Abstract
Background
There is a long-standing controversial about how parthenogenetic species can be defined in absence of a generally accepted species concept for this reproductive mode. An integrative approach was suggested, combining molecular and morphological data to identify distinct monophyletic entities. Using this approach, speciation of parthenogenetic lineages was recently demonstrated for groups of bdelloid rotifers and oribatid mites. Trhypochthonius tectorum, an oribatid mite from the entirely parthenogenetic desmonomatan family Trhypochthoniidae, is traditionally treated as a single species in Central Europe. However, two new morphological lineages were recently proposed for some Austrian populations of T. tectorum, and were described as novel subspecies (T. silvestris europaeus) or form (T. japonicus forma occidentalis). We used the morphological and morphometrical data which led to this separation, and added mitochondrial and nuclear DNA sequences and the chemical composition of complex exocrine oil gland secretions to test this taxonomical hypothesis. This is the first attempt to combine these three types of data for integrative taxonomical investigations of oribatid mites.
Results
We show that the previous European species T. tectorum represents a species complex consisting of three distinct lineages in Austria (T.tectorum, T. silvestris europaeus and T. japonicus forma occidentalis), each clearly separated by morphology, oil gland secretion profiles and mitochondrial cox1 sequences. This diversification happened in the last ten million years. In contrast to these results, no variation among the lineages was found in the nuclear 18S rDNA.
Conclusions
Our approach combined morphological, molecular and chemical data to investigate diversity and species delineation in a parthenogenetic oribatid mite species complex. To date, hypotheses of a general oribatid mite phylogeny are manifold, and mostly based on single-method approaches. Probably, the integrative approach proposed here can be used to uncover further hidden biodiversity of glandulate Oribatida and help to build up more stable phylogenetic hypotheses in the future.
Similar content being viewed by others
Background
More than twenty hypotheses try to explain the advantages of sexual reproduction over parthenogenesis or asexuality [1, 2]. Most of these theories tolerate the existence of parthenogenetic species in the short-term, but predict that there should be no radiation and long-term survival of groups lacking sexual reproduction. About 2,000 parthenogenetic species have been described among almost all groups of animals [3]. However, existence and recognition of parthenogenetic species remains a controversial topic, mostly due to the fact that the traditional biological species concept is axiomatically related to sexuality. Additionally, misunderstandings of parthenogenetic population genetics have led to the prediction that parthenogenetic organisms must form a continuum of genetic variation [4]. But this is not necessarily true - parthenogenetic lineages can split into independently evolving entities, thus speciation of parthenogens can be addressed empirically [5]. Recently, speciation of ancient parthenogenetic lineages has been demonstrated for bdelloid rotifers [4, 6–8] and several groups of oribatid mites [9–13]. High and consistent clonal diversity was also demonstrated for the putative ancient parthenogenetic Darwinula stevensoni (Ostracoda) [14], contrasting the low diversity shown earlier [15].
The existence of parthenogenetic species has been proposed in different species concepts, including the evolutionary, ecological and phylogenetic species concepts [16–18], but it remains a major concern how a parthenogenetic species can be defined in a biological meaningful context. Recently, a new evolutionary genetic species concept, based on population genetic theory and DNA sequence data, has been proposed and applied to delineate parthenogenetic species of bdelloid rotifers and oribatid mites [19, 20]. Another DNA-sequence based approach, genetic barcoding, uses a part of the mitochondrial cytochrome oxidase 1 (cox1) gene to differentiate between species on the basis of genetic distances and was proposed to be useful for the identification of undescribed species [21–23]. However, this pure molecular-based barcoding was criticized [24–27] to be a phenetic, non-cladistic approach and no general definition is available for the amount of genetic distance indicating a separation of lineages into species. Hence, an integrative approach was suggested, combining data from multiple sources for the identification and definition of new species [28–31] and such integrative approaches using molecular and morphological data were successfully used for the identification of independently evolving lineages within parthenogenetic clusters of bdelloid rotifers [6] and the parthenogenetic oribatid mite genus Tectocepheus[13]. However, it was suggested that at least three different sources of data should be included for a reliable delimitation of species boundaries [30, 31]. Besides morphological and molecular data, we included the chemical composition of oil gland secretions to investigate characteristics of Austrian populations of the oribatid mite Trhypochthonius tectorum.
Oil glands are paired opisthosomal sac-like exocrine glands characteristic of the so-called 'glandulate Oribatida' [32] and may contain complex mixtures of terpenes, aromatics, hydrocarbons [33] and alkaloids [34]. The chemical composition of oil gland secretions was shown to be a phylogenetically informative set of characters [35], allowing also differentiation between populations of parthenogenetic oribatid mite species [33].
Oribatid mites are a speciose group of chelicerates (~10.000 species, [36]) with Devonian [37], Silurian [38] or Precambrian [39] origin. Parthenogenesis is widespread among the Oribatida and several large monophyletic and parthenogenetic groups exist, consisting of 50 to 180 morphologically described species [10, 12]. One of these exclusively parthenogenetic families, the Trhypochthoniidae [40], comprises 51 species [41] with about 25 species in the genus Trhypochthonius[42]. Parthenogenetic reproduction of Trhypochthoniidae was first assumed by Grandjean in 1941, based on the rarity of males [43], and later experimentally proven for numerous species of this family [44–46]. Trhypochthonius tectorum[47] was reported from Holarctic, Oriental and Neotropic regions and a number of subspecies have been described using morphology only [41], although their identity is questionable [40]. Previously assumed as a single species, Trhypochthonius tectorum was recently hypothesized to be a species complex rather than a single species in Austria, and a new subspecies (T. silvestris europaeus) as well as a new form (T. japonicus forma occidentalis) have been differentiated from T. tectorum s. str. using morphological data [48]. Here, we expand this morphological analysis of Austrian populations by including molecular and chemical data to test the hypothesis of independent entities using an integrative approach.
We show that the three lineages proposed by [48] are independent entities, clearly separated by morphology, gland secretions and mitochondrial sequences and that completely homogeneous nuclear ribosomal DNA contrasts this separation.
Results
Chemical analyses
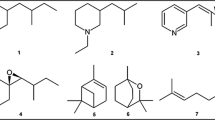
Analyses of oil gland secretion profiles led to three distinct chemical profiles (Figure 1, Table 1). One of the gas chromatographic profiles was identical to published data of T. tectorum[49] hence the lineage showing this profile was denoted as T. tectorum (TT) for morphometrical and molecular analyses. The chemical profile of TT consisted of eleven compounds with characteristic relative abundance (Table 1). The compounds were 2-hydroxy-6-methylbenzaldehyde (= 2,6-HMBD; peak 1), neral (peak 2), geranial (peak 3), 2-formyl-3-hydroxy benzaldehyde (= 2,3-FHBD, = γ-acaridial; peak 5), pentadecane (peak 7), 6,9-heptadecadiene (peak 9, identified by DMDS-derivatives), heptadecene (peak 10, double bond position not identified, probably 4-heptadecane), (Z,E)-farnesal (peak 11), (E,E)-farnesal (peak 12) and two unknown components (peaks 6, 8). The described profile was consistently found in all extracts of TT from any location (CF, SG and SB; see Methods for locations). In contrast to this already well-known profile of T. tectorum, the profiles of T. silvestris europaeus (denoted as TA) and T. japonicus forma occidentalis (denoted as TB) were considerably different. The TA profile from collection site CW (see Methods for location), lacked 2,6-HMBD, but in addition showed small amounts of neryl formate (Figure 1, peak 4). TB was syntopically found at sample site CW and the chemical profile lacked 2,6-HMBD, neral, geranial and neryl formate. Hence, the TB-profile consisted of eight compounds only (Figure 1). An outgroup comparison was done with Archegozetes longisetosus, confirming the already published ten compounds-profile of 2,6-HMBD, neral, geranial, neryl formate, γ-acaridial, pentadecene, n-pentadecane, heptadecadiene, heptadecene and, although only in trace quantities, heptadecane [50, 51].
Apart from easily visible qualitative differences, all profiles were quantified (leading to characteristic patterns of relative abundance of components in each profile (Table 1), and were subsequently subjected to multivariate statistics, forming consistent and significant clusters that do not overlap (Figure 2).
Canonical discriminant analysis of oil gland secretions. Estimation of the validity of the discriminant function is based on the significance of Wilk's Lambda and the percentage of correct assignment. Four chemical groups were verified, clearly indicating that TT, TA, TB and AL are chemically completely separated (100% of cases were correctly assigned to the four previously defined taxa).
Morphometrical analyses
Details of morphometrical measurements are given in [48]. Here, we only shortly summarize the main results that we used within this integrative framework. TT, TA and TB differed significantly in body length. With a mean body length of 643 μm, TT was larger than TA (mean: 597 μm), but smaller than TB (mean: 717 μm). Besides this, TT, TA and TB could be separated by their different numbers of genital setae and their relative length of the notogastral setae c 2 , d 1 , d 3 , e 1 and p 3 (exemplified for c 2 in Figure 3). In addition, distance-based cluster analyses of the setae types show a clear separation of the three groups with a higher similarity of TT and TA than any of these has to TB (Figure 4).
Morphometric similarities. Tree cluster on the similarities among TT, TA and TB. Distances calculated from summed differences of notogastral setae types (see [48]).
Molecular analyses
A 600 bp fragment of the cox1 gene was obtained from each three specimens of TT, TA and TB and the outgroup AL and aligned by hand without any ambiguity or gaps. No variation was found within the replicates of TT, TA, TB and AL. In total, 181 (30.2%) nucleotide positions were variable and informative. Excluding the outgroup, 127 bp (21.2%) were variable and informative among the three Trhypochthonius groups TT, TA and TB. TT was characterized by eleven apomorphic nucleotide positions, TA showed two apomorphies, and for TB there were 73 apomorphic characters. TT and TA showed 82 synapomorphies, contradicted by three positions shared by TA and TB. Not a single synapomorphy was found for TT and TB. Maximum Parsimony analyses in PAUP* resulted in a single tree with a tree-length of 218 and consistency index (CI) and rescaled consistency index (RC) of 0.99 each (Figure 5). The identical topology was found with Maximum Likelihood analyses.
Relative rate tests using AL as outgroup and all combinations of TT, TA and TB as ingroup taxa resulted in no significant rate variations (χ²(1) < 3.6, p > 0.05). In addition, a likelihood ratio test, based on the likelihoods of the corresponding branch-and-bound trees, was performed with the molecular clock enforced and not enforced (enforced: -ln L = 1679.427; not enforced: -ln L = 1674.96; χ²(10) = 0.104, p > 0.99) and showed also no rate variation. Therefore, the assumption of a molecular clock seems appropriate. A molecular divergence rate of 2.15% per million years was estimated for the cox1 gene of oribatid mites [12, 52]. Genetic pairwise p-distances were corrected by an evolutionary model (HKY, [53]; estimated by hLRTs, AIC and BIC in Modeltest 3.7, [54]) with nucleotide composition A: 0.2531, C: 0.2198, G: 0.19, T: 0.3371 and k: 1.5108 (Table 2). Evolutionary ages of the lineages were estimated based on the corrected distances: TT and TA separated about one million years ago and the last common ancestor of TB and TT/TA lived about eleven million years ago (Figure 5).
The alignment of the nuclear 18S rDNA consisted of 1700 nucleotides. All Trhypochthonius-sequences were identical, and all were identical to a published sequence of T. americanus (EF081298, [11]). Hence, no further phylogenetic analyses of these sequences were performed.
Discussion
What is a parthenogenetic species? We do not aim to give an exhaustive discussion on this difficult topic, but we want to shade light on some practical implications, i.e. methods to detect separated genotypic and phenotypic entities (whether they will be denoted as species or not). It is clear that reproductive isolation, the basis of the biological species concept [55], is meaningless for the definition of parthenogenetic species. Almost all parthenogens are described on their morphology only (morphospecies), applying at best the same subjective criteria for discriminating species as taxonomists do with sexual species. The occurrence of phenotypic plasticity or the absence of phenotypic variation despite genotypic variability ('cryptic species', see [12]) can be observed in many groups irrespective their mode of reproduction. Therefore, both may be analyzed with the same procedures. However, the delineation of species and their subsequent classification into larger taxonomic units may be somehow problematic, especially when dealing with character-poor organisms of small size and similar morphology. Many soil-dwelling arthropods belong to this group, such as some highly-conservative opilionids of different suborders [56, 57], but also many Oribatida. In these groups numerous so-called 'species-complexes' exist, i.e. assemblages of similar species or sub-species that are not clearly delineated from each other. Such 'species' may either show a high intraspecific variability of characters or may actually represent groups of closely related, cryptic (or nearly cryptic) species. In many cases, one set of characters alone - e.g. traditional characters from external morphology - fails to answer questions on this low taxonomic level. Hence, numerous approaches towards integrative taxonomy have been attempted in the last years: using a combination of methods, a more rigorous concept of the delimitation of problematic species has been introduced [30, 31]. Many examples of the successful application of combined methods meanwhile exist, and with respect to the Opiliones mentioned above, a large number of new but so far cryptic opilionid Cyphophthalmus-species on the Balkan Peninsula have been discovered using morphological and molecular characters [57, 58]. By contrast, the systematics of Oribatida suffers greatly from still uni-methodological approaches: i) the majority of taxonomic studies in the Oribatida is still exclusively based on traditional sets of data derived from external morphology; and ii) novel methods, such as molecular phylogenetic approaches, are rarely combined with morphological data. Molecular data for the delineation of the parthenogenetic oribatid mite genus Tectocepheus were presented [13] and combined with morphological data from [59] to demonstrate parthenogenetic radiation - a rare example of an integrative approach in oribatid mite systematics.
Each uni-methodological approach, including molecular techniques, is assumed to have an inherent failure rate in the delimination of species [31]. With respect to taxonomic studies in arthropods, and according to [31], the failure rate is 28% when using nuclear DNA-data alone, and 33% when using mitochondrial DNA. Failure rates arising from studies using morphological or chemical data alone show similar failure rates of 23% and 22%, respectively. Combining any two of these methods leads to a reduced failure rate of 9%, but only when three are combined, a statistically acceptable failure rate below 5% can be achieved [31].
Chemical data
With respect to glandulate Oribatida and their multicomponent secretions from the oil glands, an independent pool of characters has been made available to oribatid systematics and phylogeny in the last years [33, 35]. One model group for such studies is the Trhypochthoniidae, medium to large oribatids that i) possess largely developed oil glands, making it possible to analyze individual extracts in some species, ii) show specific combinations of chemically already characterized compounds (so-called 'Astigmata compounds' sensu[60]), and iii) generally exhibit information-rich multi-component secretion profiles. In addition, a considerable data base on their secretions has been generated, representing an important source for reference: in detail, secretion profiles of Archegozetes longisetosus[50, 51], Trhypochthoniellus crassus and three species of Trhypochthonius (T. tectorum, T. japonicus and a not determined Japanese Trhypochthonius species) have already been analyzed, each showing species-specific and interspecifically distinctive secretion profiles [49, 61, 62]. Considering these data, the profile of TT appears to be rather basal within Trhypochthoniidae, showing the full spectrum of 'Astigmata compounds' except for neryl formate. The lack of 2,6-HMBD in TA and TB, however, may be due to convergent reduction, especially when regarding the clear phylogenetic relatedness of TT and TA implied by molecular data.
Morphological data
In a morphometrical analysis that was the initiation of this integrative project (details in [48]) the three distinct European lineages within Trhypochthonius tectorum s. lat. (TT, TA, TB) were compared with T. japonicus[63] from Japan, T. americanus[64] and T. silvestris[65], both from North America. TA looked quite similar to T. silvestris, but was statistically distinct, and hence was proposed as subspecies T. silvestris europaeus[48]. TB was very similar to T. japonicus, the difference in morphometric respect was small, but partly significant, and thus TB was classified as geographically distinct T. japonicus forma occidentalis[48]. This close relationship is also supported by oil gland chemistry showing nearly identical secretion profiles of TB and T. japonicus[62]. Since there is a graduated degree of similarities with respect to the morphological characters within the lineages, these were expressed taxonomically as form or subspecies [48]. Morphologically, species of Trhypochthonius show several evolutionary lineages, one of these is the T. tectorum species complex [48], which - apart from T. tectorum - contains several other representatives from, e.g., North America and Japan.
Molecular data
Phylogenetic analyses using the maximum parsimony criterion are prone to the phenomenon of long-branch attraction, especially when molecular data are used and divergences between sequences are high [66]. Hence, if long branches occur in the data, an alternative method, such as maximum likelihood, is desired. However, if no long branches exist in the data, the data-set is small enough (less than 25 taxa) to be analyzed exhaustively (i.e. with a guarantee to find the shortest tree), only a single shortest tree is to be found, and the distribution of characters is highly congruent on this shortest tree, then we see no good reason to use other methods than maximum parsimony (however, we performed also maximum likelihood analyses with identical results). In this study, the 600 bp cox1-alignment of the Trhypochthonius-species showed 127 variable and phylogenetically informative nucleotide positions (21.2%). There were clear apomorphies for each of the three lineages, and a high number (82) of synapomorphies for the sister-taxa TT and TA, contradicted by only three nucleotide positions supporting TA+TB. This results in a high consistency (and rescaled consistency) index of 0.99, very close to complete congruence (Figure 5). However, the bootstrap-support for the monophyly of TA is only 78, which can be explained by the low (but consistent) number of only two apomorphies that define this taxon. There is not a single position that supports any other hypotheses than the monophyly of TA, but the two apomorphies simply get lost by chance in 22% of the bootstrap resampling procedure. Hence, we think that the high amount of informative positions and the high consistency index clearly support the topology given in Figure 5.
In contrast to the high divergence and information of the cox1-sequences, the 1,700 bp alignment of the nuclear ribosomal 18S sequences showed no variation at all. We included a published sequence of T. americanus (EF081298) in the alignment, and this sequence also was identical. This phenomenon is not unique among mites - Navajas et al. [67] reported 5% of divergence in the mitochondrial COI sequences and no variability in the ribosomal nuclear ITS2 sequences in the spider mite Tetranychus urticae. This was explained by a high colonization potential of this species, preventing long-term differentiation. However, T. urticae is a sexually reproducing, thus recombining pest-species, and T. tectorum is a parthenogenetic species belonging to the Desmonomata, hence presumably has an inverted meiotic sequence and no meiotic recombination [68–71]. Another ancient parthenogenetic species, Darwinula stevensoni (Ostracoda) also showed this same pattern: homogenized nuclear ribosomal sequences in contrast to divergent mitochondrial COI sequences [15]. Here, this pattern was explained by a reduced mutation rate and effective machinery for DNA repair. We do not exactly know ultimate causes for the contrasting nuclear and mitochondrial divergence in T. tectorum, but we think that besides a lower mutation rate of the nuclear genome this could be a result of the special reproductive mechanism: automixis with inverted meiosis and terminal fusion [70].
The Trhypochthonius tectorum complex was hitherto conceived as a single species in Europe. Our integrative approach shows consistently that i) the recently proposed T. silvestris europaeus and T. japonicus forma occidentalis are distinct taxonomical entities, ii) T. tectorum and T. silvestris europaeus are related taxa which separated about one million years ago, iii) T. japonicus forma occidentalis separated from T. tectorum and T. silvestris europaeus 11-12 million years ago.
Conclusions
We showed that an integrative approach, combining morphometrical, chemical, and molecular data, could be used to identify distinct lineages within a parthenogenetic oribatid mite species complex. A combination of these three methods might also help in unraveling at least some of the numerous controversies in glandulate oribatid mite phylogeny.
The two new lineages T. silvestris europaeus and T. japonicus forma occidentalis were found by taking only a few, random samples in Austria. Hence, we assume that a more thorough sampling all over the Holarctic range of distribution will probably uncover numerous additional lineages within the T. tectorum complex. Thus, unless this complex is investigated in more detail, and to avoid further confusion, the recently proposed taxonomical rank of T. silvestris europaeus (subspecies) and T. japonicus forma occidentalis (form) is presently left unchanged.
A future agreement for the definition of parthenogenetic species in an integrative context seems desirable, especially since more and more different sources of data (morphological, molecular, chemical, biochemical, physiological, ecological, behavioural) are included in integrative approaches.
Methods
Specimens
Four sites in Austria were sampled; specimens of T. tectorum were collected by hand and kept alive for individual extraction and chemical analyses of oil gland secretion profiles. Subsequently, specimens were sorted with respect to their secretion profiles and size, stored in ethanol and analyzed morphometrically and genetically. Sample sites were: (1) Carinthia, Ferlach, moss on a roof (= CF); (2) Carinthia, Waidischbach, moss and litter in a Pinus stand (= CW); (3) Styria, Graz, moss on a street pavement (= SG); (4) Styria, Bachsdorf, moss on a roof (= SB).
The laboratory lineage A. longisetosus ran (= AL, [72]), also a member of the parthenogenetic Trhypochthoniidae, originated from our laboratory culture and was used as outgroup for phylogenetic analyses of molecular data and for comparisons of oil gland chemistry.
Chemical analyses
Specimens were handled with care to avoid release of their oil gland secretions prior to extraction. Extracts were prepared by submersing living individuals in 50 μl of hexane for 30 minutes for a discharge of secretions into the solvent [73]. Crude extracts were used for chemical analyses using a Trace gas chromatograph (GC) coupled to a Voyager mass spectrometer (MS) (both from Thermo, Vienna, Austria). The GC-column (ZB-5MS fused silica capillary column: 30 m × 0.25 mm i.d., 0.25 μm film thickness; Phenomenex, Aschaffenburg, Germany) was directly connected to the ion source of the MS. The splitless Grob injector was kept at 260°C, and helium was used as a carrier gas with a constant flow rate of 1.5 ml/min. The temperature program was set to 50°C (1 min), followed by an increase of 10°C/min until 200°C were reached, then 15°C/min until 300°C were reached with a final isothermal hold (300°C) for 5 minutes. The ion source of the MS was kept at 150°C and the transfer line at 310°C. Electron impact spectra were recorded at 70 eV.
Where possible, compounds were identified on the basis of mass spectral data and comparison of retention times to authentic standards or tentatively, by interpretation and comparison of mass spectra to reference spectra from literature and the NIST-library [73].
Secretion profiles were evaluated by integration of peak areas in the chromatograms and by calculation of the relative abundance of peaks (given in % of peak area of the whole secretion). Secretion profiles, including qualitative and quantitative information, were further subjected to discriminant analyses (using SPSS 16). Compounds were treated as variables, and the profiles evaluated represented the 'cases' for analyses. Stepwise discriminant analyses were carried out to determine whether the previously (morphologically) defined groups (4 species) could be discriminated on basis of their chemical profiles and to evaluate which compounds mainly discriminated between groups. Wilk`s Lambda and the percentage of correct assignment were used to estimate validity of discrimination.
Morphometrical analyses
Specimens were macerated in lactic acid and mounted in open cavity slides covered partly by a cover glass, which allows turning each specimen for microscopic analyses from all perspectives. Details of measurements are given in [48].
Each statistical analysis for setae and notogaster lengths was performed as Kruskal-Wallis-ANOVA-test (H-test) for multiple tests over all populations. In cases of significance, subsequently a pairwise Mann-Whitney-Median-test (U-test) was used for detecting the significant differences between population pairs.
The multi-dimensional cluster analysis of the qualitative differences of notogastral setae between the populations were based on setal types where each of the 15 setae (c1 - p3) represents one dimension. The pairwise numerical differences between the Trhypochthonius populations of all notogastral setal types were used for a tree-cluster analysis (complete linkage of all Manhattan-City-Block-distances).
Molecular analyses
Total DNA was extracted from single specimens using the DNeasy Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. PCR was performed with the HotStarTaq Master Mix kit (Qiagen, Hilden, Germany); the total reaction volume of 20 μl contained 1.5 mM MgCl2, 100 pmol of each primer, 200 μM of each dNTP and 1 Unit of Taq-polymerase.
A 600 bp fragment of the mitochondrial cox1 gene, corresponding to the amino acid positions 19-218 of the Steganacarus magnus (Oribatida) cox1 protein [74] was obtained with the primers and protocol given in [12]. Nuclear sequences of the 18S rDNA (1,700 bp) were amplified using primers and procedure described in [13]. Sequencing was performed in both directions on an ABI capillary sequencer. Sequences were deposited in GenBank (18S data set: A. longisetosusHQ661379, T. silvestris europaeusHQ661380-HQ661382, T. japonicus forma occidentalisHQ711366-HQ711368, T. tectorumHQ711369-HQ711371; cox1 data set: A. longisetosusHQ711372, T. silvestris europaeusHQ711373-HQ711375, T. japonicus forma occidentalisHQ711376-HQ711378, T. tectorumHQ711379-HQ711381).
Sequences were verified to be of oribatid mite origin by comparisons with known sequences in GenBank using the BLASTN search algorithm [75] and aligned by hand in BioEdit 7 [76]. Models for sequence evolution and corresponding parameters were estimated using hierarchical likelihood ratio tests (hlrts) with Modeltest 3.7 [54]. Relative rate tests [77] were performed in MEGA4 [78] using A. longisetosus as outgroup. Phylogenetic and genetic distance analyses were performed in PAUP* [79]. We used the branch-and-bound option to ensure finding the best tree within maximum parsimony (MP) and maximum likelihood (ML) analyses.
References
Kondrashov AS: Classification of hypotheses on the advantage of amphimixis. J Hered. 1993, 84: 372-387.
Butlin R: The costs and benefits of sex: new insights from old asexual lineages. Nat Rev Genet. 2002, 3: 311-317. 10.1038/nrg749.
Milius S: Life without sex. So, how many million years has it been?. Science News. 2003, 163: 406-10.2307/4014492.
Birky CW, Wolf C, Maughan H, Hebertson L, Henry E: Speciation and selection without sex. Hydrobiologia. 2005, 546: 29-45. 10.1007/s10750-005-4097-2.
Barraclough TG, Birky CW, Burt A: Diversification in sexual and asexual organisms. Evolution. 2003, 57: 2166-2172.
Fontaneto D, Herniou EA, Boschetti C, Caprioli M, Melone G, Ricci C, Barraclough TG: Independently evolving species in asexual bdelloid rotifers. PLOS Biology. 2007, 5: e87-10.1371/journal.pbio.0050087.
Fontaneto D, Boschetti C, Ricci C: Cryptic diversification in ancient asexuals: evidence from the bdelloid rotifer Philodina flaviceps. J Evol Biol. 2008, 21: 580-587. 10.1111/j.1420-9101.2007.01472.x.
Fontaneto D, Kaya M, Herniou EA, Barraclough TG: Extreme levels of hidden diversity in microscopic animals (Rotifera) revealed by DNA taxonomy. Mol Phyl Evol. 2009, 53: 182-189. 10.1016/j.ympev.2009.04.011.
Maraun M, Heethoff M, Scheu S, Norton RA, Weigmann G, Thomas RH: Radiation in sexual and parthenogenetic oribatid mites (Oribatida, Acari) as indicated by genetic divergence of closely related species. Exp Appl Acarol. 2003, 29: 265-277. 10.1023/A:1025833814356.
Maraun M, Heethoff M, Schneider K, Scheu S, Weigmann G, Cianciolo J, Thomas RH, Norton RA: Molecular phylogeny of oribatid mites (Oribatida, Acari): evidence for multiple radiations of parthenogenetic lineages. Exp Appl Acarol. 2004, 33: 183-201. 10.1023/B:APPA.0000032956.60108.6d.
Domes K, Norton RA, Maraun M, Scheu S: Re-evolution of sexuality breaks Dollo's law. Proc Natl Acad Sci USA. 2007, 104: 7139-7144. 10.1073/pnas.0700034104.
Heethoff M, Domes K, Laumann M, Maraun M, Norton RA, Scheu S: High genetic divergences indicate ancient separation of parthenogenetic lineages of the oribatid mite Platynothrus peltifer (Acari, Oribatida). J Evol Biol. 2007, 20: 392-402. 10.1111/j.1420-9101.2006.01183.x.
Laumann M, Norton RA, Weigmann G, Scheu S, Maraun M, Heethoff M: Speciation in the parthenogenetic oribatid mite genus Tectocepheus (Acari, Oribatida) as indicated by molecular phylogeny. Pedobiologia. 2007, 51: 111-122. 10.1016/j.pedobi.2007.02.001.
van Doninck K, Schön I, Martens K, Backeljau T: Clonal diversity in the ancient asexual ostracod Darwinula stevensoni assessed by RAPD-PCR. Heredity. 2004, 93: 154-160. 10.1038/sj.hdy.6800486.
Schön I, Butlin RK, Griffiths HI, Martens K: Slow molecular evolution in an ancient asexual ostracod. Proc R Soc Lond B. 1998, 265: 235-242.
Templeton A: The meaning of species and speciation: a population genetics approach. Speciation and its Consequences. Edited by: Otte D, Endler J. 1989, Sunderland: Sinauer Associates, 3-27.
Mayden RL: A hierarchy of species concepts: the denouement in the saga of the species problem. Species: The Units of Biodiversity. Edited by: Claridge MF, Dawah HA, Wilson MR. 1997, London: Chapman and Hall, 381-424.
Cracraft J: Species concepts in theoretical and applied biology: a systematic debate with consequences. Species Concepts and Phylogenetic Theory. Edited by: Wheeler QD, Meier R. 2000, New York: Columbia University Press, 3-14.
Birky CW, Barraclough TG: Asexual speciation. Lost Sex - The Evolutionary Biology of Parthenogenesis. Edited by: Schön I, Martens K, van Dijk P. 2009, Dordrecht: Springer Press, 201-216. full_text.
Birky CW, Adams G, Gemmel M, Perry J: Using population genetic theory and DNA sequences for species detection and identification in asexual organisms. PLoS one. 2010, 5: 5-10.1371/journal.pone.0010609.
Hebert PDN, Cywinska A, Ball SL, deWaard JR: Biological identifications through DNA barcodes. Proc R Soc Lond B. 2003, 270: 313-321. 10.1098/rspb.2002.2218.
Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM: Identification of birds through DNA barcodes. PLOS Biology. 2004, 2: 1657-1663. 10.1371/journal.pbio.0020312.
Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S, Kamoun S, Sumlin WD, Vogler AP: Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol. 2006, 55: 595-609. 10.1080/10635150600852011.
Rubinoff D, Holland BS: Between two extremes: mitochondrial DNA is neither the panacea nor the nemesis of phylogenetic and taxonomic inference. Syst Biol. 2005, 54: 952-961. 10.1080/10635150500234674.
Will LW, Mishler BD, Wheeler QD: The perils of DNA barcoding and the need for integrative taxonomy. Syst Biol. 2005, 54: 844-851. 10.1080/10635150500354878.
Rubinoff D, Cameron S, Will K: A genomic perspective on the shortcomings of mitochondrial DNA for "barcoding" identification. J Heredity. 2006, 97: 581-594. 10.1093/jhered/esl036.
Birky CW: Workshop on barcoded DNA: application to rotifer phylogeny, evolution and systematic. Hydrobiologia. 2007, 593: 175-183. 10.1007/s10750-007-9052-y.
Dayrat B: Towards integrative taxonomy. Biol J Linn Soc. 2005, 85: 407-415. 10.1111/j.1095-8312.2005.00503.x.
Valdecasas AG, Williams D, Wheeler QD: 'Integrative taxonomy' then and now: a response to Dayrat (2005). Biol J Linn Soc. 2008, 93: 211-216. 10.1111/j.1095-8312.2007.00919.x.
Padial JM, Miralles A, de la Riva I, Vences M: The integrative future of taxonomy. Front Zool. 2010, 7: 16-10.1186/1742-9994-7-16.
Schlick-Steiner BC, Steiner FM, Seifert B, Stauffer C, Christian E, Crozier RH: Integrative taxonomy: a multiscore approach to exploring biodiversity. Annu Rev Entomol. 2010, 55: 421-438. 10.1146/annurev-ento-112408-085432.
Norton RA: Morphological evidence for the evolutionary origin of Astigmata. Exp Appl Acarol. 1998, 22: 559-594. 10.1023/A:1006135509248.
Raspotnig G, Stabentheiner E, Föttinger P, Schaider M, Krisper G, Rechenberger G, Leis HJ: Opisthonotal glands in the Camisiidae (Acari, Oribatida): evidence for a regressive evolutionary trend. J Zool Syst Evol Res. 2009, 47: 77-87. 10.1111/j.1439-0469.2008.00486.x.
Saporito R, Donnelly MA, Norton RA, Garraffo HM, Spande TF, Daly JW: Oribatid mites as a major dietary source for alkaloids in poison frogs. Proc Natl Acad Sci USA. 2007, 104: 8885-8890. 10.1073/pnas.0702851104.
Raspotnig G: Characterisation of monophyletic oribatid groups by oil gland chemistry - a novel systematic approach in Oribatida (Acari). Abh Ber Naturkundemuseum Görlitz. 2006, 78: 31-46.
Schatz H: Die Oribatidenliteratur und die beschriebenen Oribatidenarten (1758-2001) - Eine Analyse. Abh Ber Naturkundemuseum Görlitz. 2002, 72: 37-45.
Shear WA, Bonamo M, Grierson JD, Rolfe WDI, Smith EL, Norton RA: Early land animals on North America: evidence from Devonian age arthropods from Gilboa, New York. Science. 1984, 224: 492-494. 10.1126/science.224.4648.492.
Lindquist EE: Current theories on the evolution of major groups of Acari and on their relationships with other groups of Arachnida, with consequent implications for their classification. Acarology VI. Edited by: Griffith DA, Bowman CE. 1984, Chichester: Ellis Horwood Publ, 28-62.
Schaefer I, Norton RA, Scheu S, Maraun M: Arthropod colonization of land - Linking molecules and fossils in oribatid mites (Acari, Oribatida). Mol Phyl Evol. 2010, 57: 113-121. 10.1016/j.ympev.2010.04.015.
Willmann C: Moosmilben oder Oribatiden (Cryptostigmata). Die Tierwelt Deutschlands. Edited by: Dahl F. 1931, Jena: Fischer Verlag, 79-200.
Subias LS: Listado sistimatico, sininimico y biogeografico de los Acaros Oribatidos (Acariformes, Oribatida) del mundo (1748-2002). Graellsia. 2004, 60: 3-305.
Berlese A: Acari nuovi. Manipulus III. Redia. 1904, 2: 10-32.
Grandjean F: Statistique sexuelle et parthénogénèse chéz les Oribates (acariens). C. R. Séanc. Ac. Sci. 1941, 212: 463-467.
Taberly G: Demonstration de la parthénogénèse chèz Trhypochthonius tectorum Berl. (Acariens, Oribates). C. R. Séanc. Ac. Sci. 1951, 233: 1226-1228.
Palmer SC, Norton RA: Taxonomic, geographic and seasonal distribution of thelytokous parthenogenesis in Desmonomata (Acari: Oribatida). Exp. Appl. Acarol. 1991, 12: 67-81. 10.1007/BF01204401.
Palmer SC, Norton RA: Further experimental proof of thelytokous parthenogenesis in oribatid mites (Acari: Oribatida: Desmonomata). Exp. Appl. Acarol. 1990, 8: 149-159. 10.1007/BF01194176.
Berlese A: Acari, Myriapoda et Scorpiones hucusque in Italia reperta. Ordo Cryptostigmata (Oribatidae) (Cryptostigmata II). 1896, Padova, Portici
Weigmann G, Raspotnig G: Comparative morphological and biometrical studies on Trhypochthonius species of the tectorum species group (Acari: Oribatida: Trhypochthoniidae). Zootaxa. 2009, 2269: 1-31.
Raspotnig G, Krisper G, Schuster R: Oil gland chemistry of Trhypochthonius tectorum (Acari: Oribatida) with reference to the phylogenetic significance of secretion profiles in the Trhypochthoniidae. Internat J Acarol. 2001, 30: 369-374. 10.1080/01647950408684407.
Sakata T, Norton RA: Opisthonotal gland chemistry of a middle-derivative oribatid mite, Archegozetes longisetosus (Acari: Trhypochthoniidae). Int J Acarol. 2003, 29: 345-350. 10.1080/01647950308684351.
Raspotnig G, Föttinger P: Analysis of individual oil gland secretion profiles in oribatid mites (Acari: Oribatida). Int J Acarol. 2008, 34: 409-417. 10.1080/17088180809434785.
Salomone N, Emerson BC, Hewitt GM, Bernini F: Phylogenetic relationships among the Canary Island Steganacaridae (Acari, Oribatida) inferred from mitochondrial DNA sequence data. Mol Ecol. 2002, 11: 79-89. 10.1046/j.0962-1083.2001.01421.x.
Hasegawa M, Kishino H, Yano TA: Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985, 22: 160-174. 10.1007/BF02101694.
Posada D, Crandall KA: ModelTest: testing the model of DNA substitution. Bioinformatics. 1998, 14: 817-818. 10.1093/bioinformatics/14.9.817.
Mayr E: Speciation phenomena in birds. Am Nat. 1940, 74: 49-278. 10.1086/280892.
Schönhofer AL, Martens J: Hidden Mediterranean diversity: Assessing species taxa by molecular phylogeny within the opinioni family Trogulidae. Mol Phyl Evol. 2010, 54: 59-75.
Karaman IM: The taxonomical status and diversity of Balkan sironids (Opiliones, Cyphophthalmi) with descriptions of twelve new species. Zool J Linn Soc. 2009, 156: 260-318. 10.1111/j.1096-3642.2009.00446.x.
Boyer SL, Karaman I, Giribet G: The genus Cyphophthalmus (Arachnida, Opiliones, Cyphophthalmi) in Europe: A phylogenetic approach to Balkan Peninsula biogeography. Mol Phyl Evol. 2005, 36: 554-567. 10.1016/j.ympev.2005.04.004.
Weigmann G: Morphological variability between and within populations of Tectocepheus (Acari, Oribatida, Tectocepheidae) from the velatus-complex in central Europe. Acarid phylogeny and evolution: Adaptation in mites and ticks. Edited by: Bernini F, Nanelli R, Nuzzaci G, deLillo E. 2002, Dordrecht: Kluwer Academic Publisher, 141-152.
Sakata T, Norton RA: Opisthonotal gland chemistry of early-derivative oribatid mites (Acari) and its relevance to systematic relationships of Astigmata. Int J Acarol. 2001, 27: 281-292. 10.1080/01647950108684268.
Sakata T, Tagami K, Kuwahara Y: Chemical ecology of oribatid mites I. Oil gland components of Hydronothrus crispus Aoki. J Acarol Soc Japan. 1995, 4: 69-75.
Sakata T, Shimano S, Kuwahara Y: Chemical ecology of oribatid mites III. Chemical composition of oil gland exudates from two oribatid mites, Trhypochthoniellus sp. and Trhypochthonius japonicus (Acari: Trhypochthoniidae). Exp Appl Acarol. 2003, 29: 279-291. 10.1023/A:1025882214375.
Aoki JI: The Oribatid mites of the Islands of Tsushima. Bull Nat Sci Mus Tokyo. 1970, 13: 395-442.
Ewing HE: A new genus and species of Oribatidae. Entomological News. 1908, 19: 243-245.
Jacot AP: Journal of North American moss-mites. J NY Entomol Soc. 1937, 45: 353-375.
Bergsten J: A review of long-branch attraction. Cladistics. 2006, 21: 163-193. 10.1111/j.1096-0031.2005.00059.x.
Navajas M, Lagnel J, Guitierrez J, Boursot P: Species-wide homogeneity of nuclear ribosomal ITS2 sequences in the spider mite Tetranychus urticae contrasts with extensive mitochondrial COI polymorphism. Heredity. 1998, 80: 742-752. 10.1046/j.1365-2540.1998.00349.x.
Taberly G: Recherches sur la parthénogenèse thélytoque de deux espèces d'acariens oribates: Trhypochthonius tectorum (Berlese) et Platynothrus peltifer (Koch). III. Etude anatomique, histologique et cytologique des femelles parthenogenetiques II. Acarologia. 1987, 28: 389-403.
Wrensch DL, Kethley JB, Norton RA: Cytogenetics of holokinetic chromosomes and inverted meiosis: keys to the evolutionary success of mites, with generalizations on eukaryotes. Mites: Ecological and Evolutionary Analyses of Life-history Pattern. Edited by: Houck MA. 1994, New York: Chapman and Hall, 282-343.
Heethoff M, Bergmann P, Norton RA: Karyology and sex determination of oribatid mites. Acarologia. 2006, 46: 127-131.
Schaefer I, Domes K, Heethoff M, Schneider K, Schön I, Norton RA, Scheu S, Maraun M: No evidence for the 'Meselson effect' in parthenogenetic oribatid mites (Acari, Oribatida). J Evol Biol. 2006, 19: 184-193. 10.1111/j.1420-9101.2005.00975.x.
Heethoff M, Laumann M, Bergmann P: Adding to the reproductive biology of the parthenogenetic oribatid mite Archegozetes longisetosus (Acari, Oribatida, Trhypochthoniidae). Turk J Zool. 2007, 31: 151-159.
Raspotnig G, Schuster R, Krisper G, Fauler G, Leis HJ: Chemistry of the oil gland secretion of Collohmannia gigantea (Acari: Oribatida). Exp Appl Acarol. 2001, 25: 933-946. 10.1023/A:1020634215709.
Domes K, Maraun M, Scheu S, Cameron SL: The complete mitochondrial genome of the sexual oribatid mite Steganacarus magnus: genome rearrangements and loss of tRNAs. BMC Genomics. 2008, 9: 532-10.1186/1471-2164-9-532.
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman J: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25: 3389-3402. 10.1093/nar/25.17.3389.
Hall TA: BioEdit, a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999, 41: 95-98.
Tajima F: Simple methods for testing molecular clock hypothesis. Genetics. 1993, 135: 599-607.
Tamura K, Dudley J, Nei M, Kumar S: MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007, 24: 1596-1599. 10.1093/molbev/msm092.
Swofford D: PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland
Acknowledgements
We thank Roy Norton for helpful discussions and Heinrich Schatz for comparative Material. MH is funded by a DFG-Forschungsstipendium (HE4593/3-1). Three anonymous reviewers provided helpful comments.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
GR provided the initial idea for the study; GR, MH, GW and ML designed the study; GR performed chemical analyses; ML performed molecular data acquisition; GW performed morphometrical analyses; MH performed molecular data analyses, combined all results and drafted the manuscript. All authors read, discussed and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Heethoff, M., Laumann, M., Weigmann, G. et al. Integrative taxonomy: Combining morphological, molecular and chemical data for species delineation in the parthenogenetic Trhypochthonius tectorum complex (Acari, Oribatida, Trhypochthoniidae). Front Zool 8, 2 (2011). https://doi.org/10.1186/1742-9994-8-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1742-9994-8-2