Abstract
Background
Proteins of human and animal viruses are frequently expressed from RNA polymerase II dependent expression cassettes to study protein function and to develop gene-based vaccines. Initial attempts to express the G protein of vesicular stomatitis virus (VSV) and the F protein of respiratory syncytial virus (RSV) by eukaryotic promoters revealed restrictions at several steps of gene expression.
Results
Insertion of an intron flanked by exonic sequences 5'-terminal to the open reading frames (ORF) of VSV-G and RSV-F led to detectable cytoplasmic mRNA levels of both genes. While the exonic sequences were sufficient to stabilise the VSV-G mRNA, cytoplasmic mRNA levels of RSV-F were dependent on the presence of a functional intron. Cytoplasmic VSV-G mRNA levels led to readily detectable levels of VSV-G protein, whereas RSV-F protein expression remained undetectable. However, RSV-F expression was observed after mutating two of four consensus sites for polyadenylation present in the RSV-F ORF. Expression levels could be further enhanced by codon optimisation.
Conclusion
Insufficient cytoplasmic mRNA levels and premature polyadenylation prevent expression of RSV-F by RNA polymerase II dependent expression plasmids. Since RSV replicates in the cytoplasm, the presence of premature polyadenylation sites and elements leading to nuclear instability should not interfere with RSV-F expression during virus replication. The molecular mechanisms responsible for the destabilisation of the RSV-F and VSV-G mRNAs and the different requirements for their rescue by insertion of an intron remain to be defined.
Similar content being viewed by others
Background
Eukaryotic cells differ from prokaryotic cells by increased compartmentalisation of the intracellular environment to facilitate complex enzymatic reactions required for efficient protein expression and modification, cell metabolism and/or cell division. Adaptation to the host cell and particularly to its expression machinery is the key requirement for the replication of any virus. Several RNA viruses only replicate in the cytoplasm of their eukaryotic host cell. These viruses possess their own transcription machinery involving a viral RNA-dependent RNA polymerase which allows cytoplasmic mRNA synthesis from the viral genomic RNA. Therefore, these viruses are not adapted to the complex nuclear milieu of the eukaryotic host cell. Inefficient expression of genes from RNA viruses by RNA polymerase II (Pol II) dependent cellular promoters might be explained by lack of critical elements required for pre-mRNA stabilisation, mRNA processing and/or nuclear export. However, problems that occur during Pol II dependent expression of RNA virus proteins can be overcome by changing the codons of viral genes to those most frequently used by the genes of the host cells [1–3]. Since the codon optimised genes should also lack defined RNA elements directing mRNA processing and/or transport, the nucleotide sequence or composition of the viral wild type sequences might actually be inhibitory in nature or be targeted by innate viral defence mechanisms.
The precise reason why genes of RNA viruses are inefficiently expressed is still poorly understood. For lentiviruses, which were studied in more detail, expression of viral structural genes is regulated at the level of nuclear export and these viruses have a regulatory protein (Rev) involved in shuttling the mRNA for the structural proteins from the nucleus to the cytoplasm [4]. Retention of these lentiviral mRNAs in the nucleus has been attributed to cis-repressive sequences or regions of instability but these sequences could not be narrowed down to well-defined nucleotide motifs. The unusual low GC content has also been reported to be responsible for the nuclear instability of lentiviral structural mRNAs [5]. Whether similar mechanisms govern the fate of recombinant Pol II mRNAs of viruses replicating in the cytoplasm is unclear.
Instead of using cellular RNA polymerases for expression of viral proteins in eukaryotic cells, cytoplasmic expression systems based on RNA polymerases from vaccinia viruses, alpha-viruses or phages have been developed. The latter are also used for generation of recombinant vesicular stomatitis virus (VSV) [6, 7] and respiratory syncytial virus (RSV) [8] by reverse genetics. These systems are based on cytoplasmic transcription of viral cDNA by coexpression of phage T7 RNA polymerase. Recovery of infectious viruses was achieved by cotransfection of T7 RNA polymerase dependent expression plasmids for full-length antigenomic RNA and viral helper proteins which are necessary and sufficient for both RNA-replication and transcription. Expression of these viral helper proteins and/or the antigenomic RNA transcripts by eukaryotic promoters might facilitate and improve strategies for production of such recombinant viruses.
Additionally, the lack of eukaryotic expression systems not depending on coexpressed cytoplasmic polymerases hampered DNA vaccine development for several RNA viruses. This is a particular problem for the development of RSV vaccines, since immunisation with whole inactivated virus particles led to enhancement of RSV disease in children not protected from RSV infection [9, 10]. An aberrant T-helper cell type 2 response to the G protein of RSV and excessive CD4+ and CD8+ T cell responses to the F protein of RSV might be responsible for the enhanced airway inflammation underlying the detrimental effect of vaccination [11]. Expression of a single viral protein by a DNA vaccine triggering T-helper cell type 1 responses might overcome vaccine-induced enhancement of RSV disease.
The potential of DNA vaccines and techniques used for reverse genetics has sparked our interest to better understand the requirements for expression of heterologous genes not adapted to the nuclear environment. Using the open reading frames of the G protein of VSV and the F protein of RSV as representatives of the rhabdovirus and paramyxovirus family, respectively, we analysed expression efficiency on mRNA and protein levels. We also attempted to rescue expression of these viral ORF by more subtle changes than codon optimisation to get hints on mechanisms responsible for inefficient expression of these viral genes.
Results
Expression of the VSV-G protein can be rescued by insertion of the CMV-IE 5'-untranslated region independent of splicing
Heterologous genes are commonly expressed in eukaryotic cells by cloning the ORF into a Pol II dependent expression vector containing a strong constitutive promoter and a polyadenylation signal (poly(A) signal). For expression of the G protein of VSV expression plasmids pGwt and pGsyn, containing either the wild type or codon optimised ORF under the control of the human cytomegalovirus immediate early promoter and enhancer (CMV-IE, [12]), were transfected into 293T cells (Fig. 1A). Neither mRNA nor protein expression could be detected after transfection of the wild type constructs (Fig. 1B, C). By contrast, the codon optimised construct led to efficient expression of both VSV-G mRNA and protein. Since the probe used for the Northern blot analysis targets the transcribed region of the bovine growth hormone polyadenylation signal (BGH poly(A) [13]) present in the codon optimised and the wild type expression plasmid, the intensity of the bands should directly reflect mRNA expression levels. Since the amino acid sequences encoded by the codon optimised and the wild type ORF are identical, both proteins should be detected with the same sensitivity in Western blot analysis. Detection of VSV-G after transfection of the codon optimised expression plasmid also excludes the possibility that lack of detectable levels of VSV-G after transfection of the wild type construct is due to instability of the protein. The VSV-G expression plasmids were cotransfected with lentiviral gag-pol expression plasmids and a lentiviral vector construct to assess expression levels of VSV-G by a sensitive VSV-G dependent transduction assay. Vector titers obtained with the wild type construct were at least 100-fold lower than those obtained with the codon optimised expression plasmid (data not shown). Since the cotransfected gag-pol expression plasmid also contained the BGH poly(A), the Northern blot analyses also detected the encoded gag-pol mRNA migrating at a size of approximately 5 kb. Similar gag-pol mRNA levels (Fig. 1B) confirm that the differences observed in VSV-G expression are not due to experimental variations.
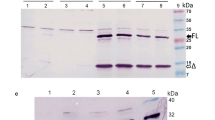
Characterisation of VSV-G expression plasmids. A) Map of VSV-G expression plasmids. Wild type (wt) or codon optimised (syn) open reading frames of VSV-G are flanked by the human cytomegalovirus immediate early promoter/enhancer region (CMV) and the bovine growth hormone poly(A) signal (pA). Angled black arrows mark the transcriptional start point. The pIGwt vector contains intron A and flanking untranslated exonic regions E1 and E2 of the cytomegalovirus immediate early gene. In pIΔIGwt the exon boundaries were precisely fused by deleting the intron. B) Northern blot analysis. Cells were cotransfected with the indicated VSV-G expression plasmids, a codon optimised HIV-1 gag-pol expression plasmid (Hgpsyn) and the lentiviral vector construct VICGΔBH containing a GFP expression cassette. Poly(A) RNA was isolated from transfected cells and analysed by Northern blot with a probe spanning the transcribed region of the BGH poly(A) signal present on all VSV-G transcripts and the positive 5 kb HIV-1 gag-pol transcript. C) Western blot analyses. Cells were cotransfected with the indicated VSV-G expression plasmids, an SIV gag-pol expression plasmid (SgpΔ2) and the lentiviral vector construct VICGΔBH containing a GFP expression cassette. Monoclonal antibodies to HIV-1 p24 capsid protein, which is cross reactive to SIV p27, or to VSV-G, respectively, were used for detection of the viral proteins in lysates of transfected cells.
Inserting the first intron of CMV-IE gene including exonic flanking regions restored VSV-G expression from the wild type ORF to levels comparable to those obtained by the codon optimised expression plasmid. Despite a lower transfection efficiency, as evident from the Northern blot analysis (Fig. 1B), VSV-G mRNA expression was clearly detectable (pIGwt in Fig. 1B). Protein expression levels were comparable to those obtained with the codon optimised expression plasmid (pIGwt vs pGsyn in Fig. 1C, left panel). However, splicing was not required for this rescue, since a DNA expression plasmid, in which the CMV intron had been deleted by fusing the splice sites and retaining the exonic sequences also led to efficient expression of the protein (compare pIGwt to pIΔIGwt in Fig. 1C, right panel). Thus, correctly fused exons were sufficient to enhance VSV-G expression levels.
Further deletion analyses revealed that the first 106 nucleotides of the 5'-exon are mediating most of the effect (data not shown).
Expression of the RSV-F mRNA is dependent on splicing
An analogous expression plasmid containing the wild type RSV-F ORF under the control of the CMV-IE promoter-enhancer (pFwt in Fig. 2A) also failed to express detectable levels of RSV-F mRNA and protein (Fig. 2B,C). After insertion of the first intron of CMV-IE gene with the exonic flanking regions into the wild type RSV-F expression plasmid, RSV-F mRNA could be detected in the cytoplasm of transfected cells (pIFwt in Fig. 2B). However, RSV-F protein remained undetectable. In contrast to VSV-G, splicing seemed to be required, as selective removal of the intronic sequences (pIΔIFwt) abolished detection of cytoplasmic RSV-F mRNA (Fig. 2B). To exclude the possibility that our failure to detect RSV-F protein is due to instability of RSV-F or poor antibody reagents, we also analysed a codon optimised RSV-F expression plasmid encoding the same RSV-F amino acid sequence as the wild type constructs. Expression of RSV-F was readily detectable after transfection of the codon optimised expression plasmid independent of the presence or absence of the intron (pFsyn and pIFsyn in Fig. 2C).
Characterisation of RSV-F expression plasmids. A) Map of RSV-F expression plasmids. Wild type (wt) or codon optimised (syn) open reading frames of RSV-F are flanked by the human cytomegalovirus immediate early promoter/enhancer region (CMV) and the bovine growth hormone poly(A) signal (pA). Angled black arrows mark the transcriptional start point. The pIFwt and pIFsyn plasmids contain intron A and flanking untranslated exonic regions E1 and E2 of the cytomegalovirus immediate early gene. In pIΔIFwt the exon boundaries were precisely fused by deleting the intron. B) Northern blot analysis. 293T cells were transfected with the indicated plasmids containing the RSV-F ORF. As a negative control the empty vector (pcDNA3.1) was also transfected and processed in parallel. A total RNA extract from RSV-infected HEp2-cells served as a positive control. Cytoplasmic (C) or total (T) RNA was isolated from transfected cells, separated by agarose gel electrophoresis and used for subsequent Northern blot analysis. Size separated RNA was stained with ethidiumbromide (EtBr) revealing non-degraded 18S and 28S ribosomal RNA bands (18S shown as representative). A DIG-labelled probe spanning 780 bp of the RSV-F ORF was used for hybridisation. C) Western blot analysis. 293T cells were transfected with the indicated plasmids. Equal amounts of protein were separated on an acrylamide gel for subsequent detection of RSV-F expression in Western blot analysis using a monoclonal antibody against the F protein. As a positive control, RSV-infected HEp2 cells were processed in parallel.
Pol II mediated expression of the wild type RSV-F ORF results in premature polyadenylation
Undetectable levels of RSV-F protein in the presence of cytoplasmic RSV-F mRNA suggested an additional block at the translational level. We noticed that the mRNA species detected in the Northern blot analysis (Fig. 2B) migrated faster than the viral RSV-F mRNA, although they should be slightly larger due to the extended 5'- and 3'-UTR.
To analyse correct splicing of the RSV-F mRNA, cytoplasmic RNA of 293T cells transfected with pIFwt was isolated and reverse transcribed by an oligo-dT primer. A PCR spanning the splice sites in the 5'-untranslated region was performed (see Fig. 3A). Size and sequence analysis (not shown) of the PCR product revealed correct splice junctions and no other deviation from the expected transcript (Fig. 3B).
Analysis of RSV-F mRNA processing. A) Map of exon-intron structure and poly(A) signals of the precursor mRNA encoded by pIFwt. Arrows indicate location of primers used for the PCR analyses. The scale indicates the distance to the transcriptional start site. AATAAA: consensus signal for polyadenylation. B) Characterisation of splicing. 293T cells were transfected with pIFwt. Cytoplasmic RNA was isolated from transfected cells and reverse transcribed by oligo-dT priming (pIFwtcDNA). A PCR spanning the splice sites was performed with primers: 5'UTR-s and RSV-F-ia. The size of the PCR-products was compared to the size obtained in parallel PCR using pIΔIFwt and pIFwt plasmid-DNA (pDNA) as templates. H2O: negative control. C) Characterisation of poly(A) signal usage. 293T cells were transfected with the indicated plasmids. Cytoplasmic RNA was isolated from transfected cells and reverse transcribed by priming with Oligo(dT)Add-a. As a control for DNA contamination, the reverse transcription reaction was also performed without the enzyme (-). The cDNA was amplified by PCR with primers 5' UTR-s and Oligo(dT)Add-a and the size of the PCR products was determined by agarose gel electrophoresis. D) PCR products from pIFwt transfected cells were cloned and sequenced. The 3' end of the sequence obtained in 9 of 10 clones (RSV-F mRNA exp.) is shown aligned to the RSV-F sequence of the parental plasmid (RSV-F mRNA theor.).
However, inspection of the RSV-F sequence revealed four potential polyadenylation consensus signals (AATAAA) [14] within the coding region (Fig. 3A). Using a PCR approach with an antisense primer anchored at the poly(A) tail (Oligo(dT)Add-a, Fig. 3A) and a pcDNA3.1+ specific sense primer at the 5'-UTR of the transcript (5' UTR-s, Fig. 3A), the entire mRNA transcript was reverse transcribed and amplified. Size and sequence analysis revealed that the second consensus poly(A) signal was used in 9 of 10 clones analysed (Fig. 3C, D) resulting in a mRNA with an RSV-F ORF truncated at position 1295. Since the absence of a stop codon has been shown to lead to degradation of such prematurely terminated proteins by cellular quality control pathways [15, 16], this might explain the absence of detectable levels of a truncated protein.
Deletion of the poly(A) consensus signal rescues RSV-F expression
Mutation of the consensus poly(A) signal 2 by a point mutation not affecting the protein sequence at position 1278 of the ORF (1st mutation: AATAAA → AACAAA in pIFwtΔ2) led to detectable full-length transcripts (Fig. 4A) and a faint RSV-F band became detectable in the Western blot analysis (Fig. 4C). Despite detection of full length transcripts, RSV-F protein levels after transfection of pIFwt Δ2 were more than 100-fold lower than those obtained after transfection of the codon optimised expression plasmid. In addition to full length transcripts a second band consistent in size with polyadenylation at the fourth consensus poly(A) signal was obtained by PCR analysis of RSV-F transcripts (Fig. 4A), which might be responsible for the poor expression observed at the protein level. Therefore, the second and fourth poly(A) signal were inactivated simultaneously by introducing an additional silent point mutation at position 1425 of the RSV-F ORF (2nd mutation: AATAAA → AATCAA: pIFwtΔ24). This led to detection of only full length mRNA transcripts (Fig. 4A, B) and expression of RSV-F protein was increased relative to the pIFwt Δ2 construct. However, despite detection of full-length mRNA in the cytoplasm, protein expression was at least 50-fold less efficient than expression from the codon optimised plasmid (Fig. 4C).
Characterisation of poly(A) signal mutants. A) Poly(A) signal usage after transfection of the indicated plasmids into 293T cells was characterised as described in figure legend 3C. Plasmids pIFwt Δ2 and pIFwt Δ24 contain mutations in the second or the second and forth consensus poly(A) signal, respectively. B) Northern blot analysis of cytoplasmic RNA of 293T cells transfected with the indicated plasmids or HEp2 cells infected with RSV. Size separated RNA was stained with ethidiumbromide (EtBr) revealing non-degraded 18S and 28S ribosomal RNA bands (18S shown as representative). A DIG-labelled probe from the RSV-F ORF was used for hybridisation. C, D) Western blot analysis. 293T cells were cotransfected with an EGFP expression plasmid and the indicated RSV-F expression plasmids and analysed by Western blot using an anti-RSV-F monoclonal antibody. The lysate of pIFsyn transfected cells was diluted from 1:10 to 1:104, while the lysates from the other transfected cell were loaded at a 1:1 dilution. Similar transfection efficiencies were controlled for by measuring the fluorescence activity of cell lysates (data not shown).
Virus infection does not enhance Pol II dependent RSV-F expression
Since the wild type ORF of RSV-F is readily translated in the context of the virus, this inefficient expression does not seem to be a general deficiency of the translation machinery or a consequence of rare codon usage. Due to the cytotoxicity of RSV-F we hypothesised that unidentified cis-repressive sequences in the wild type ORF might participate in regulation of RSV-F protein expression during the viral replication cycle, and that another viral protein might activate RSV-F protein expression at the translational level. To test this possibility, cells transfected with the wild type RSV-F expression plasmid with inactivated premature poly(A) signals were infected with RSV. To distinguish between RSV-F protein expressed from the expression plasmid or the virus, a 10 amino acid myc-tag was added to the C-terminus of the RSV-F ORF in pIFwtΔ24 resulting in pIFwtΔ24myc. Recombinant RSV expressing GFP was used and infection efficiency was controlled by fluorescence microscopy. Expression levels of myc-tagged RSV-F in transfected cells were not enhanced by viral infection (Fig. 5), thus failing to provide evidence for upregulation of RSV-F expression by another viral factor.
Influence of RSV infection on RNA Pol II dependent RSV-F expression levels. 293T cells were cotransfected with a Gaussia luciferase expression plasmid and the indicated RSV-F expression plasmids containing an inframe 3'-terminal myc-tag. Similar transfection efficiencies were controlled by measuring the Gaussia luciferase activity in cell supernatants (not shown). Six hours following transfection cells were infected with the GFP expressing recombinant RSV at an MOI of 2 (+). As negative control, cells were also left uninfected (-). GFP expression analysis revealed similar infection efficiencies (data not shown). Equal amounts of protein were analysed in non-reducing Western blot analysis using a monoclonal antibody to the myc-tag 48 h after transfection.
Chimeric ORF of RSV-F revealed strong dependency of protein expression on codon usage
To dissect the functional relevance of codon optimisation and premature polyadenylation more precisely, we also replaced the first 466 nt (pIFc1) or last 679 nt (pIFc3) of the wild type sequence with the codon optimised form (Fig. 6A). In the presence of functionally active premature poly(A) signals (pIFc1) replacement of the first third of the wild type ORF by the codon optimised version did not restore protein expression. Increased expression levels were obtained with the chimeric construct in which the relevant poly(A) signals were deleted (pIFc1Δ24) relative to the wild type sequence containing the mutated polyadenylation sites (pIFwt Δ24). A comparable increase of expression was observed, if the last third of the wild type ORF was exchanged by the codon optimised sequence. Codon optimisation of the entire sequence even led to at least 10-fold higher expression levels compared to the chimeric constructs, indicating that codon optimisation seems to be the major reason for the strong enhancement of protein levels once premature poly(A) signals are inactivated.
Expression levels of chimeric ORFs. A) Map of RSV-F expression plasmids containing wild type, codon optimised, or chimeric ORFs. Numbers in boxes indicate mutated consensus poly(A) signals. B) Western blot analysis of RSV-F protein levels after transfection of the indicated expression plasmids. An expression plasmid for EGFP was cotransfected. Undiluted lysates had equal protein content and a similar amount of fluorescent activity revealing constant transfection efficiency. Numbers above the lanes indicate the dilution at which the cell lysates were loaded on the gel.
Discussion
The results demonstrate striking differences in the requirements for expression of genes of cytoplasmic RNA viruses by DNA expression plasmids. The use of codon optimised expression plasmids allowed exclusion of the possibility that protein instabilities or degradation is responsible for undetectable levels of the respective viral proteins. Insertion of intron A of the CMV-IE gene resulted in mRNA levels comparable to those obtained by codon optimised expression plasmids in case of VSV-G or by those obtained in natural infection for RSV. This was not surprising since splicing has been repeatedly shown to enhance expression levels [17, 18]. However, in case of VSV-G splicing was not the critical factor, since simple addition of the exonic sequences of the CMV-IE gene were sufficient to rescue VSV-G expression even at the protein level. This suggests that the exonic sequences somehow stabilise and/or contribute to nuclear export of the VSV-G mRNA. The same exonic sequences were not sufficient to rescue expression of RSV-F mRNA. Including the intron, however, allowed cytoplasmic expression of the RSV-F mRNA. Similar findings have been reported for mRNAs of the Simian Virus 40, where intronless RNA was retained and degraded in the nucleus, while the same transcript generated by splicing reached the cytoplasm [19]. For other genes this has been attributed to recognition of the pre-mRNA by the exon junction complex (EJC), which has been found to be linked to nuclear export by direct binding to the heterodimer transport protein Tap-Nxt [20–22]. It is therefore likely that similar mechanisms are responsible for the rescue of cytoplasmic RSV-F mRNA levels by the first intron of the CMV-IE gene.
Another block to RSV-F protein synthesis was found to be premature polyadenylation. The second of the four consensus sites initiated the predominant premature polyadenylation of the RSV-F mRNA. The lack of a stop codon preventing an accurate translation termination results in synthesis of defective ribosomal products (DRiPs) which enter a pathway of proteasomal or other cytosolic decay mechanisms coupled to MHC class I presentation [23, 24]. This might explain why DNA vaccines encoding the wild type RSV-F ORF induced immune responses, although expression of full length protein was probably not very efficient [25–29]. The small amount of protein which could be detected despite that (Fig. 4D), might be the result of a rare skipping of poly(A) signals.
Mutagenesis of the recognised consensus sequence for polyadenylation led to usage of the last downstream consensus signal. However, even after mutagenising both used poly(A) signals, protein expression was around 50-fold lower than expression from codon optimised plasmids despite substantial amounts of correctly processed RSV-F mRNA in the cytoplasm. The fact that expression of other viral proteins by superinfection of the transfected cell with RSV did not increase RSV-F expression from the plasmid suggests that poor expression levels are not the consequence of a repressive RSV-F specific regulatory RNA element that can be overcome by an activating second viral factor. It rather seems that reducing the AU content of the RSV-F mRNA in general contributes to increased expression levels. Consistently, replacement of a third of the wild type nucleotide sequence by the codon optimised fragment resulted in intermediate expression levels and not in an all or none phenomenon. In summary, these findings indicate that premature polyadenylation is the major mechanism responsible for failure of protein expression from the original RSV-F wild type construct and that codon optimisation can further enhance expression of RSV-F.
Polyadenylation consensus signals were not only found in a single RSV strain but could be detected in all RSV-F sequences deposited in GenBank database. Other members of the paramyxovirus family, such as measles virus and parainfluenzaviruses, also harbour such consensus sites for polyadenylation. Since these consensus poly(A) signals are not expected to be of any functional relevance for the viruses due to their cytoplasmic replication, they are probably just the accidental result of the unusual high AU content of the viral genomes. The latter fact also leads to the presence of potential U-rich downstream elements that are also required for polyadenylation [30, 31].
Conclusion
Expression of genes of RNA viruses by Pol II dependent expression plasmids can be impaired at several steps. For VSV-G, a splicing-independent mechanism can lead to stabilisation of Pol II transcribed VSV-G mRNA, while splicing seems to be necessary for Pol II dependent expression of RSV-F mRNA. Premature polyadenylation is a second major block to expression of RSV-F protein from the wild type ORF. All these restrictions were efficiently overcome by codon optimisation providing a straightforward approach for the generation of Pol II dependent expression cassettes needed for development and production of antiviral vaccines and recombinant RNA viruses.
Methods
Viruses and infection
RSV based on the A2 long strain was kindly provided by B. Schweiger from the Robert Koch Institute, Berlin, Germany. GFP expressing recombinant rgRSV [32] was obtained by M. E. Peeples and P. L. Collins, Maryland, USA. RSV was passaged on Hep2 cells and stored at -80°C. Hep2 or 293T cells were infected at an MOI of 10 by adding RSV containing cell supernatant. Two hours following addition of the virus, supernatants were removed and cells were supplied with DMEM medium containing 0,5% FCS and 100 μg/ml penicillin G and streptomycin sulphate.
Expression plasmids
The ORF of VSV-G was amplified from pHIT-G [33] and cloned into pcDNA3.1 (Invitrogen, Karlsruhe, Germany) via BamHI/EcoRI (pGwt, kindly provided by R. Wagner, Regensburg). A Kozak consensus sequence (gccgccacc) [34] was inserted directly upstream of the start codon. For codon optimisation, viral codons were replaced by those most frequently used in human cells [35]. Synthesis of the optimised VSV-G encoding nucleotide sequence was performed by Geneart (Regensburg, Germany) based on the amino acid sequence of GenBank database entry J02428 (pGsyn). Amino acid 57 and 96 were mutated from L to I and H to Q, respectively, to match the amino acid sequence of the wild type VSV-G precisely. The CMV-IE intron A was added into both vectors by inserting the SnaBI/HindIII fragment (GenBank database entry BK000394, nt 174903–173696) of the VSV-G expression plasmid pHIT-G resulting in pIGwt and pIGsyn.
Deletion of the 828 nt intron and exact fusion of the exon boundaries was achieved by replacement of a SacII/HindIII fragment by the annealed oligonucleotides (Sigma, Munich, Germany) Is (5'-gg ccgggaacggtgcattggaacgcggattccccgtgccaagagtgactcaccgtccttgacacga) and Ia (5'-agctt cgtgtcaaggacggtgagtcactcttggcacggggaatccgcgttccaatgcaccgttcccggccgc) resulting in pIΔIGwt. Nucleotides involved in generation of restriction sites are printed bold. VSV-G expression analyses included studies on the functional incorporation of VSV-G into lentiviral vector particles. Therefore, lentiviral gag-pol expression plasmids Hgpsyn [2] for HIV-1 gag-pol and SgpΔ2 [36] for SIV gag-pol were cotransfected with VSV-G expression plasmids and the lentiviral vector construct VICGΔBH. VICGΔBH is based on the lentiviral vector VIGΔBH [36], containing a murine leukemia virus promoter driven GFP expression cassette. This cassette was excised via BglII/XhoI and replaced by the BamHI/XhoI fragment of HIV-CS-CG [37] containing a CMV-GFP expression cassette.
For the construction of the RSV-F expression plasmids, viral RNA was isolated from RSV containing cell supernatants using the QIAamp® viral RNA Mini Kit. After reverse transcription (ThermoScript™ RT-PCR System, Invitrogen) the RSV-F cDNA was amplified by PCR (Primers (Sigma): sense: 5'-gatccaagctt ccaccatggagttgccaatcctcaaa; antisense: 5'-tcgacctcgag ttagttactaaatgcaatattatttatacc) using the Platinum® Taq DNA-polymerase (Invitrogen). The 1.7 kb fragment including a Kozak sequence upstream of the ORF (ccacc) was subcloned into pcDNA3.1 (Invitrogen, Karlsruhe, Germany), or used to replace the VSV-G sequence pIGwt and pIΔI by digestion with HindIII/XhoI. Codon optimisation of the wild type ORF was performed by Geneart. The codon optimised ORF (GenBank database entry EF566942), also including a Kozak sequence (gccacc), was subcloned into pcDNA3.1 (Invitrogen) and pI vector by HindIII/XhoI restriction.
Deletion of the stop codon of the RSV-F ORF was achieved by PCR-directed mutagenesis. The RSV-Fsyn ORF without the stop codon was then subcloned into the pcDNA3.1(+) vector and the myc-tag was fused to the C-terminus of RSV-F by ligating annealed primers (Sigma) mycTAAs: 5'-tcgag gaacaaaaactcatctcagaagaggatctgtaat and mycTAAa: 5'-ctaga ttacagatcctcttctgagatgagtttttgttcc into the expression plasmid containing the RSV-F ORF lacking the stop codon via XhoI and XbaI sites.
Point mutations were introduced to the RSV-F ORF by overlap extension PCR and ligation of PpuMI/XhoI fragments.
Chimeric ORFs were produced by amplification of portions of the synthetic ORF and subcloning via HindIII/PpuMI or BsaBI/XhoI into the wild type expression vector pIFwt. All plasmids were confirmed by sequence analysis (Genterprise, Mainz, Germany).
Cells and transfection
293T and HEp2 cells were cultured in Dulbecco s modified Eagle s medium (Invitrogen) supplemented with 10% fetal calf serum (Invitrogen), penicillin G and streptomycin sulphate in a final concentration of 100 μg/ml each. Cells were transfected in 25 cm2 flasks with 5 μg plasmid-DNA by the calcium phosphate coprecipitation method as described elsewhere [38].
Control of transfection efficiency
To control transfection levels and guarantee comparable amounts of protein in lysates of transfected cells, plasmids for expression of reporter proteins were transfected additionally to VSV-G and RSV-F expression plasmids. In case of VSV-G expression analyses, cotransfection of lentiviral gag-pol expression plasmids Hgpsyn [2] for HIV-1 gag-pol and SgpΔ2 [36] for SIV gag-pol served as control in Western and Northern blot analyses, respectively. Cotransfection of the lentiviral vector VICG3ΔBH containing a GFP-expression cassette directly monitored transfection efficiency in treated cells. For RSV-F expression analyses cotransfection of an EGFP expression plasmid (pEGFP-C1, BD Biosciences Clontech, Heidelberg, Germany) and quantitative measurement of fluorescence activity in cell lysates guaranteed similar transfection efficiency. In transfected cells subsequently infected with rgRSV, transfection efficiency was controlled by transfection of an expression plasmid for Gaussia luciferase (pCMV-GLuc1; Targeting Systems, Santee, USA) and measurement of its activity in cell supernatants.
Western blot analysis
Transfected 293T cells were lysed 48 h following transfection. Equal amounts of total protein measured by Bradford-Assay (Biorad, Munich, Germany) were loaded on sodium dodecyl sulphate 8–12% polyacrylamide gels in reducing (500 mM TrisHCl pH 6,8; SDS; 20 v/v β-mercaptoethanol; 40 v/v Glycerin; 0,04% (w/v) PyroninY) or non reducing (without β-mercaptoethanol) Laemmli buffer. After protein separation and blotting on nitrocellulose membrane, proteins were incubated at 4°C over night with monoclonal antibody against either VSV-G (P5D4, Sigma-Aldrich, Munich, Germany), HIV-p24 (AIDS Research and Reference Reagent Program, Dr. Jonathan Allan [39]), RSV-F (18F12 [40]) or the myc-tag (9E10 [41]). After washing, the membrane was incubated with horseradish peroxydase-linked goat-anti-mouse-Fc antibody (SantaCruz, Heidelberg, Germany) and detected proteins were visualised by enhanced chemiluminescence reaction (Chemiglow®, Biozym, Hamburg, Germany).
Northern blot analysis
Total or cytoplasmic RNA was isolated from transfected 293T cells by RNeasy® Mini Kit (Qiagen, Hilden, Germany), mRNA was isolated by Fast Track 2.0 kit (Invitrogen, Karlsruhe, Germany). Concentration of purified RNA was determined by measuring absorbance at 260 nm. Five μg RNA was separated on an 1% agarose gel and blotted on nylon membrane. DIG-labelled probes where synthesised by PCR using the DIG synthesis kit (Roche, Mannheim, Germany). Transcripts were detected by hybridisation to a probe directed to either the BGH-poly(A) signal of the pcDNA3.1(+) (length: 130 bp; Primers: BGHs: 5'-gagtctagagggcccgtttaa; BGHa: 5'-aggaaaggacagtgggagtg) or the RSV-F ORF (length: 780 bp bp; Primers: RSV-Fis: 5'-ggtcctgcacttagaaggag; RSV-Fia: 5'-catgacacaatggctcctag). Oligonucleotides for probe synthesis PCR were derived from Sigma.
DIG-labelled nucleic acids were visualised by an alkaline phosphatase coupled anti-DIG antibody and CSPD substrate (Roche).
References
Haas J, Park EC, Seed B: Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol 1996, 6: 315-324. 10.1016/S0960-9822(02)00482-7
Wagner R, Graf M, Bieler K, Wolf H, Grunwald T, Foley P, Uberla K: Rev-independent expression of synthetic gag-pol genes of human immunodeficiency virus type 1 and simian immunodeficiency virus: implications for the safety of lentiviral vectors. Hum Gene Ther 2000, 11: 2403-2413. 10.1089/104303400750038507
Morton CJ, Cameron R, Lawrence LJ, Lin B, Lowe M, Luttick A, Mason A, Kimm-Breschkin J, Parker MW, Ryan J, Smout M, Sullivan J, Tucker SP, Young PR: Structural characterization of respiratory syncytial virus fusion inhibitor escape mutants: homology model of the F protein and a syncytium formation assay. Virology 2003, 311: 275-288. 10.1016/S0042-6822(03)00115-6
Sodroski J, Goh WC, Rosen C, Dayton A, Terwilliger E, Haseltine W: A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature 1986, 321: 412-417. 10.1038/321412a0
Schwartz S, Felber BK, Pavlakis GN: Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J Virol 1992, 66: 150-159.
Barr JN, Whelan SP, Wertz GW: Transcriptional control of the RNA-dependent RNA polymerase of vesicular stomatitis virus. Biochim Biophys Acta 2002, 1577: 337-353.
Lawson ND, Stillman EA, Whitt MA, Rose JK: Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci USA 1995, 92: 4477-4481. 10.1073/pnas.92.10.4477
Collins PL, Hill MG, Camargo E, Grosfeld H, Chanock RM, Murphy BR: Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5' proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA 1995, 92: 11563-11567. 10.1073/pnas.92.25.11563
Collins PL, Murphy BR: Respiratory syncytial virus: reverse genetics and vaccine strategies. Virology 2002, 296: 204-211. 10.1006/viro.2002.1437
Openshaw PJ, Culley FJ, Olszewska W: Immunopathogenesis of vaccine-enhanced RSV disease. Vaccine 2001,20(Suppl 1):S27-S31. 10.1016/S0264-410X(01)00301-2
Openshaw PJ, Tregoning JS: Immune responses and disease enhancement during respiratory syncytial virus infection. Clin Microbiol Rev 2005, 18: 541-555. 10.1128/CMR.18.3.541-555.2005
Boshart M, Weber F, Jahn G, Dorsch-Hasler K, Fleckenstein B, Schaffner W: A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell 1985, 41: 521-530. 10.1016/S0092-8674(85)80025-8
Pfarr DS, Sathe G, Reff ME: A highly modular cloning vector for the analysis of eukaryotic genes and gene regulatory elements. DNA 1985, 4: 461-467.
Pfarr DS, Rieser LA, Woychik RP, Rottman FM, Rosenberg M, Reff ME: Differential effects of polyadenylation regions on gene expression in mammalian cells. DNA 1986, 5: 115-122.
Moore MJ: From birth to death: the complex lives of eukaryotic mRNAs. Science 2005, 309: 1514-1518. 10.1126/science.1111443
Maquat LE: Nonsense-mediated mRNA decay in mammals. J Cell Sci 2005, 118: 1773-1776. 10.1242/jcs.01701
Le HH, Nott A, Moore MJ: How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci 2003, 28: 215-220. 10.1016/S0968-0004(03)00052-5
Sleckman BP, Gorman JR, Alt FW: Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu Rev Immunol 1996, 14: 459-481. 10.1146/annurev.immunol.14.1.459
Ryu WS, Mertz JE: Simian virus 40 late transcripts lacking excisable intervening sequences are defective in both stability in the nucleus and transport to the cytoplasm. J Virol 1989, 63: 4386-4394.
Le HH, Gatfield D, Izaurralde E, Moore MJ: The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 2001, 20: 4987-4997. 10.1093/emboj/20.17.4987
Cullen BR: Nuclear RNA export. J Cell Sci 2003, 116: 587-597. 10.1242/jcs.00268
Le HH, Moore MJ, Maquat LE: Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev 2000, 14: 1098-1108.
Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR: Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 2000, 404: 770-774. 10.1038/35004754
Yewdell JW, Schubert U, Bennink JR: At the crossroads of cell biology and immunology: DRiPs and other sources of peptide ligands for MHC class I molecules. J Cell Sci 2001, 114: 845-851.
Li X, Sambhara S, Li CX, Ewasyshyn M, Parrington M, Caterini J, James O, Cates G, Du RP, Klein M: Protection against respiratory syncytial virus infection by DNA immunization. J Exp Med 1998, 188: 681-688. 10.1084/jem.188.4.681
Bembridge GP, Rodriguez N, Garcia-Beato R, Nicolson C, Melero JA, Taylor G: DNA encoding the attachment (G) or fusion (F) protein of respiratory syncytial virus induces protection in the absence of pulmonary inflammation. J Gen Virol 2000, 81: 2519-2523.
Bembridge GP, Rodriguez N, Garcia-Beato R, Nicolson C, Melero JA, Taylor G: Respiratory syncytial virus infection of gene gun vaccinated mice induces Th2-driven pulmonary eosinophilia even in the absence of sensitisation to the fusion (F) or attachment (G) protein. Vaccine 2000, 19: 1038-1046. 10.1016/S0264-410X(00)00344-3
Tree JA, Bembridge G, Hou S, Taylor G, Fashola-Stone E, Melero J, Cranage MP: An assessment of different DNA delivery systems for protection against respiratory syncytial virus infection in the murine model: gene-gun delivery induces IgG in the lung. Vaccine 2004, 22: 2438-2443. 10.1016/j.vaccine.2003.11.069
Park EK, Soh BY, Jang YS, Park JH, Chung GH: Immune induction and modulation in mice following immunization with DNA encoding F protein of respiratory syncytial virus. Mol Cells 2001, 12: 50-56.
Wilusz J, Shenk T: A uridylate tract mediates efficient heterogeneous nuclear ribonucleoprotein C protein-RNA cross-linking and functionally substitutes for the downstream element of the polyadenylation signal. Mol Cell Biol 1990, 10: 6397-6407.
Chou ZF, Chen F, Wilusz J: Sequence and position requirements for uridylate-rich downstream elements of polyadenylation signals. Nucleic Acids Res 1994, 22: 2525-2531. 10.1093/nar/22.13.2525
Hallak LK, Collins PL, Knudson W, Peeples ME: Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology 2000, 271: 264-275. 10.1006/viro.2000.0293
Fouchier RA, Meyer BE, Simon JH, Fischer U, Malim MH: HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J 1997, 16: 4531-4539. 10.1093/emboj/16.15.4531
Kozak M: Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res 1984, 12: 857-872. 10.1093/nar/12.2.857
Deml L, Bojak A, Steck S, Graf M, Wild J, Schirmbeck R, Wolf H, Wagner R: Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J Virol 2001, 75: 10991-11001. 10.1128/JVI.75.22.10991-11001.2001
Schnell T, Foley P, Wirth M, Munch J, Uberla K: Development of a self-inactivating, minimal lentivirus vector based on simian immunodeficiency virus. Hum Gene Ther 2000, 11: 439-447. 10.1089/10430340050015905
Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM: Development of a self-inactivating lentivirus vector. J Virol 1998, 72: 8150-8157.
DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP: Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol 1987, 7: 379-387.
Simm M, Shahabuddin M, Chao W, Allan JS, Volsky DJ: Aberrant Gag protein composition of a human immunodeficiency virus type 1 vif mutant produced in primary lymphocytes. J Virol 1995, 69: 4582-4586.
Arnold R, Werner F, Humbert B, Werchau H, Konig W: Effect of respiratory syncytial virus-antibody complexes on cytokine (IL-8, IL-6, TNF-alpha) release and respiratory burst in human granulocytes. Immunology 1994, 82: 184-191.
Evan GI, Lewis GK, Ramsay G, Bishop JM: Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol 1985, 5: 3610-3616.
Acknowledgements
We thank B. Schweiger from the Robert Koch Institute (Berlin, Germany) for the RSV A2 strain. Recombinant GFP expressing RSV was generously provided by M. E. Peeples and P. L. Collins (NIH, Maryland, USA). R. Wagner (University of Regensburg, Germany) kindly provided pGwt and pGsyn expression plasmids. NT was granted a scholarship from the "Allgemeines Promotionskolleg" of the Ruhr-Universität Bochum. The study was supported by "FoRUM" grant F467-2005 of the Ruhr-Universität Bochum.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
KÜ and TG have filed a patent application on the use of the codon optimised RSV-F gene.
Authors' contributions
Cloning of RSV-F expression plasmids and RSV-F expression studies were performed by NT. DS analysed expression of VSV-G and synthesised required plasmids. SK supervised and participated in VSV-G experimental setups. TG and KÜ supervised and attributed to study design and planning. KÜ and TG revised the manuscript written by NT. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ternette, N., Stefanou, D., Kuate, S. et al. Expression of RNA virus proteins by RNA polymerase II dependent expression plasmids is hindered at multiple steps. Virol J 4, 51 (2007). https://doi.org/10.1186/1743-422X-4-51
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-4-51