towbin H, Staehelin T, Gordon J: Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proceedings of the National Academy of Sciences 1979, 76: 4350-4354. 10.1073/pnas.76.9.4350
Livak KJ, D ST: Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25: 402-408. 10.1006/meth.2001.1262
Guo Y, Cheng A, Wang M, Zhou Y: Purification of anatid herpesvirus 1 particles by tangential-flow ultrafiltration and sucrose gradient ultracentrifugation. Journal of Virological Methods 2009, 161: 1-6. 10.1016/j.jviromet.2008.12.017
Wen M, Cheng A, Wang M, Wu T: Comparison of purification methods of duck plague virus. Journal of Mountain Agriculture and Biology 2006, 25: 115-120.
Acknowledgements
The research was supported by Changjiang Scholars and Innovative Research Team in University (PCSIRT0848), the earmarked fund for Modern Agro-industry Technology Research System (nycytx-45-12).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AS and GM carried out most of the experiments and wrote the manuscript. AC and MW critically revised the experiment design and the manuscript. DL, LL, TZ, DZ, QL, RJ, ZC, YZ and XC helped with the experiment. All the authors read and approved the final manuscript.
Ai-Mei Shen, Guang-Peng Ma contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Shen, AM., Ma, GP., Cheng, AC. et al. Transcription phase, protein characteristics of DEV UL45 and prokaryotic expression, antibody preparation of the UL45 des-transmembrane domain. Virol J 7, 232 (2010). https://doi.org/10.1186/1743-422X-7-232
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-7-232