Abstract
Background
Nipah virus (NiV) is a highly pathogenic zoonotic agent in the family Paramyxoviridae that is maintained in nature by bats. Outbreaks have occurred in Malaysia, Singapore, India, and Bangladesh and have been associated with 40 to 75% case fatality rates. There are currently no vaccines or postexposure treatments licensed for combating human NiV infection.
Methods and results
Four groups of ferrets received a single vaccination with different recombinant vesicular stomatitis virus vectors expressing: Group 1, control with no glycoprotein; Group 2, the NiV fusion protein (F); Group 3, the NiV attachment protein (G); and Group 4, a combination of the NiV F and G proteins. Animals were challenged intranasally with NiV 28 days after vaccination. Control ferrets in Group 1 showed characteristic clinical signs of NiV disease including respiratory distress, neurological disorders, viral load in blood and tissues, and gross lesions and antigen in target tissues; all animals in this group succumbed to infection by day 8. Importantly, all specifically vaccinated ferrets in Groups 2-4 showed no evidence of clinical illness and survived challenged. All animals in these groups developed anti-NiV F and/or G IgG and neutralizing antibody titers. While NiV RNA was detected in blood at day 6 post challenge in animals from Groups 2-4, the levels were orders of magnitude lower than animals from control Group 1.
Conclusions
These data show protective efficacy against NiV in a relevant model of human infection. Further development of this technology has the potential to yield effective single injection vaccines for NiV infection.
Similar content being viewed by others
Background
Nipah virus (NiV) and Hendra virus (HeV) represent the highly pathogenic zoonotic agents in the paramyxovirus genus Henipavirus with human case fatality rates ranging between 40 and 75% [1]. These viruses are categorized as biosafety level 4 (BSL4) pathogens due to the significant morbidity and mortality associated with disease and the lack of approved vaccines and therapeutics for human use. The primary reservoir for henipaviruses are bats of the genus Pteropus[2]; however; the viruses can be transmitted to many mammalian species including humans. Currently, there are two distinct strains of NiV: 1) the Malaysia strain (NiVM) discovered in 1999 during an outbreak on pig farms which resulted in spread to humans [3]; and 2) the Bangladesh strain (NiVB), which was discovered in India and Bangladesh during 2001 [4]. NiVB has been linked to direct transmission from bats to humans and evidence suggests human to human transmission is possible [5].
The near annual outbreaks of NiVB with high case fatality rates [6] underscores the urgent need for effective vaccines and therapeutics. To date, there have been four experimental preventive candidate vaccines against henipaviruses evaluated in animal models. Vaccinia and canarypox viruses encoding the NiVM glycoproteins have shown protection against NiVM in hamsters and pigs [7, 8]. A recombinant adeno-associated vaccine expressing the NiVM G protein completely protected hamsters against homologous NiVM challenge and protected 50% of animals against heterologous HeV infection [9]. In addition, a recombinant subunit vaccine based on the HeV G protein (sGHeV) completely protects small animals against lethal HeV and NiVM infection [10–13] and more recently was shown to be efficacious in the robust African green monkey model of NiVM infection [14]. Though very promising, the sGHeV vaccine requires a prime-boost strategy to confer protection whereas a single-injection vaccine would be particularly beneficial during outbreaks where there is little time to employ lengthy vaccination regimens.
Single-injection recombinant vesicular stomatitis virus (rVSV) vectors have been developed as vaccine candidates against many important human pathogens such as papillomavirus [15, 16], human immunodeficiency virus (HIV) [17–19], influenza virus [20], measles virus [21, 22], respiratory syncytial virus [23, 24], severe acute respiratory syndrome coronavirus [25], chikungunya virus [26], and hemorrhagic fever viruses such as Lassa, Ebola, and Marburg [27]. Single-cycle replication rVSVs have been developed against NiV and have shown strong immunogenicity in mice vaccinated with rVSVs expressing either the NiVM fusion protein (F) or the NiVM attachment protein (G) as high neutralizing antibody titers were generated [28]. These vaccine vectors were just recently shown to provide homologous protection in the hamster model of NiVM infection [29].
Here, we developed alternative rVSV vaccine vectors expressing either the NiVB F or NiVB G proteins. These vaccines were evaluated 28 days after a single dose vaccination in the NiVM ferret model, which along with the African green monkey, most faithfully recapitulates human disease [30–32]. Each group of specifically vaccinated ferrets were protected from NiVM-induced disease while the non-specifically vaccinated ferrets succumbed to NiVM infection. To date, this is the first study to protect ferrets from NiV infection using a single-injection vaccine.
Results
Recovery of rVSVΔG-NiVB/glycoprotein vectors
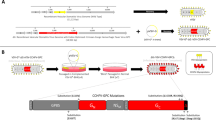
To investigate the protective efficacy of rVSV NiVB vaccine vectors against heterologous NiVM challenge in ferrets, we first developed and recovered two rVSVΔG constructs expressing the NiVB F protein rVSV-ΔG-NiVB/F-GFP (Figure 1A, blue) or NiVB G protein rVSV-ΔG-NiVB/G-GFP (Figure 1A, yellow) using reverse genetics. Propagation of these vectors requires VSV glycoprotein (GInd) complementation (GInd*) of viruses where GInd is provided in trans during infection [33]. GInd* complementation allows for single-cycle replication of vectors and results in expression of the NiVB glycoproteins and the production non-infectious virions containing either glycoprotein. As seen previously with similar NiVM rVSV vectors [28], co-infection with GInd* rVSV-ΔG-NiVB/F-GFP and GInd* rVSV-ΔG-NiVB/G-GFP produced infectious virions (rVSV-ΔG-NiVB/F/G-GFP) containing either genome with the NiVB F and NiVB G proteins incorporated in the virion envelope (Figure 1A, Group 4 green-spiked virion) as evidenced by the ability of these virus preparations to infect Vero cells (Figure 1B, *) and the inability of the rVSV-ΔG-NiVB/F-GFP and rVSV-ΔG-NiVB/G-GFP to infect cells without GInd* complementation (negative data not shown). The rVSV-ΔG-NiVB/F/G-GFP virus stocks were able to reach titers of up to 3 × 108 PFU/ml and infection of Vero cells with these stocks resulted in syncytia formation (Figure 1B). While this vaccine preparation could undergo a single round of replication, it could not be passaged further without GInd* complementation as was observed with the rVSV-ΔG-NiVB/F-GFP and rVSV-ΔG-NiVB/G-GFP vaccine preparations.
Schematic rVSV NiV B vaccine genomes and groups. (A) Group 1: rVSV-ΔG-GFP expressing no glycoprotein; produces ‘bald’ virions, Group 2 GInd*rVSV-NiVB /F-GFP expressing the NiVB F protein (blue); produces virions with F on the surface, Group 3 GInd*rVSV-NiVB /G-GFP expressing the NiVB G protein (yellow); produces virions with G on the surface, Group 4 made from co-infection in Vero cells resulting in a combination of rVSV-NiVB /F-GFP and rVSV-NiVB /G-GFP virions expressing the NiVB F and G proteins in the same cells; produces single-cycle infectious virions with NiVB F and G proteins on the cell surface (green). (B) Phase and fluorescence microscopy of Vero cells infected with virions from Group 4 displaying multinucleated syncytia cells * and cells positive for GFP expression.
Immunization of ferrets and examination of the humoral immune response
Four groups of ferrets (Figure 1A) received a single vaccination of rVSV vectors as follows: Group 1 served as the nonspecific vaccine control group and received GInd* rVSV-ΔG-GFP; Group 2 received GInd* rVSV-NiVB/F-GFP; Group 3 received GInd* rVSV-NiVB /G-GFP; and Group 4 received the rVSV-ΔG-NiVB/F/G-GFP (Figure 2A, triangles). Serum collected from each animal on the day of vaccination (day -28) and just before challenge (day 0) was analyzed for circulating IgG to the NiV F and NiV G proteins by microsphere assay [30]. As expected, we did not detect NiV F-specific Ig in sera before vaccination (Figure 2B, day -28). We were able to detect Ig directed at NiV F in sera of the Group 2 and 4 vaccinated cohorts 28 days after vaccination but not in the ferrets from Groups 1 and 3 (Figure 2B, day 0). Similar to the analysis of NiV F-specific circulating IgG in vaccinated ferrets, we were unable to detect IgG directed to NiV G before vaccination (Figure 2C, day -28) and were able to detect anti-NiV G IgG 28 days post vaccination (Figure 2C, day -28). Animals in Group 3 had higher levels of circulating anti-NiV G IgG when compared to Group 4 and surprisingly we were able to detect anti-NiV G IgG in Group 2 animals although not to the levels of the NiVB G-specifically vaccinated Group 3 animals (Figure 2C, day -0). These results suggested that the animals in Groups 2-4 and not Group 1 had generated a humoral immune response to the rVSV-ΔG-NiVB antigens delivered in a single injection.
NiV vaccine study design and circulating IgG post vaccination. (A) Flow chart showing the days of vaccination (triangles), days of sampling (arrows), day of challenge (*),terminal succumbing to infection (^, day 7 or 8), and day of necropsy (^, day 22). Black triangle, Group 1; blue triangle Group 2; yellow triangle Group 3; green triangle Group 4 vaccination. (B and C) Detection of specific anti-NiV F (B) and anti-NiV G (C) IgG antibodies circulating in vaccinated ferrets. Mean fluorescence intensities (Mean FI) are shown on the y-axis and represent binding of specific IgG. Error bars represent the s.d. of fluorescence intensity across 100 beads for each sample.
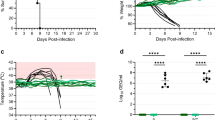
NiV challenge of vaccinated ferrets
To determine whether vaccination with the rVSV-ΔG-NiVB vectors could prevent NiVM disease course in ferrets, we challenged ferrets intranasally with a lethal challenge dose of NiVM on day 0 (Figure 2A, *). The animals were closely monitored over the course of 22 days post challenge (p.c.) for clinical signs of illness. The specifically vaccinated animals in Groups 2-4 did not lose weight over the course of the study (Figure 3A) and were 100% protected against NiVM (Figure 3B), while the animals in the non-specifically vaccinated Group 1 succumbed to infection on days 7 or 8 (Figure 2A, ^), respectively (Figure 3B, Table 1). Clinical scores were recorded each day after challenge for each animal using a scoring system based on coat grooming, social behavior, and provoked behavior. The clinical scores for each animal correlated with the survival data as seen with the mean clinical score for each animal in the vaccinated groups having no score on any day p.c. versus the animals in Group 1 having clinical scores on days 5 to 8 p.c. Clinical signs in response to NiVB infection were more dramatic for the animals in Groups 1 when compared to the animals in the other three groups (Table 1). In all animals that succumbed to NiVM infection, the gross pathologic findings included varying severity of dehydration, ventral cervical subcutaneous hemorrhage with edema (Figure 4A) and crusting serous nasal discharge (Figure 4B). The internal gross pathologic findings from animals in Group 1 included varying severity of heavy, wet, diffusely mottled with dark pin point foci throughout the pulmonary parenchyma, multifocal pin point raised foci throughout the renal parenchyma, mottling of the spleen with diffuse splenomegaly, and diffuse reticulation of the liver. Each animal had diffuse hemorrhagic interstitial pneumonia (Figure 4C) and splenomegaly with multifocal necrosis (pic 4D, *). Additionally, diffuse reticulation of the liver (Figure 4D, +) and multifocal renal hemorrhage (Figure 4D, arrow) were noted. There were no external or internal gross pathologic findings of note in any of the Group 2-4 animals at the study endpoint (Figure 2A, day 22 ^).
Gross pathology of ferrets succumbing to NiV M mediated disease. Representative gross pathology from animals in Group 1 challenged with NiVM. (A) Ventral cervical subcutaneous hemorrhage with edema. (B) Crusting serous nasal discharge. (C) Diffuse hemorrhagic interstitial pneumonia. (D) Splenomegaly with multifocal necrosis (*), diffuse reticulation of the liver (+), and multifocal renal hemorrhage (white arrow).
Histopathological and immunohistochemical analysis of NiVM-infected ferrets
Tissues examined from animals in Groups 2-4 had no significant histologic findings (Figure 5A,E,I, and M) and were devoid of NiV antigen (Figure 5B,F,J, and N). In contrast, tissues examined from ferrets in control Group 1 had substantial histologic findings which included mild to moderate interstitial pneumonia with marked congestion, endothelial syncytial cell formation, and respiratory epithelial syncytial cell formation (Figure 5C, arrow head). Severe lymphoid depletion, necrosis, hemorrhage, fibrin deposition and syncytial cell formation were observed throughout the spleen (Figure 5G). Glomerular tufts in the kidney had multifocal segmental to global fibrin deposition and endothelial syncytial cell formation (Figure 5K). Ferrets in control Group 1 also had detectable NiV antigen systemically, with the endothelium, scattered mononuclear cells, and syncytial cells (endothelial and epithelial) of the lung (Figure 5D), spleen (Figure 5H), kidney (Figure 5L), and brain (Figure 5P) having strong immunolabeling for NiV antigen.
H&E and immunohistochemistry of ferret tissues. Representative H&E stained tissues (A , C , E, G, I, K, M, and O) and immuno-histochemistry of tissues labeled with a NiV N protein-specific polyclonal rabbit antibody (B, D, F, H, J, L, N, and P). Tissues from representative vaccinated ferrets from groups 2-4; lung (A, B), spleen (E, F), kidney (I, J), and brain, choroid plexus (M, N). Tissues from representative control Group 1 ferrets; lung (C, D), spleen (G, H), kidney (K, L), and brain, choroid plexus (O, P). C, (arrow head) Respiratory epilthelial syncytia; D, (arrow) NiV antigen-positive respiratory epithelial syncytia, G, (white asterisk) lymphoid depletion in spleen; K, (*) fibrin deposition in glomerular tuft of kidney. Images taken: lung 40x, spleen 20x, kidney 40x, brain 40x.
NiV load
To determine if there was NiVM replication in animals p.c., viremia was assessed by virus isolation from serum (negative data not shown) and by qRT-PCR on whole blood samples (Figure 6A). NiVM genome equivalents (GEq) were detected in all blood samples from day 6 p.c. While we detected NiVM GEq for all animals on day 6, the Group 1 animals had over 100 fold more detectable GEq and an increase in GEq from terminal bleeds on day 7 or 8 (Figure 6A). However, none of the specifically vaccinated animals had any detectable NiVM RNA in terminal blood samples on day 21 (Figure 6A). NiVM RNA was also detected systemically in the tissues of all animals that succumbed to infection in control Group 1 and in the spleen of one animal from Group 4 on the study end date (Figure 6B), though no NiVM antigen was detected by immunohistochemistry (data not shown), whereas NiVM RNA was not detected in the tissues of the remaining animals in Groups 2-4. Additionally, we were able to isolate NiVM from all tissues sampled in all animals from control Group 1, with the exception of the liver for animal 1-3 (Figure 6C). Overall, the level of detection of NiVM RNA in tissues and blood correlated with outcome gross pathology, and histology (Figure 4, Figure 5, and Table 1) for each animal.
Viral load in NiV M challenged ferrets. Viral load in ferrets as detected by genome equivalents (GEq) by qRT-PCR from (A) blood as GEq/ml and (B) from tissues a GEq/g (Animal 4-1 was negative for NiVM antigen in spleen). Right upper (R.U.), right middle (R.M.), right lower (R.L.), left upper (L.U.), left middle (L.M.), left lower (L.L.), lymph node (LN). (C) PFU/g of NiVM isolated from tissues of ferrets in Group 1. Liver (L), spleen (S), kidney (K), adrenal gland (A). Error bars represent the s.d.
Neutralizing NiVM antibody titers pre and post challenge with NiVM
To further address the antibody response to rVSV-ΔG-NiVB vaccination and after NiVM challenge, we characterized the circulating antibodies before and after vaccination for their neutralizing activity using a plaque reduction neutralization titer (PRNT50) assay. All four groups lacked neutralizing antibody titers before vaccination (Figure 7A). On the day of challenge, animals from control Group 1 did not have neutralizing antibody titers against NiVM whereas all ferrets from Groups 2-4 had neutralizing antibodies against NiVM (Figure 7B), although it appeared that Groups 2 and 3 had higher titers than Group 4. Neutralizing antibody titers were also assessed for all animals from terminal bleeds on the day of death (day 7 or 8) or at day 21. All animals that succumbed to NiVM challenge in Group 1 had consistently lower neutralizing antibody titers against NiVM when compared to the neutralizing antibodies found in animals from Groups 2-4 (Figure 7C).
Reciprocal NiV M serum neutralizing antibody titers. Neutralizing antibody titers which reduce virus infectivity are shown for all groups on the (A) day of vaccination showing a lack of neutralizing antibody for all groups, (B) on the day of challenge showing neutralizing antibodies for all groups except Group 1, and (C) on the terminal bleed days showing high levels of neutralizing antibodies for Groups 2-4 and low neutralizing antibodies for Group 1. Error bars represent the s.d.
Discussion
While significant progress on a veterinary vaccine for one henipavirus, HeV, has been made [34], the development of effective human vaccines and antiviral drugs for high consequence pathogens such as NiV is a much slower and complicated process. In particular, the restriction of infectious NiV work to BSL-4 containment has hampered progress. Conventional clinical trials with viruses such as NiV are not practical. To address the development of countermeasures for exotic pathogens such as NiV the FDA implemented the Animal Efficacy Rule in 2002. This rule specifically applies to the development of countermeasures when human efficacy studies are not possible or ethical. Briefly, this rule permits the evaluation of vaccines or therapeutics using data generated from studies performed in animal models that faithfully recapitulate human disease. There are currently at least eight animal models for NiV including the IFNAR-KO mouse model [35]; the guinea pig model [36]; the pig model [8, 37–39]; the cat model [10, 11]; the golden Syrian hamster model [7, 40–42]; the ferret model [30, 43]; and two nonhuman primate models, one in squirrel monkeys [44] and the other in African green monkeys [32]. While each of these models shares at least one or more aspects seen in human pathogenesis and can contribute to vaccine and therapeutic testing, the two animal models which completely emulate the pathogenesis seen in human cases to date are the ferret model and the African green monkey model. These models recapitulate what is seen during human infection with severe respiratory and neurologic disease and a generalized vasculitis.
Based on this knowledge, the ferret model was chosen for the initial evaluation of our rVSV-ΔG-NiVB-F/G-GFP vectors. This model should be considered as the initial small animal screening model for vaccines against NiV, especially since the recombinant sGHeV protein vaccine has been shown to be efficacious in ferrets [12] and African green monkeys [14]. While sGHeV vaccine has proven to have utility against NiV infection in these two animal models, the vaccine regimen requires two doses using a prime-boost strategy with the two adjuvants CpG oligodeoxynucleotide (ODN) 2006 and Alhydrogel included in the vaccine formulation [14]. Here, we evaluated rVSV-ΔG-NiVB-F/G-GFP vectors as single-injection vaccines against lethal NiVM challenge in ferrets.
Non-specifically vaccinated control animals in Group 1 did not produce anti-NiV F or anti-NiV G IgGs, did not produce significant neutralizing antibody titers against NiVM, and thus were not protected from NiVM-induced disease as they displayed clinical symptoms and pathology consistent with NiVM infection, and had viral RNA and infectious virus isolated systemically. Animals in Group 2 vaccinated with GInd* rVSV-ΔG-NiVB/F-GFP generated anti-NiV F IgGs (and possible cross-reactive NiV G IgG, Figure 2C) and neutralizing antibody titers against NiVM and were protected from NiVM-induced disease with one animal (Table 1, 2-5) having a low-grade fever on day 7 p.c. Similarly, ferrets in Group 3 vaccinated with GInd* rVSV-ΔG-NiVB/G-GFP generated anti-NiV G IgGs and neutralizing antibody titers against NiVM and were protected from NiVM-induced disease. Using vectors that were independent of GInd* complementation, ferrets in Group 4 vaccinated with rVSV-ΔG-NiVB-F/G-GFP generated anti-NiV F and anti-NiV G IgGs and neutralizing antibody titers against NiVM and were also protected from NiVM-induced disease. The observation that the percent weight growth was less than Group 2 and 3 (Figure 3A) could have been due to the social housing of these animals where it is possible that certain members of the cohort were out competed for extra food as some animals in this group reached percent weight growth of animals from the other groups. While the protection afforded to Groups 2-4 did not appear to be sterile as virus replication could be detected by the presence of viral RNA in whole blood at day 6 p.c., it was only detected in two animals per group versus all animals in Group 1 (Table 1) and the mean virus replication was 100 times lower than what was detected in animals from Group 1 (Figure 6A).
The observation of anti-NiV F antibodies circulating in the rVSV-ΔG-NiVB/G-GFP vaccinated ferrets was interesting considering our experience with the Bioplex assay and sGHeV vaccinated ferrets and African green monkeys where we have never detected cross-reactivity against NiVM F from G vaccinated animals. One possible explanation is that the antibodies produced in response to the NiVB G results in the formation of some antibodies that can recognize NiVM F since the target antigen in the Bioplex assay is NiVM F. Our experience with the sGHeV vaccine may not be applicable here as this cross-reactivity may be a single-cycle replicating vaccine observation. While not within the scope of this study, the further examination of the avidity of these antibodies toward NiVM F is warranted as this rVSV-NiV vaccine platform is developed.
As it is difficult to determine which vaccine platform would be best for the human population against deadly pathogens such as NiV, it is important to develop and test a number of platforms to combat a potential outbreak. A preventive vaccine would be important for several populations: 1) the general population during NiV outbreaks particularly in endemic areas in India and Bangladesh; 2) healthcare workers and family members involved in patient care and management in endemic regions; 3) personnel involved in outbreak response missions; 4) laboratory workers conducting research on NiV; and 5) military and other service personnel susceptible to the use of NiV as a biological weapon. While multi-dose vaccine regimens would be feasible for laboratory and healthcare workers and some military personnel in stable settings with defined risk, an outbreak setting or a case of deliberate release would require rapidly conferred protection with a single administration. Here we describe the protection of ferrets from NiVM-induced disease using single-injection rVSV vaccine vectors expressing the NiVB F and/or NiVB G proteins. To date, this is the first study to show efficacy of a vaccine against NiV using a single administration in the ferret model. Based on the success of this study, these vectors should be evaluated further in the African green monkey model against NiVB challenge as this strain appears to be the most pathogenic in humans [6].
Methods
Plasmid design and construction
RNA was isolated from NiVB virus stocks (NiVB #200401066 was obtained from a patient from the 2004 outbreak in Bangladesh (kindly provided by Dr. Thomas Ksiazek) using approved protocols at biosafety level 4 (BSL-4) in the Galveston National Laboratory (GNL) at the University of Texas Medical Branch (UTMB). The RNA was used to make cDNA with gene specific primers and the genes were amplified using gene specific primers (NiVB F or G) containing MluI and NheI restriction sites at the 5′ and 3′ ends respectively. The PCR amplified products and pΔG-VSV-2.6 plasmid were restriction digested with MluI and NheI and gel purified. Purified products and vector were ligated and positive colonies were screened and sequenced for positive constructs.
rVSV vaccine vectors and challenge virus
The rVSV NiVB vaccines (rVSV-ΔG-NiVB/F-GFP and rVSV-ΔG-NiVB/G-GFP) and rVSV-ΔG-GFP were recovered using methods previously described [33]. The rVSVΔG viruses were propagated on BHK-21 cells transfected with 2 μg of pCAGGS-GInd expressing the VSV glycoprotein (GInd) and titered as previously described [33]. Viruses complemented with VSV GInd are denoted as GInd* rVSV-ΔG-GFP, G*rVSV-ΔG-NiVB/F-GFP, and G* rVSV-ΔG-NiVB/G-GFP. To make the vaccine in Group 4 (Figure 1A), Vero cells were co-infected with G*rVSV-ΔG-NiVB/F-GFP and G* rVSV-ΔG-NiVB/G-GFP at MOI 5 for each virus. Supernatants were collected 24 h.p.i. and titered on BHK-21 cells complemented with VSV GInd.
NiVM #1999011924 was obtained from a patient from the 1999 outbreak in Malaysia (kindly provided by Dr. Thomas Ksiazek). NiVM was chosen for challenge in ferrets based on our lethality data with this particular stock at the time of the study. The virus was propagated on Vero E6 cells in Eagle’s minimal essential medium (EMEM) supplemented with 10% fetal calf serum. The NiVM challenge virus stock was assessed for the presence of endotoxin using The Endosafe®-Portable Test System (PTS) (Charles River, Wilmington, MA). Virus preparations were diluted 1:10 in Limulus Amebocyte Lysate (LAL) Reagent Water (LRW) per manufacturer’s directions and endotoxin levels were tested in LAL Endosafe®-PTS cartridges as directed by the manufacturer. Each preparation was found to be below detectable limits while positive controls showed that the tests were valid.
Statistics
Conducting animal studies in BSL-4 severely restricts the number of animal subjects, the volume of biological samples that can be obtained and the ability to repeat assays independently and thus limit statistical analysis. Consequently, data are presented as the mean calculated from replicate samples, not replicate assays, and error bars represent the standard deviation across replicates.
Animals
Animal studies were performed in BSL-4 biocontainment at the GNL at the UTMB at Galveston and were approved by the UTMB Institutional Animal Care and Use Committee (IACUC). Animal research was conducted in compliance with the Animal Welfare Act and other Federal statutes and regulations relating to animals and experiments involving animals and adheres to the principles stated in the eighth edition of the Guide for the Care and Use of Laboratory Animals, National Research Council, 2013. The facility where this research was conducted is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Twenty female ferrets weighing 0.75-1 kg were housed in groups of 3 and 2 animals per vaccine group. Before vaccination, subjects were anesthetized by i.m. injection with ketamine-acepromazine-xylazine (KAX) cocktail and had transponder chips (BioMedic Data Systems, Seaford, DE) implanted subcutaneously for animal identification and temperature monitoring. For procedures, animals were anesthetized with KAX and vaccinated with ~ 1 × 107 PFU by i.m. injection on day -28 (Figure 2A). Animals were inoculated intranasally (i.n.) with ~ 5 × 103 pfu of NiVM in 1 ml of Dulbecco’s minimal essential medium (DMEM) (Sigma-Aldrich, St Louis, MO) 28 days after vaccination (Figure 2A, *). Animals were anesthetized for clinical examination including temperature, respiration quality, blood collection, and on days 0, 6, and 21 p.c. Before and after challenge animals were assessed daily for weight, temperature, and scored on a scale of 0 of 9 for clinical observations based on coat appearance, social behavior, and provoked behavior; animals scoring 7 or greater were euthanized per IACUC protocol. Subjects in the vaccine cohorts were euthanized at the study endpoint on day 22 p.c. whereas the subjects in Group 1 had to be euthanized according to approved humane end points on day 7 or 8 p.c. All other subjects survived until the end of the study.
Measurement of serum or plasma NiV F and G specific antibodies
Ferret serum collected at indicated time points was tested for IgG antibodies against NiV F and G using previously developed multiplexed microsphere assays [30]. 96-well filter plates were primed with PBS. Test sera were diluted in PBS at 1:10 for pre vaccination time points and 1:10,000 for time points after vaccination. Biotinylated goat anti-ferret IgG and streptavidin-phycoerythrin (strep-PE) were also diluted in PBS. Coupled microspheres (sG-HeV, sG-NiV, sF-Hev, sF-NiV) were prepared by sonication for 1 minute followed by vortex mixing for 1 minute each and then diluted in PBS. Priming liquid was removed from plates using a Bio-Plex Pro II Wash Station (Bio-Rad, Hercules, CA) and 100 μL containing 1500 of each coupled microsphere was added to each well. The microsphere mixture was removed by vacuum, 100 μL of diluted test sera was added to appropriate wells and incubated at room temperature (RT) for 30 minutes while shaking in the dark. Diluted test samples were removed by vacuum and 100 μL of diluted biotinylated goat anti-ferret (1:500) (Pierce, ThermoScientific, Rockford, IL) was added to each well and incubated as previously described above. Liquid was removed by vacuum and 100 μL of strep-PE (1:1000) (Qiagen, Valencia, CA) was added to each well and again incubated for 30 minutes. All liquid was removed from plates with a vacuum manifold and washed twice with 300 μL PBS, removing liquid between wash steps. Finally, 125 μL of PBS was added to each well and incubated for 2 minutes as described above. Samples were assayed for mean fluorescence intensity (MFI) across at least a 100 bead region performed on the BioPlex-200 machine and analyzed using Bio-Plex Manager Software (v 6.1) (Bio-Rad). MFI and the standard deviation (s.d.) of fluorescence intensity across 100 beads were determined for each sample and plotted.
NiVM serum neutralization assays
PRNTs were determined using a conventional serum neutralization assay. Briefly, sera were serially diluted twofold, and incubated with ~ 100 pfu of NiV for 1 hour at 37°C. Virus and antibodies were then added to individual wells of 6-well plates of confluent Vero cell monolayers. Plates were stained with neutral red 2 days after infection and plaques were counted 24 hours after staining. The 50% neutralization titer (PRNT50) was determined as the serum dilution at which there was a 50% reduction in plaque counts versus control wells.
Specimen collection and processing in NiV-infected ferrets
Blood was collected and placed in MiniCollect EDTA tubes or serum tubes (Greiner Bio One, Monroe, NC). Immediately following sampling, 100 μl of blood was added to 600 μl of AVL viral lysis buffer (Qiagen) for RNA extraction. For tissues, approximately 100 mg was stored in 1 ml RNAlater (Qiagen) for 7 days to stabilize RNA. RNAlater was completely removed, and tissues were homogenized in 600 μl RLT buffer (Qiagen) in a 2-ml cryovial using a tissue lyser (Qiagen) and stainless steel beads. The tissues sampled included right lung upper lobe, right lung middle lobe, right lung lower lobe, left lung upper lobe, left lung middle lobe, left lung lower lobe, liver, spleen, kidney, adrenal gland, pancreas, and brain (frontal cortex). All blood samples were inactivated in AVL viral lysis buffer, and tissue samples were homogenized and inactivated in RLT buffer prior to removal from the BSL-4 laboratory. Subsequently, RNA was isolated from blood and swabs using the QIAamp viral RNA kit (Qiagen) and from tissues using the RNeasy minikit (Qiagen) according to the manufacturer’s instructions supplied with each kit.
Hematology and serum biochemistry
Prior to the study, baseline blood and sera were collected via the anterior vena cava from all 20 ferrets. On days -28, 0, 6, and 21 blood was collected from all animals. Complete blood counts of total white blood cell counts, white blood cell differentials, red blood cell counts, platelet counts, hematocrit values, total hemoglobin concentrations, mean cell volumes, mean corpuscular volumes, and mean corpuscular hemoglobin concentrations were analyzed from blood collected in MiniCollect EDTA tubes (Greiner Bio One) using a Hemavet HV950FS instrument per manufacturer’s instructions (Drew Scientific, Oxford, CT). Serum analysis of blood chemistries was performed using a VetScan classic analyzer and comprehensive diagnostic profile rotors measuring of albumin (ALB), amylase, alanine aminotransferase (ALT), alkaline phosphatase (ALP), calcium, glucose, total protein, total bilirubin (TBIL), blood urea nitrogen (BUN), creatinine (CRE), phosphorus, sodium, and total protein (Abaxis, Union City, CA). All blood and serum samples were processed and analyzed immediately after collection.
Histopathology and immunohistochemistry
Necropsy was performed on all subjects. Tissue samples of all major organs were collected for histopathologic and immunohistochemical examination and were immersion-fixed in 10% neutral buffered formalin for at least 21 days in BSL-4. Subsequently, formalin was changed; specimens were removed from BSL-4, processed in BSL-2 by conventional methods and embedded in paraffin and sectioned at 5 μm thickness. For immunohistochemistry, specific anti-NiV immunoreactivity was detected using an anti-NiV N protein rabbit primary antibody (kindly provided by Dr. Christopher Broder) at a 1:5000 dilution for 30 minutes. The tissue sections were processed for immunohistochemistry using the Dako Autostainer (Dako, Carpinteria, CA). Secondary antibody used was biotinylated goat anti-rabbit IgG (Vector Laboratories, Burlingame, CA) at 1:200 for 30 minutes followed by Dako LSAB2 streptavidin-HRP (Dako) for 15 minutes. Slides were developed with Dako DAB chromagen (Dako) for 5 minutes and counterstained with hematoxylin for one minute. Non-immune rabbit IgG was used as a negative staining control.
Detection of NiV load
RNA was isolated from blood or tissues and analyzed using primers/probe targeting the N gene and intergenic region between N and P of NiV for quantitative real-time PCR (qRT-PCR) with the probe used here being 6-carboxyfluorescein (6FAM)-5′ CGT CAC ACA TCA GCT CTG ACG A 3′-6 carboxytetramethylrhodamine (TAMRA) (Life Technologies, Carlsbad, CA). NiV RNA was detected using the CFX96 detection system (Bio-Rad) in One-step probe qRT-PCR kits (Qiagen) with the following cycle conditions: 50°C for 10 minutes, 95°C for 10 seconds, and 40 cycles of 95°C for 10 seconds and 59°C for 30 seconds. Threshold cycle (CT) values representing NiV genomes were analyzed with CFX Manager Software, and data are shown as genome equivalents (GEq). To create the GEq standard, RNA from NiV challenge stocks was extracted and the number of NiV genomes was calculated using Avogadro’s number and the molecular weight of the NiV genome. Virus titration was performed by plaque assay with Vero cells from all serum and control tissue samples. Briefly, increasing 10-fold dilutions of the samples were adsorbed to Vero cell monolayers in duplicate wells (200 μl); the limit of detection was 25 pfu/ml.
References
Wang L, Harcourt BH, Yu M, Tamin A, Rota PA, Bellini WJ, Eaton BT: Molecular biology of Hendra and Nipah viruses. Microbes Infect 2001, 3: 279-287. 10.1016/S1286-4579(01)01381-8
Halpin K, Hyatt AD, Fogarty R, Middleton D, Bingham J, Epstein JH, Abdul Rahman S, Hughes T, Smith C, Field HE: Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am J Trop Med Hyg 2011, 85: 946. 10.4269/ajtmh.2011.10-0567
Chua KB, Goh KJ, Wong KT, Kamarulzaman A, Tan PS, Ksiazek TG, Zaki SR, Paul G, Lam SK, Tan CT: Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 1999, 354: 1257-1259. 10.1016/S0140-6736(99)04299-3
Chadha MS, Comer JA, Lowe L, Rota PA, Rollin PE, Bellini WJ, Ksiazek TG, Mishra A: Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis 2006, 12: 235-240. 10.3201/eid1202.051247
Gurley ES, Montgomery JM, Hossain MJ, Bell M, Azad AK, Islam MR, Molla MA, Carroll DS, Ksiazek TG, Rota PA, et al.: Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis 2007, 13: 1031-1037. 10.3201/eid1307.061128
Luby SP, Gurley ES: Epidemiology of henipavirus disease in humans. Curr Top Microbiol Immunol 2012, 359: 25-40.
Guillaume V, Contamin H, Loth P, Georges-Courbot MC, Lefeuvre A, Marianneau P, Chua KB, Lam SK, Buckland R, Deubel V, Wild TF: Nipah virus: vaccination and passive protection studies in a hamster model. J Virol 2004, 78: 834-840. 10.1128/JVI.78.2.834-840.2004
Weingartl HM, Berhane Y, Caswell JL, Loosmore S, Audonnet JC, Roth JA, Czub M: Recombinant nipah virus vaccines protect pigs against challenge. J Virol 2006, 80: 7929-7938. 10.1128/JVI.00263-06
Ploquin A, Szecsi J, Mathieu C, Guillaume V, Barateau V, Ong KC, Wong KT, Cosset FL, Horvat B, Salvetti A: Protection against henipavirus infection by use of recombinant adeno-associated virus-vector vaccines. J Infect Dis 2013, 207: 469-478. 10.1093/infdis/jis699
McEachern JA, Bingham J, Crameri G, Green DJ, Hancock TJ, Middleton D, Feng YR, Broder CC, Wang LF, Bossart KN: A recombinant subunit vaccine formulation protects against lethal Nipah virus challenge in cats. Vaccine 2008, 26: 3842-3852. 10.1016/j.vaccine.2008.05.016
Mungall BA, Middleton D, Crameri G, Bingham J, Halpin K, Russell G, Green D, McEachern J, Pritchard LI, Eaton BT, et al.: Feline model of acute nipah virus infection and protection with a soluble glycoprotein-based subunit vaccine. J Virol 2006, 80: 12293-12302. 10.1128/JVI.01619-06
Pallister JA, Klein R, Arkinstall R, Haining J, Long F, White JR, Payne J, Feng YR, Wang LF, Broder CC, Middleton D: Vaccination of ferrets with a recombinant G glycoprotein subunit vaccine provides protection against Nipah virus disease for over 12 months. Virol J 2013, 10: 237. 10.1186/1743-422X-10-237
Pallister J, Middleton D, Wang LF, Klein R, Haining J, Robinson R, Yamada M, White J, Payne J, Feng YR, et al.: A recombinant Hendra virus G glycoprotein-based subunit vaccine protects ferrets from lethal Hendra virus challenge. Vaccine 2011, 29: 5623-5630. 10.1016/j.vaccine.2011.06.015
Bossart KN, Rockx B, Feldmann F, Brining D, Scott D, LaCasse R, Geisbert JB, Feng YR, Chan YP, Hickey AC: A Hendra virus G glycoprotein subunit vaccine protects African green monkeys from Nipah virus challenge. Sci Transl Med 2012, 4: 146ra107.
Reuter JD, Vivas-Gonzalez BE, Gomez D, Wilson JH, Brandsma JL, Greenstone HL, Rose JK, Roberts A: Intranasal vaccination with a recombinant vesicular stomatitis virus expressing cottontail rabbit papillomavirus L1 protein provides complete protection against papillomavirus-induced disease. J Virol 2002, 76: 8900-8909. 10.1128/JVI.76.17.8900-8909.2002
Roberts A, Reuter JD, Wilson JH, Baldwin S, Rose JK: Complete protection from papillomavirus challenge after a single vaccination with a vesicular stomatitis virus vector expressing high levels of L1 protein. J Virol 2004, 78: 3196-3199. 10.1128/JVI.78.6.3196-3199.2004
Egan MA, Chong SY, Rose NF, Megati S, Lopez KJ, Schadeck EB, Johnson JE, Masood A, Piacente P, Druilhet RE, et al.: Immunogenicity of attenuated vesicular stomatitis virus vectors expressing HIV type 1 Env and SIV Gag proteins: comparison of intranasal and intramuscular vaccination routes. AIDS Res Hum Retroviruses 2004, 20: 989-1004. 10.1089/aid.2004.20.989
Rose NF, Marx PA, Luckay A, Nixon DF, Moretto WJ, Donahoe SM, Montefiori D, Roberts A, Buonocore L, Rose JK: An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 2001, 106: 539-549. 10.1016/S0092-8674(01)00482-2
Rose NF, Roberts A, Buonocore L, Rose JK: Glycoprotein exchange vectors based on vesicular stomatitis virus allow effective boosting and generation of neutralizing antibodies to a primary isolate of human immunodeficiency virus type 1. J Virol 2000, 74: 10903-10910. 10.1128/JVI.74.23.10903-10910.2000
Roberts A, Buonocore L, Price R, Forman J, Rose JK: Attenuated vesicular stomatitis viruses as vaccine vectors. J Virol 1999, 73: 3723-3732.
Schlereth B, Buonocore L, Tietz A, Meulen Vt V, Rose JK, Niewiesk S: Successful mucosal immunization of cotton rats in the presence of measles virus-specific antibodies depends on degree of attenuation of vaccine vector and virus dose. J Gen Virol 2003, 84: 2145-2151. 10.1099/vir.0.19050-0
Schlereth B, Rose JK, Buonocore L, ter Meulen V, Niewiesk S: Successful vaccine-induced seroconversion by single-dose immunization in the presence of measles virus-specific maternal antibodies. J Virol 2000, 74: 4652-4657. 10.1128/JVI.74.10.4652-4657.2000
Johnson JE, McNeil LK, Megati S, Witko SE, Roopchand VS, Obregon JH, Illenberger DM, Kotash CS, Nowak RM, Braunstein E, et al.: Non-propagating, recombinant vesicular stomatitis virus vectors encoding respiratory syncytial virus proteins generate potent humoral and cellular immunity against RSV and are protective in mice. Immunol Lett 2013, 150: 134-144. 10.1016/j.imlet.2012.12.005
Kahn JS, Roberts A, Weibel C, Buonocore L, Rose JK: Replication-competent or attenuated, nonpropagating vesicular stomatitis viruses expressing respiratory syncytial virus (RSV) antigens protect mice against RSV challenge. J Virol 2001, 75: 11079-11087. 10.1128/JVI.75.22.11079-11087.2001
Kapadia SU, Rose JK, Lamirande E, Vogel L, Subbarao K, Roberts A: Long-term protection from SARS coronavirus infection conferred by a single immunization with an attenuated VSV-based vaccine. Virology 2005, 340: 174-182. 10.1016/j.virol.2005.06.016
Chattopadhyay A, Wang E, Seymour R, Weaver SC, Rose JK: A chimeric vesiculo/alphavirus is an effective alphavirus vaccine. J Virol 2013, 87: 395-402. 10.1128/JVI.01860-12
Garbutt M, Liebscher R, Wahl-Jensen V, Jones S, Moller P, Wagner R, Volchkov V, Klenk HD, Feldmann H, Stroher U: Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J Virol 2004, 78: 5458-5465. 10.1128/JVI.78.10.5458-5465.2004
Chattopadhyay A, Rose JK: Complementing defective viruses that express separate paramyxovirus glycoproteins provide a new vaccine vector approach. J Virol 2011, 85: 2004-2011. 10.1128/JVI.01852-10
Lo MK, Bird BH, Chattopadhyay A, Drew CP, Martin BE, Coleman JD, Rose JK, Nichol ST, Spiropoulou CF: Single-dose replication-defective VSV-based Nipah virus vaccines provide protection from lethal challenge in Syrian hamsters. Antiviral Res 2013., 101C: 26-29. doi: 10.1016/j.antiviral.2013.10.012
Bossart KN, Zhu Z, Middleton D, Klippel J, Crameri G, Bingham J, McEachern JA, Green D, Hancock TJ, Chan YP, et al.: A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute nipah virus infection. PLoS Pathog 2009, 5: e1000642. 10.1371/journal.ppat.1000642
Clayton BA, Middleton D, Bergfeld J, Haining J, Arkinstall R, Wang L, Marsh GA: Transmission routes for Nipah virus from Malaysia and Bangladesh. Emerg Infect Dis 2012, 18:. (12):1983-93. doi: 10.3201/eid1812.120875
Geisbert TW, Daddario-DiCaprio KM, Hickey AC, Smith MA, Chan YP, Wang LF, Mattapallil JJ, Geisbert JB, Bossart KN, Broder CC: Development of an acute and highly pathogenic nonhuman primate model of Nipah virus infection. PLoS One 2010, 5: e10690. 10.1371/journal.pone.0010690
Whitt MA: Generation of VSV pseudotypes using recombinant DeltaG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J Virol Methods 2010, 169: 365-374. 10.1016/j.jviromet.2010.08.006
Mendez D, Buttner P, Speare R: Response of Australian veterinarians to the announcement of a Hendra virus vaccine becoming available. Aust Vet J 2013, 91: 328-331. 10.1111/avj.12092
Dhondt KP, Mathieu C, Chalons M, Reynaud JM, Vallve A, Raoul H, Horvat B: Type I interferon signaling protects mice from lethal henipavirus infection. J Infect Dis 2013, 207: 142-151. 10.1093/infdis/jis653
Middleton DJ, Morrissy CJ, van der Heide BM, Russell GM, Braun MA, Westbury HA, Halpin K, Daniels PW: Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus). J Comp Pathol 2007, 136: 266-272. 10.1016/j.jcpa.2007.03.002
Berhane Y, Weingartl HM, Lopez J, Neufeld J, Czub S, Embury-Hyatt C, Goolia M, Copps J, Czub M: Bacterial infections in pigs experimentally infected with Nipah virus. Transbound Emerg Dis 2008, 55: 165-174. 10.1111/j.1865-1682.2008.01021.x
Middleton DJ, Westbury HA, Morrissy CJ, van der Heide BM, Russell GM, Braun MA, Hyatt AD: Experimental Nipah virus infection in pigs and cats. J Comp Pathol 2002, 126: 124-136. 10.1053/jcpa.2001.0532
Weingartl H, Czub S, Copps J, Berhane Y, Middleton D, Marszal P, Gren J, Smith G, Ganske S, Manning L, Czub M: Invasion of the central nervous system in a porcine host by Nipah virus. J Virol 2005, 79: 7528-7534. 10.1128/JVI.79.12.7528-7534.2005
Georges-Courbot MC, Contamin H, Faure C, Loth P, Baize S, Leyssen P, Neyts J, Deubel V: Poly(I)-poly(C12U) but not ribavirin prevents death in a hamster model of Nipah virus infection. Antimicrob Agents Chemother 2006, 50: 1768-1772. 10.1128/AAC.50.5.1768-1772.2006
Guillaume V, Contamin H, Loth P, Grosjean I, Courbot MC, Deubel V, Buckland R, Wild TF: Antibody prophylaxis and therapy against Nipah virus infection in hamsters. J Virol 2006, 80: 1972-1978. 10.1128/JVI.80.4.1972-1978.2006
Rockx B, Brining D, Kramer J, Callison J, Ebihara H, Mansfield K, Feldmann H: Clinical outcome of henipavirus infection in hamsters is determined by the route and dose of infection. J Virol 2011, 85: 7658-7671. 10.1128/JVI.00473-11
Pallister J, Middleton D, Crameri G, Yamada M, Klein R, Hancock TJ, Foord A, Shiell B, Michalski W, Broder CC: Chloroquine administration does not prevent Nipah virus infection and disease in ferrets. J Virol 2009, 83: 11979-11982. 10.1128/JVI.01847-09
Marianneau P, Guillaume V, Wong T, Badmanathan M, Looi RY, Murri S, Loth P, Tordo N, Wild F, Horvat B, Contamin H: Experimental infection of squirrel monkeys with Nipah virus. Emerg Infect Dis 2010, 16: 507-510. 10.3201/eid1603.091346
Acknowledgements
We thank Joan B. Geisbert and Daniel J. Deer for study support, Jessica Graber for expert technical assistance with the ABSL-2 and ABSL-4 sample collection during the vaccination and infection studies, and the staff of the UTMB Animal Resources Center for animal care. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by UTMB.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CEM designed the vaccines and vaccination study, carried out the animal vaccination and infection studies, processed animal tissues and blood, analyzed data, and drafted the manuscript. KMV cloned, recovered, characterized, and propagated the vaccines, processed animal tissues and blood, and participated in manuscript preparation. RWC carried out animal vaccination and infection studies, processed animal tissues and blood, analyzed data, and participated in manuscript preparation. KNA participated in animal infection studies, processed animal tissues and blood, performed serology, performed virus isolation, performed qRT-PCR, analyzed data, and participated in manuscript preparation. KAF participated in animal infection studies, provided veterinary pathology expertise, analyzed data, and participated in manuscript preparation. MAW participated in design of vaccine construction and study design, analyzed data, and edited manuscript. TWG conceived the study, supported the work with research funds from the National Institutes of Health (U01 AI082121) and UTMB, analyzed data, and edited the manuscript. All authors read and approved the final manuscript.
Chad E Mire, Krista M Versteeg contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Mire, C.E., Versteeg, K.M., Cross, R.W. et al. Single injection recombinant vesicular stomatitis virus vaccines protect ferrets against lethal Nipah virus disease. Virol J 10, 353 (2013). https://doi.org/10.1186/1743-422X-10-353
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-10-353