Abstract
Background
There has been a recent resurgent interest in bacteriophage biology. Research was initiated to examine Campylobacter jejuni-specific bacteriophage in the Russian Federation to develop alternative control measures for this pathogen.
Results
A C. jejuni flagellum-specific phage PV22 from Proteus vulgaris was identified in sewage drainage. This phage interacted with C. jejuni by attachment to flagella followed by translocation of the phage to the polar region of the bacterium up to the point of DNA injection. Electron microscopic examination revealed adsorption of PV22 on C. jejuni flagella after a five minute incubation of the phage and bacteria. A different phenomenon was observed after incubating the mix under the same conditions, but for twenty minutes or longer. Phage accumulated primarily on the surface of cells at sites where flagella originated. Interestingly, PV22 did not inject DNA into C. jejuni and PV22 did not produce lytic plaques on medium containing C. jejuni cells. The constant of velocity for PV22 adsorption on cells was 7 × 10-9 ml/min.
Conclusion
It was demonstrated that a bacteriophage that productively infects P. vulgaris was able to bind C. jejuni and by a spot test that the growth of C. jejuni was reduced relative to control bacteria in the region of phage application. There may be two interesting applications of this effect. First, it may be possible to test phage PV22 as an antimicrobial agent to decrease C. jejuni colonization of the chicken intestine. Second, the phage could potentially be utilized for investigating biogenesis of C. jejuni flagella.
Similar content being viewed by others
Background
Campylobacter spp. are commensal bacteria in chickens and can cause a significant proportion of food-borne disease [1]. The high colonization incidences of poultry by campylobacters and the resultant clinical infections in humans have prompted a number of investigations focused upon identifying and subsequently eliminating Campylobacter spp. from poultry. Phage typing for Campylobacter spp. was developed [2–5] and compared to other classification schemes to trace these bacteria [6]. More recently, the presence of bacteriophage among chickens has been investigated [7, 8] along with examining their presence among specified commercial poultry flocks relative to isolates of C. jejuni [9]. Dramatic increases in isolation of fluoroquinolone resistant C. jejuni have been reported [10] and treatment of chickens with fluoroquinolones can induce rapid selection of ciprofloxacin-resistant campylobacters [11]. Consequently, reduction of Campylobacter spp. populations on chicken skin with bacteriophage has been attempted as an alternative control measure to antibiotics with varying degrees of success [7, 8, 13, 14].
There has been a resurgent interest in bacteriophage biology and their use or use of phage gene products as antibacterial agents [15–19]. During ongoing collaborative investigations between our laboratories, a collection of bacteriophages that attach to and/or infect C. jejuni were isolated in the Russian Federation to address the issue of utilizing bacteriophage for bacterial control. Interestingly, electron micrographs of a bacteriophage that attaches to C. jejuni, but productively infected Proteus vulgaris were identified from drainage water samples in the Moscow region. Bacteriophages that infect P. vulgaris, as in the case of other bacteria, have been utilized for typing schemes [20–22] and are structurally similar to phage from other bacteria [22–25]. Several of the Proteus-phages were shown to attach to the flagella of these bacteria [26, 27]. Herein we report the isolation and phage attachment kinetics of a bacteriophage that productively infects P. vulgaris, but which attaches to the flagella of C. jejuni.
Results and discussion
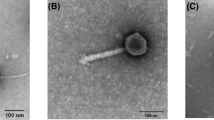
During research examining bacteriophage from the Moscow region by purifying material from sewage drainage a C. jejuni flagellum-specific phage PV22 from P. vulgaris was identified (Fig. 1) that structurally most closely resembled members of the Siphoviridae [28, 29]. The icosohedral head of phage PV22 measured from 56 to 58 nm with a non-contractile tail of greater than 200 nm in length. This phage, PV22, had a wide spectrum of lytic activity to P. vulgaris isolates (data not shown), but was subsequently propagated on a single isolate designated 1922. Members of the Myoviridae, Podoviridae and Siphoviridae have been isolated from P. vulgaris and utilized as a typing tool for this bacterium [22, 25].
The adsorption of phage PV22 on the surface of C. jejuni flagella was visualized utilizing three different isolates, with the illustration of attachment to C. jejuni strain L4 (Fig. 2a, b). Bacteriophage PV22 interacted with C. jejuni by attachment followed by translocation of the phage to the polar region of the bacterium up to the point of DNA injection. Electron microscopic examination revealed adsorption of PV22 on C. jejuni flagella after a five minute incubation of the phage and bacteria. A different phenomenon was observed when the mix was incubated at the same conditions but for a period of 20 min. or greater. Phage PV22 subsequently accumulated on cell surfaces mainly near areas where flagella originated on C. jejuni (Fig. 2c). Interestingly, PV22 did not appear to inject its DNA into C. jejuni.
The constant of velocity of PV22 adsorption on cells was determined to be 7 × 10-9 ml/min. Phage PV22 did not produce lytic cells in medium containing C. jejuni strains. At the same time, it was demonstrated by a spot test that the growth of C. jejuni was reduced relative to control bacteria in the region of phage application. Another observation was that PV22-treated C. jejuni cells appeared to lose their capability for chemotaxis (data not shown). Based on preliminary observations it was hypothesized that phage PV22 interacted with H. pylori in a similar manner (data not shown).
Our results suggest that particles of the phage PV22 are interacting with a C. jejuni cell on the same lines as infecting a P. vulgaris cell wherein certain phage interact by attaching to the flagella [26, 27]. However, it should be noted that phage PV22 failed to replicate in C. jejuni. Negative results from conventional titration of the phage in the presence of campylobacter cultures provide evidence for this conclusion. Also, phage PV22 did not generate plaque lysis on the surface of lawns produced by C. jejuni test cultures. Nevertheless, adsorption of the phage on flagella and in polar areas of the cell may influence C. jejuni replication as the cultures had reduced growth within the areas of phage application following spotting on a lawn of C. jejuni. It is currently unknown how PV22 fits in the scheme of Proteus spp. phage typing [22, 25], although structurally it can be classified as a Siphoviridae member based on structural characteristics [28, 29]. There consequently may be two interesting applications of this effect. First, it may be possible to test phage PV22 as an antimicrobial agent to control C. jejuni colonization of the chicken intestine. Second, the phage could potentially be utilized for investigating biogenesis of Campylobacter jejuni flagella.
Methods
Bacteriophage purification, propagation and bacterial culture
Bacteriophage PV22 was isolated by sampling drainage sewage waters in the Moscow region of the Russian Federation by standard procedures utilizing a Proteus vulgaris strain as a host [22, 25]. Bacterial cultures of P. vulgaris strain 1922 were supplied by the Tarasevich Institute of Standardization and Control of Medicinal Biological Preparations (Moscow) and propagated in meat-peptone broth [21]. Phage PV22was isolated according to the method of Snustad & Dean [30] as described in detail [21] by first clarifying drainage samples by low-speed centrifugation (5,000 × g for 20 min.) followed by filtration of the supernatant through 0.45 and then 0.22 um filters. Resultant filtered supernatants were cultured with P. vulgaris strain 1922 for 18 hrs followed by limit-dilution cloning to isolate individual viruses lytic for P. vulgaris utilizing standard techniques. C. jejuni isolates L4, 11168 and F2 were propagated in Brucella FBP agar and incubated at 42°C for 36–48 hours in microaerobic atmosphere (5% O2, 10% CO2 and 85% N2) as described previously [31].
In order to provide supportive evidence of the interaction between phage PV22 and C. jejuni, cultures of phage PV22 were sequentially centrifuged at 7,000 g for 20 min and 30,000 g for 120 min. Pellets obtained were suspended in 0.01 M Tris – HCl buffer (pH 7.0) to 4.75 ml of Tris – HCl buffer, 7 g of CsCl and 0.25 ml of phage suspension were then added to the vial. This was centrifuged in a SW-50 rotor at 35,000 rpm for 48 hours to produce fractions. An aliquot of purified phage was then dialyzed against 0.01 M Tris – HCl buffer (pH 7.0).
Bacteriophage attachment for electron microscopy and binding assay to C. jejuni
Cells of C. jejuni were suspended in 0.01 M Tris – HCl buffer (pH 7.0) containing 0.1 M MgSO4 and 0.001 M CaCl2 were mixed with purified preparation of phage PV22 (MOI of 10) and incubated at 40C in microaerobic conditions for 5 or 20 min. The suspension was centrifuged at 7,000 × g for 5 min., placed onto colloidal supporting films and treated with 1% uranyl acetate for further examination by electron microscopy (Hitachi H-300) utilizing standard methods [32]. The number of phage that bound to C. jejuni L4 was determined by first titration of PV22 with P. vulgaris strain 1922 and a constant velocity of adsorption was determined by the formula of Adams [33] by titration of the PV22 with its host. A preliminary chemotaxis assay was conducted as described by Adler [34] utilizing a capillary method with chicken epithelial cecal cells as the attractant with phage PV22 at an MOI of 10 with C. jejuni or C. jejuni alone.
References
Newell DG, Fearnley C: Sources of Campylobacter colonization in broiler chickens. Appl Environ Microbiol 2003, 69: 4343-4351. 10.1128/AEM.69.8.4343-4351.2003
Grajewski BA, Kusek JW, Gelfand HM: Development of a bacteriophage typing system for Campylobacter jejuni and Campylobacter coli . J Clin Microbiol 1985, 22: 13-18.
Khakhria R, Lior H: Extended phage-typing scheme for Campylobacter jejuni and Campylobacter coli . Epidemiol Infect 1992, 108: 403-414.
Sails AD, Wareing DR, Bolton FJ, Fox AJ, Curry A: Characterisation of 16 Campylobacter jejuni and C. coli typing bacteriophages. J Med Microbiol 1998, 47: 123-128.
Frost JA, Kramer JM, Gillanders SA: Phage typing of Campylobacter jejuni and Campylobacter coli and its use as an adjunct to serotyping. Epidemiol Infect 1999, 123: 47-55. 10.1017/S095026889900254X
Hopkins KL, Desai M, Frost JA, Stanley J, Logan JM: Fluorescent amplified fragment length polymorphism genotyping of Campylobacter jejuni and Campylobacter coli strains and its relationship with host specificity, serotyping, and phage typing. J Clin Microbiol 2004, 42: 229-235. 10.1128/JCM.42.1.229-235.2004
Atterbury RJ, Connerton PL, Dodd CE, Rees CE, Connerton IF: Isolation and characterization of Campylobacter bacteriophages from retail poultry. Appl Environ Microbiol 2003, 69: 4511-4518. 10.1128/AEM.69.8.4511-4518.2003
Atterbury RJ, Connerton PL, Dodd CE, Rees CE, Connerton IF: Application of host-specific bacteriophages to the surface of chicken skin leads to a reduction in recovery of Campylobacter jejuni . Appl and Environ Microbiol 2003, 69: 6302-6306. 10.1128/AEM.69.10.6302-6306.2003
Connerton PL, Loc Carrillo CM, Swift C, Dillon E, Scott A, Rees CE, Dodd CE, Frost J, Connerton IF: Longitudinal study of Campylobacter jejuni bacteriophages and their hosts from broiler chickens. Appl Environ Microbiol 2004, 70: 3877-3883. 10.1128/AEM.70.7.3877-3883.2004
Nachamkin I, Ung H, Li M: Increasing fluoroquinoloneresistance in Campylobacter jejuni , Pennsylvania, USA, 1982–2001. Emerg Infect Dis 2002, 8: 1501-1503.
McDermott PF, Bodeis SM, English LL, White DG, Walker RD, Zhao S, Simjee S, Wagner DD: Ciprofloxacin resistance in Campylobacter jejuni evolves rapidly in chickens treated with fluoroquinolones. J Infect Dis 2002, 185: 837-840. 10.1086/339195
Goode D, Allen VM, Barrow PA: Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl Environ Microbiol 2003, 69: 5032-5036. 10.1128/AEM.69.8.5032-5036.2003
Atterbury RJ, Dillon E, Swift C, Connerton PL, Frost JA, Dodd CE, Rees CE, Connerton IF: Correlation of Campylobacter bacteriophage with reduced presence of hosts in broiler chicken ceca. Appl Environ Microbiol 2005, 71: 4885-4887. 10.1128/AEM.71.8.4885-4887.2005
Wagenaar JA, Van Bergen MA, Mueller MA, Wassenaar TM, Carlton RM: Phage therapy reduces Campylobacter jejuni colonization in broilers. Vet Microbiol 2005, 109: 275-283. 10.1016/j.vetmic.2005.06.002
Merril CR, Biswas B, Carlton R, Jensen NC, Creed GJ, Zullo S, Adhya S: Long-circulating bacteriophage as antibacterial agents. Proc Natl Acad Sci USA 1996, 93: 3188-92. 10.1073/pnas.93.8.3188
Sulakvelidze A, Alavidze Z, Morris JG Jr: Bacteriophage therapy. Antimicrob Agents Chemother 2001, 45: 649-59. 10.1128/AAC.45.3.649-659.2001
Schuch R, Nelson D, Fischetti VA: A bacteriolytic agent that detects and kills Bacillus anthracis . Nature 2002, 418: 884-889. 10.1038/nature01026
Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, Callejo M, Ferretti V, Ha N, Kwan T, McCarty J, Srikumar R, Williams D, Wu JJ, Gros P, Pelletier J, DuBow M: Antimicrobial drug discovery through bacteriophage genomics. Nat Biotechnol 2004, 22: 185-191. 10.1038/nbt932
Greer GG: Bacteriophage control of foodborne bacteria. J Food Prot 2005, 68: 1102-1111. Erratum in: J. Food Prot 2005, 68: 1334
France DR, Markham NP: Epidemiological aspects of Proteus infections with particular reference to phage typing. J Clin Path 1968, 21: 97-102.
Schmidt WC, Jeffries CD: Bacteriophage typing of Proteus mirabilis , Proteus vulgaris , and Proteus morganii . Appl Microbiol 1974, 27: 47-53.
Sekaninova G, Hofer M, Rychlik I, Pillich J, Kolarova M, Zajicova V, Kubickova D: A new phage typing scheme for Proteus mirabilis and Proteus vulgaris strains. 1. Morphological analysis. Folia Microbiol (Praha) 1994, 39: 381-6.
Prozesky OW, De Klerk HC, Coetzee JN: The morphology of Proteus bacteriophages. J Gen Microbiol 1965, 41: 29-36.
Coetzee HL, De Klerk HC, Coetzee JN, Smit JA: Bacteriophage-tail-like particles associated with intra-species killing of Proteus vulgaris . J Gen Virol 1968, 2: 29-36.
Sekaninova G, Rychlik I, Kolarova M, Pillich J, Semenka J, Zajicova V: A new bacteriophage typing scheme for Proteus mirabilis and Proteus vulgaris strains. 3. Analysis of lytic properties. Folia Microbiology (Praha) 1998, 43: 136-140.
Appelbaum PC, Hugo N, Coetzee JN: A flagellar phage for the proteus-providence group. J Gen Virol 1971, 13: 153-162.
Krizsanovich K, Appelbaum PC, Coetzee JN: Ecology of Proteus flagellar phages. Arch Mikrobiol 1973, 89: 241-245. 10.1007/BF00422204
Ackermann HW: Bacteriophage observations and evolution. Res Microbiol 2003, 154: 245-51. 10.1016/S0923-2508(03)00067-6
Ackermann HW, DuBow MS, Gershman M, Karska-Wysocki B, Kasatiya SS, Loessner MJ, Mamet-Bratley MD, Regue M: Taxonomic changes in tailed phages of enterobacteria. Arch Virol 1997, 142: 1381-1390. 10.1007/s007050050167
Snustad SA, Dean DS: Genetic Experiments with Bacterial Viruses. W.H. Freeman & Co. San Francisco; 1971.
Rothenberg PJ, Stern NJ, Westhoff DC: Selected enrichment broths for recovery of Campylobacter jejuni from foods. Appl Environ Microbiol 1984, 48: 78-80.
Brenner S, Horne RW: A negative staining method for high resolution electron microscopy of viruses. Bioch Biophys Acta 1959, 34: 103-110. 10.1016/0006-3002(59)90237-9
Adams MH: Methods of Study of Bacterial Viruses. In Bacteriophages. Interscience Publishers, Inc. New York; 1959:443-457.
Adler J: A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli . J Gen Microbiol 1973, 74: 77-91.
Acknowledgements
Funding was provided from the International Science and Technology Center (ISTC) grant no. 1720 administered through the Office of International Research Programs (OIRP), Agricultural Research Service (ARS), USDA, the ARS, USDA CRIS project no. 6612-3200-046-00D and the Russian Federation State Research Center for Applied Microbiology (SRCAM).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
The research was completed at the State Research Center for Applied Microbiology, Obolensk, Russian Federation in the laboratory of E. L. Zhilenkov under the laboratory unit direction of E. A. Svetoch. N. J. Stern is a co-principle investigator for funding with collaborator B. S. Seal at the Poultry Microbiological Safety Research Unit, ARS, USDA in Athens, GA, USA who completed writing and final editing of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Zhilenkov, E., Popova, V., Popov, D. et al. The ability of flagellum-specific Proteus vulgaris bacteriophage PV22 to interact with Campylobacter jejuni flagella in culture. Virol J 3, 50 (2006). https://doi.org/10.1186/1743-422X-3-50
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-3-50