Abstract
Introduction
Many marine planktonic crustaceans such as copepods have been considered as widespread organisms. However, the growing evidence for cryptic and pseudo-cryptic speciation has emphasized the need of re-evaluating the status of copepod species complexes in molecular and morphological studies to get a clearer picture about pelagic marine species as evolutionary units and their distributions. This study analyses the molecular diversity of the ecologically important Paracalanus parvus species complex. Its seven currently recognized species are abundant and also often dominant in marine coastal regions worldwide from temperate to tropical oceans.
Results
COI and Cytochrome b sequences of 160 specimens of the Paracalanus parvus complex from all oceans were obtained. Furthermore, 42 COI sequences from GenBank were added for the genetic analyses. Thirteen distinct molecular operational taxonomic units (MOTU) and two single sequences were revealed with cladistic analyses (Maximum Likelihood, Bayesian Inference), of which seven were identical with results from species delimitation methods (barcode gaps, ABDG, GMYC, Rosenberg’s P(AB)). In total, 10 to 12 putative species were detected and could be placed in three categories: (1) temperate geographically isolated, (2) warm-temperate to tropical wider spread and (3) circumglobal warm-water species.
Conclusions
The present study provides evidence of cryptic or pseudocryptic speciation in the Paracalanus parvus complex. One major insight is that the species Paracalanus parvus s.s. is not panmictic, but may be restricted in its distribution to the northeastern Atlantic.
Similar content being viewed by others
Introduction
Species delimitation and DNA barcoding
Species delimitation is a necessary process to study the life history and ecology of marine planktonic organisms, but its preciseness is dependent on the prior taxonomic knowledge. Detailed taxonomic keys may not be available for a specific region making it difficult to evaluate whether the studied specimen belongs to an already described species or an unknown species. In copepods identification is often based on only a few diagnostic characters due to the high abundances and the necessity to classify thousands of organisms [1]. These characteristics are mostly developed only in adult organisms often making it nearly impossible to identify juveniles, which are regularly more abundant than adults. Furthermore, morphological differences between sibling species may only be inconspicuous or non-existent (pseudocryptic and cryptic speciation) and thus, species may be overlooked. This phenomenon has been observed in many marine organisms (e.g. [2–4]). Cryptic speciation may be more prevalent in the marine realm than in terrestrial habitats [5].
These observations imply that traditional species concepts based on morphologically identified marine taxa may have greatly underestimated species richness [6]. Also, genetically divergent yet morphologically similar species may differ in their ecological and behavioural adaptations [7]. In general, barriers to gene flow in marine pelagic systems can often not be clearly identified (e.g. [8, 9]). To overcome these obstacles, DNA barcoding with mitochondrial gene fragments has been successfully used for species discrimination in marine plankton (e.g. [10–12]). Initially, DNA analysis using “barcoding gaps” was based on genetic distances between a priori defined groups and did not take into account differences in divergence times between species or other taxa and thus is questioned to be useful for DNA taxonomy (e.g. [13, 14]). However, a number of methods to measure species delimitation including DNA barcoding without defining prior groups have been published (e.g. [15–18]) These methods will be applied in the present study.
Paracalanus parvus species complex
Species of the Paracalanus parvus complex are abundant in many marine ecosystems from temperate to tropical regions (e.g. [19–21]). Extensive research has provided valuable information on the feeding and reproduction biology of Paracalanus parvus (e.g. [22–24]). However, the taxonomy and species distribution of this species complex is not well understood. Currently the P. parvus complex consists of seven species: P. parvus, P. indicus, P. quasimodo, P. nanus, P. intermedius, P.tropicus, and P. serrulus. The latter may possibly belong to the Paracalanus aculeatus species complex [25], and P. intermedius may be a junior synonym of P. parvus[26]. The circumglobal distribution of P. parvus has also been questioned e.g. [24].
In the present study mitochondrial marker genes (cytochrome c oxidase subunit I (COI), cytochrome b (Cytb)) will be applied to investigate the genetic diversity of this species complex. Genetic markers are favourable as a tool to distinguish between species compared to microscopy considering that exact morphological identifications can be nearly impossible if morphological characteristics, such as antennules or exopods of swimming legs, are missing due to net sampling. We aim to define molecular operational taxonomic units (MOTUs; [27]) within the P. parvus species complex to establish a framework for future studies, and to elucidate their geographic boundaries.
Material and methods
Preservation and morphological identification
A total of 162 females of the Paracalanus parvus species complex from 44 samples were analysed (Additional file 1). Specimens were preserved in 96% pure ethanol with a change in ethanol after 24 hours of the initial fixation. Individuals of the P. parvus species complex were separated from other Paracalanus species such as P. aculeatus or P. denudatus due to the differences in segmentation and length of the antennules, the form of the spermatheca and the length of the inner setae on the caudal rami. Furthermore, the total length (TL), the prosome:urosome ratio (P:U), the length of the antennules (A1) relative to TL, and the shape of the forehead were noted prior to the DNA extraction (Figure 1, Additional file 2). However, some important diagnostic morphological characters can only be seen using light microscopy. Therefore, specimens from each sampling location were set aside as paratypes for detailed morphological analysis (according to e.g. [28–30]) and as para-vouchers preserved in ethanol. The latter are stored in the cooling facilities of the Alfred-Wegener-Institut. Specimens from Chinese coastal waters (Yellow Sea) were available from samples preserved in formalin and used for morphological identification only, while sequences from this region were obtained from GenBank (Table 1).
Five species were identified combining the measurements of each individual prior to DNA extraction and the detailed morphological analysis of paratypes from each clade after the genetic analysis according to the morphological characteristics summarised by [34]. Paracalanus nanus can be distinguished from the other species by its small size and short antennules (barely reaching the end of the prosome), and the distal edges of the exopod segment 3 (Exp3) of the swimming legs 2 – 4 (P2-P4) are not serrated (Figure 1, Additional file 2). Paracalanus tropicus has very short urosome segments and as such a high P:U ratio. Paracalanus indicus, Paracalanus parvus, and Paracalanus quasimodo are distinguished by differences in the serration of the distal outer edge of the Exp3 of the P2-P4. P. parvus has a vaulted forehead, while a dorsalic hump on the prosome is present in P. quasimodo. P. indicus is characterised by posterior-dorsal spines on the female genital segment. Due to the process of net sampling, specimens were often lacking distal parts of the antennules and the swimming legs and, thus complicating the identification of the morphospecies.
DNA extraction and amplification
DNA was extracted using the QIAamp DNA Mini Kit (Qiagen) from whole individuals and were eluted in 200 μl elution buffer (AE). DNA samples were stored at −20°C until further analysis. The mitochondrial protein coding genes COI and Cytb were sequenced using the primer sets UCYTB151F and UCYTB270R for Cytb [35], and LCO1490 and HCO2198 for COI [36], and the reverse COI primer C1-N-2191 (alias Nancy, 5`-CCCGGTAAAATTAAAATATAAACTTC-3`; [37]) for difficult specimens. PCR amplifications were performed in 25 μl reaction volumes. For Cyt b it included 5 μl 5x KAPA2G Buffer B, 5 μl 5x KAPA Enhancer 1, 0.125 μl of 100 μM each primer, 0.5 μl of 10 mM dNTPs, 0.15 μl of KAPA2G DNA Polymerase (5 Units/μl) and 2 μl DNA template. PCR reactions for Cytb consisted of 35 cycles of denaturation at 94°C for 40 sec, annealing at 50°C for 45 sec and extension at 72°C for 45 sec. For COI the reaction volume included 5 μl of 5x Colorless GoTaq® Flexi Buffer (Promega), 2.5 μl of 25 mM MgCl2, 0.1 μl of 100 μM each primer, 0.5 μl of 10 mM dNTPs, 0.13 μl GoTaq® Flexi DNA Polymerase (Promega) and 2 μl of DNA template. PCR reactions for COI consisted of 35 cycles of denaturation at 94°C for 40 sec, annealing at 45°C for 45 sec and extension at 69°C for 45 sec. PCR products were run on a 1% agarose/TBE gel and afterwards stained with ethidium bromide for band characterization. Positive results were purified with ExoSap-IT (0.25 μl exo, 1 μl SAP) and subsequently used for cycle sequencing with Big Dye Terminator Ver. 3.1 (Applied Biosystems Inc., ABI) and the same primers as for the PCR amplifications. The sequences were run on an ABI 3130xl DNA sequencer.
Sequence editing
In CodonCode Aligner Vers. 3.7.1.1 (CodonCode Corporation) both strands were assembled into contigs, aligned and visually inspected for sequencing errors. 42 COI sequences named Paracalanus parvus, Paracalanus indicus or Paracalanus quasimodo from Genbank were included with the present data (Table 2). No additional COI sequences of other species from the P. parvus complex were found in GenBank (until June 30, 2013). One COI sequence from GenBank named Paracalanus parvus did not match with the other sequences of this species complex but showed close resemblance to Paracalanus aculeatus sequences [32]. It was excluded from the present analysis.
Identical sequences were merged to unique haplotypes (Table 2) with Mesquite Ver. 2.75 [38]. Out of the 162 individuals analysed, 122 (COI) or 131 (Cytb) specimens could be extracted and amplified. The 165 sequences of COI (including 43 sequences from Genbank) could be placed in 87 haplotypes and the 131 Cytb sequences could be attributed to 49 haplotypes (Table 2, Additional file 3). Sequences are published in GenBank (KF715875 – KF715996; KF715999 – KF716129; Additional file 1). All COI sequences had a minimum length of 400 bp, and 91.2% had more than 600 bp.
Tests for pseudogenes
Some COI sequences could not be sequenced due to double bands in the agarose gel. For a few other COI sequences, no consensus sequence could be built. Others produced highly divergent sequences (single sequences that differ extraordinarily from the alignment). These could be signs of either heteroplasmy, the presence of pseudogenes, contaminations, or the nonspecific binding of at least one primer under less stringent PCR conditions. All outlier sequences were excluded from the analyses. These difficulties were not found in Cytb. In the final alignments, no stop codons or indels (insertions-deletions) could be detected which would be indications for pseudogenes or incomplete lineage sorting [39, 40]. The diversity for each codon position separately was also checked [41] using MEGA 5.2.2 [42]. In mitochondrial genes the diversity should be higher in the third codon position, while in pseudogenes the diversity would be equally distributed in all three codon positions. For COI the diversity varied between all three positions (1st position: 0.0579 ± 0.0091; 2nd position: 0.0006 ± 0.0002; 3rd position: 0.3319 ± 0.0136) and similar variations were found for Cytb (1st position: 0.0602 ± 0.0128; 2nd position: 0.0195 ± 0.0088; 3rd position: 0.3165 ± 0.0191). Furthermore, 392 of 647 positions of the COI data set and 214 of 351 positions of the Cytb data set were conserved. All these tests indicate the absence of pseudogenes in the final alignment. However, pseudogenes may be very similar to the targeted mitochondrial gene and thus be overlooked.
Sequence analysis
The alignments of the haplotypes were used to infer Maximum Likelihood (ML) phylogenetic trees carried out with RAxML Vers. 7.2.8 [43] and Bayesian Inference (BI) calculated with MrBayes Vers. 3.2 [44]. The analyses were completed with un-partitioned nucleotides sequences, and with partitioned datasets accounting for the different substitution rates in the three codon positions. RAxML was run under the option GTRGAMMA and a complete random starting tree for the 10000 bootstrap replicates [45]. Then, a best-known likelihood tree search (500 inferences) was performed under GTRMIX and a completely random starting tree. The final tree topology was evaluated under GTRGAMMA to yield stable likelihood values. For the Bayesian Inference the nucleotide substitution model was selected with jModeltest Vers. 2.3 [46, 47] using the Akaike information criterion (AIC) for small sample sizes. The best-fitted model (GTR + I + G) was then implemented in MrBayes. The analysis was run for 3,000,000 generations for the unpartitioned data set and for 10,000,000 generations for the codon model with a sample frequency of 1,000 generations. The first 500 trees were discarded as burn-in. Clade support is shown on the nodes of the trees as the Bayesian Posterior Probability (BPP) when BPP > 0.90.
As outgroup Delibus spp. from the family Paracalanidae were chosen (COI: Accession numbers JQ911978 (Delibus nudus); JQ911979, KF715873, KF715874 (Delibus sp.); Cytb: KF715997, KF715998 (Delibus sp.)). The analysis of the phylogeny of the family Paracalanidae has shown that the genus Delibus has the closest relationship to the Paracalanus parvus complex [32].
For presumably recently diverged lineages that were separated with some species delimitation methods haplotype networks were created with TCS Vers. 1.2.1 [48] to demonstrate the geographic structure of the haplotypes.
Species delimitation methods
Two independent methods of species delimitation were applied to propose a first species hypothesis for the Paracalanus parvus complex: Automated Barcoding Gap Discovery (ABGD) and the generalized mixed Yule coalescent model (GMYC).
Automatic Barcode Gap Discovery (ABGD) is an automated iterative process to sort sequences into putative species based on pairwise distances without an a priori species hypothesis [17]. This algorithm automatically detects significant differences between intra- and inter-specific variations (i.e. barcoding gap). Aligned sequences of all haplotypes were uploaded to the web interface at http://wwwabi.snv.jussieu.fr/public/abgd/abgdweb.html and were run with the default settings. P (prior limit to intraspecific diversity) had a minimum value of 0.001 and a maximum value of 0.1. The relative gap width (X) had a value of 1. All available models (Jukes-Cantor (JC86), Kimura (K80)) were tested.
The generalized mixed Yule coalescent model (GMYC) infers species boundaries by measuring the transition from intra- to inter-species branching patterns [16]. This method combines models of stochastic lineage growth (Yule models) with coalescence theory. The analysis is implemented in R as part of the package “splits” (SPecies LImits by Threshold Statistics; [49]). The explanatory power of a model assuming a transition from population-specific to more phylogeny-like branching patterns is compared to a null model (all specimens are derived from a single species). The GMYC method was applied allowing a single threshold [50]. Prior to the analysis, outgroups were removed and the unique haplotypes were used to render an ultrametric consensus tree as starting point for the GMYC model (BEAST Vers. 1.7.4 [51]). A relaxed uncorrelated log-normal clock was chosen with a mean substitution rate fixed at 1 and estimated branch length with a coalescent prior. MCMC chains were run for 10 million generations sampling every 1,000 steps after a burn-in period of 1,000 trees.
As the both methods are sensitive to intra-species sampling we additionally analysed species delimitation for an alignment of all 165 specimens.
To test whether the MOTUs derived from the ABGD and GMYC analyses represent putative species, several species delimitation methods were applied. The resulting MOTUs were similar for the two gene fragments. Therefore, only the COI results are presented, since this gene fragment is frequently used to identify species (i.e. DNA barcoding; [52]), and distance thresholds can be compared to those of other copepods. Cytb results are found in the supplementary material. The methods all calculated species delimitation without a priori defined groups and are all based on uncorrected pairwise distance calculated with MEGA Vers. 5.2.2 unless otherwise indicated, since the use of K2P distance for DNA barcoding analysis is under debate [53]. Colour heat maps representing the distances between all haplotypes were plotted in MATLAB (base installation of R2013b, The MathWorks Inc.).
Barcode gaps between well-supported clades of haplotypes identified by Maximum Likelihood and Bayesian Inferences were taken as an indication of separate MOTUs. To find the optimal thresholds for intra-specific p-distances, the function localMinima of the SPIDER (SPecies IDentity and Evolution in R) Vers. 1.2 package for R (http://www.R-project.org) was used [54]. Based on the concept of the barcoding gap, this method indicates the transition between intra- and interspecific genetic distances from a dip in the density of the uncorrected p-distances without prior knowledge of species identity [54] and provides thresholds. These were used to cluster the sequences with the software jMOTU [15].
Rosenberg’s P(AB) examines whether monophyly has been produced by evolutionary processes or by insufficient sampling and calculates the probability that a MOTU with by A haplotypes is monophyletic to its closest relative with B haplotypes [18]. Significance of nodes was visualized with the R package SPIDER using a consensus ultrametric tree built in BEAST.
Results
Molecular species identification
The primary species delimitation analysis based on 87 haplotypes with ABGD and GMYC resulted in 12 or 14 MOTUs, and two or three single sequences (Figures 2 and 3). The two methods were largely congruent in 10 MOTUs (NWP, SWP, NEP, SEP, PQ, PI, PT, PA, NEA, SWA). The two remaining MOTUs detected with ABGD were each divided in two groups with the GMYC analysis. One of the MOTUs (PN) included only individuals that were previously identified as Paracalanus nanus. This species was not the main target of this study and thus, we combined the two groups found in GMYC in one MOTU as suggested by ABGD analysis. The second MOTU included two groups separated in GMYC by their geographic distribution. These were counted as two separate MOTUs (SEA/NZ, NWA) to be tested with other methods. Thus, the species delimitation with these two methods resulted in 13 MOTUs, which were evaluated with other species delimitation methods. To test whether the use of haplotypes influenced the analysis of ABGD and GMYC we also analysed a data set with all 165 specimens. The resulting MOTUs of the two data sets were congruent with two exceptions. With the ABGD analysis the MOTUs PT and PA were fused, while with GMYC the MOTU PI was divided in three groups (Figure 3).
Results of species delimitation methods (Maximum Likelihood analysis of Cytb (ML Cyt b), jMOTU analyses with 2, 4 and 6% thresholds, ABGD, GMYC and Rosenberg) with resulting putative species. White columns in ABGD and GMYC reveal differences in species delimitation using an alignment with all 165 specimens instead of haplotypes (black columns). Colors of morphospecies match with the original described species name; transverse lines mark when identification and literature records disagree.
The MOTUs were named either according to their geographic occurrence (NEA, SWA, SEA/NZ, NWA, NWP, NEP, SEP, SWP, SEI, PA) or for lineages with a wider spread distribution a potential species name, derived from morphological observations was used as abbreviation (PT, PQ, PI; Table 2). The MOTU NWP was exclusively built by sequences obtained from GenBank. Other sequences obtained from Genbank were placed in the MOTUs PQ, PI, PT and NWA (Table 1, Figures 3 and 4). These sequences could not be inspected for sequencing errors as no raw data were available.
RAxML Maximum Likelihood tree for haplotypes (COI). Numbers show the percentage bootstrap support from two analyses: unmodified nucleotide sequences/ sequences separated by codon position, and bayesian posterior probability (BPP): unmodified nucleotide sequences/codon model. Haplotypes from GenBank are marked with red color, if only some sequences of a haplotype were taken from GenBank they are marked with *. Black bars display defined MOTUs.
The single sequences (SEI, SWP1, PI04) will not be considered as MOTUs at present until further sampling at their location (northern Australia) will either confirm or contradict their existence. The sequence PI04 had a p-distance of 2.8% to all other PI haplotypes and was thus singled out in GMYC.
The cladistic analyses of the COI haplotypes yielded 13 monophyletic clades and two single sequences (Figure 4), which were conform to the results from the ABGD and GMYC analyses. Twelve clades were well supported (>80% BS, >0.9 BPP) and one (PT) was moderately well supported (74% BS, <0.9 BPP). The cladistic analyses of Cytb haplotypes yielded twelve clades, identical to the clades for COI (Additional files 4 and 5). However, NWA and PI were not retrieved as monophyletic, but p-distances within these MOTUs were much lower than to their sister-taxon (Additional file 5). NWP and SEI were not found in the Cytb tree since they were based on GenBank sequences, and SWP1 could not be sequenced.
The mean uncorrected p-distances between MOTUs were generally higher than the divergence within the MOTUs (Table 2, Figure 5). Within MOTU sequence divergences varied between 0.2 (NEA) and 3.4% (PI), while differences between MOTUs varied between 3.2 – 14.8%. NEA shared haplotypes with eight recently published COI sequences from the North Sea and the Gullmarsfjord, Sweden [55]; GenBank Accession numbers: JX995215 - JX995222) reaching uncorrected p-distances of 0.6%.
Three thresholds between intra- and inter-specific distances were detected with the SPIDER package of R (0.0202 (2% jMOTU), 0.0441(4% jMOTU), 0.0648 (6%jMOTU)). These thresholds were used in jMOTU to separate clusters yielding between 11 (jMOTU, 6% pairwise intra-specific distance) and 15 MOTUs (jMOTU, 2% pairwise intra-specific distance). Two (SEI, SWP1) or three (SEI, SWP1, PI04) single sequences were found (Figure 3). At 2%jMOTU PN and PI were separated each in two groups. At 4%jMOTU and 6%MOTU SEA/NZ and NWA were fused and at 6%jMOTU also PT and PA. Rosenberg’s P(AB) showed significant nodes that would result in 13 to 15 MOTUs (Figure 3).
All methods identified seven congruent MOTUs (NWP, SWP, NEP, SEP, PQ, NEA and SWA). PN and PI were in some analyses split into two or three groups (jMOTU 2%, GMYC, Rosenberg (Figures 2 and 3)). Four other PI haplotypes (PI03, PI06, PI17, PI18) were separated from PI in jMOTU 2% and significantly distinct from the other sequences in Rosenberg (Figures 2, 3 and 6). The heatmap and the haplotype network visualized close connections based on uncorrected p-distances between PT and PA, NEA, NWA and SEA/NZ, and PI and PQ (Figures 3 and 6). The latter were, however, separated in all methods. The MOTUs PT and PA were combined in jMOTU 6% (Figures 3 and 5). The MOTUs SEA/NZ and NWA showed the least divergence and were combined in jMOTU 4%, jMOTU 6%, and ABGD (Table 2, Figures 3 and 5).
Phylogeography
Three categories of MOTUs were discovered regarding their geographic distribution (Figure 7): (1) Eight MOTUs occurred mainly in temperate waters and were restricted to one geographic region (NWA, NEA, SWA, NWP, NEP, SEP, SWP). Only SEA/NZ was found in two regions, Southeast Atlantic and Southwest Pacific waters, also sharing a haplotype (Figure 6). (2) Three MOTUs had a wider geographic distribution and some also occurred sympatrically. PQ seemed to be refined to the Atlantic and adjacent waters. PA was only retrieved from Atlantic Ocean samples while single PT specimens were found in the Indopacific, Red Sea and Southeast Atlantic. (3) PI was found in all oceans, mainly in the Indian and Pacific Ocean but also in the Atlantic Ocean and Mediterranean Sea. The two single sequences (SEI, SWP1) were found in the Northwest and Northeast off Australia.
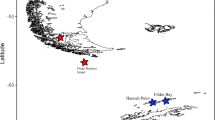
Overview of sampling locations for specimens of the Paracalanus parvus complex (for exact information on latitude/longitude etc. see Additional file 1 ) and geographic distribution of MOTUs.
Two locations for PN were also included in the analyses but we did not search particularly for this species on a global scale and thus its distribution cannot be evaluated here.
Discussion
This study presents the first step to disentangling the genetic diversity of the ecologically important Paracalanus parvus species complex by using mitochondrial genes. Cleary this complex is composed of more MOTUs than morphologically described species and may thus be subject to cryptic and pseudocryptic speciation. The results provide a “global” framework for scientists identify individuals from the Paracalanus parvus complex according to their genetic affiliation (MOTU). It can also serve as a basis for future morphological taxonomy to test the validity of the found MOTUs.
Genetic species delimitation
Independent methods without a priori defined groups are used to investigate the Paracalanus parvus complex (ABGD, GMYC, cladistic analyses (ML, BI), classical barcoding, Rosenberg’s P(AB)). These methods are generally congruent. They differ in the way that in some methods clades are subdivided or merged. In classical taxonomy this problem is well known and then nomenclature is helpful. In other words, one author defines two taxa as subspecies while another author defines the same taxa as separate species. It is important to note that the methods do not contradict each other in the subdivision but only in their assignment to hierarchy. Thus, the number of MOTUs varies between 11 and 15 MOTUs but not all of these may represent reproductively isolated species.
The seven MOTUs that are congruent in all analysis have genetic distances between MOTUS greater than 8% and therefore represent putative species. Speciation in marine copepods is assumed to have occurred when sequence divergences are approximately 8 – 9% (e.g. [56, 57]. Some methods separate lineages within the level of intraspecific variety of copepods (1 – 4%, e.g. [56, 58]. This accounts for the MOTUs SEA/NZ and NWA (uncorrected p-distance 3.2 – 4.2%) and for PA and PT (4.8 – 5.7% uncorrected p-distance). These two MOTUs may be recently diverged conspecific lineages, due to their geographic isolation, however, they show evidence of the possible existence of two species which would be in concordance with the unified species concept of [59]. In total, 10 to 12 putative species are found within the present genetic data set of the Paracalanus parvus species complex.
For this species complex, COI has provided a rapid and sufficient support for the evidence of cryptic and pseudocryptic speciation. The resulting putative species are often separated according to their geographic habitat, which provides additional support for the results of the species delimitation methods.
However, it is well known that single locus analysis of species delimitation may under- or overestimate the number of species due to e.g. pseudogenes, incomplete lineage sorting (e.g. [40]). An independent nuclear marker with a different level of gene flow will be needed in the future to validate the number of putative species found with a mitochondrial marker, which has been suggested by many authors e.g. [60, 61]. The usage of a nuclear marker could also clarify whether the low genetic divergences between sister-lineages such as the geographically distinct NEA and SEA/NZ are a result from recent speciation events or from continuing gene flow between two populations. Thus, future studies should include not only more sampling locations and detailed morphological analysis, but also further molecular markers with an independent evolutionary history compared to mitochondrial genes and possibly interbreeding studies that could also help to distinguish between species. It has been shown that geographicallly isolated populations [62] or even groups with low COI sequence divergence [63] can be reproductively isolated.
Molecular and morphological species identification
Morphological and molecular identification are not congruent. Morphospecies are found in more than one of the 10 to 12 putative species, except for Paracalanus nanus, which is conform with the MOTU PN and clearly identified by its small size and short antennules. In thoroughly revised calanoid genera such as Clausocalanus the morphological taxonomy is congruent with the molecular taxonomy [58]. Furthermore, for oncaeid copepods it has been shown that even the smallest morphological detail is significant in species identification [64]. An indication that this could also be important for Paracalanus species is that morphological variability within species has been noted previously (e.g. [24, 34]; McKinnon (personal communication)). There is also evidence that speciation in copepods can occur without apparent morphological speciation (e.g. [65–67]) and morphological similarities may also emerge after genetic differentiation due to adaptation to a similar habitat (convergent evolution). Especially species in coastal systems are known to show strong genetic differentiation [68–70]. A thorough morphological revision of the taxon Paracalanus may reveal differences whether differences between MOTUs have been overlooked previously or whether genetic differentiation is due to behavioural adaptation [66].
Putative species
Some NEA specimens are collected from the type locality of P. parvus (Helgoland, North Sea; [71]). They closely resemble the original description of Claus ([71]; vaulted forehead, lack of spinules on the posterior surfaces of the coxae of P2-P4) and are suggested to represent P. parvus s.s. (Figure 4). NWA and SEA/NZ populations are a sister-species to or a subspecies of P. parvus s.s. (NEA) but they are separated in all analysis. Hence, NWA and SEA/NZ are referred to as Paracalanus sp. F. Specimens from the Northwest Atlantic (NWA) and Southeast Atlantic (SEA) have been identified as P. parvus (e.g. [29, 72]) and closely resemble P. parvus from Helgoland, while specimens from New Zealand have been described as P. indicus[34]. However, the specimens from New Zealand lack the typical postero-lateral spines on the genital segment and are also only little ornamented on the posterior surfaces of the swimming legs.
Specimens from the type locality of Paracalanus indicus (Maldive Islands, [73]) could not be obtained for the present study. However, specimens from the Andaman Sea (Indian Ocean) belonging to PI are morphologically congruent with the description of Wolfenden [73]. Hence, this MOTU is preliminary named P. indicus. Bowman [29] redescribed P. indicus from samples off Cape Hatteras (Northwest Atlantic, USA) but his drawings (Page 27 Figure twenty two c) show comparatively short urosomal segments 2 and 3 (resulting in a high P:U ratio). This is characteristic for P. tropicus[28], and present in specimens of PA and PT (Additional file 2). PA specimens are found near in the Northwest Atlantic near Cape Hatteras. These observations suggest that P. indicus described by Bowman [29] could be identical with PA. Due to low genetic divergence PA and PT are considered to be subspecies of P. tropicus.
P. quasimodo is distinguished from other species due to the presence of spinules at the distal outer edge of Exp3 of swimming leg four and many spinules on the posterior surfaces of the coxae of P2-P4. These characteristics are found in specimens of PQ, NWP, and NEP but only PQ is found near Cape Hatteras, the type locality of P. quasimodo[29]. Hence, it is suggested that PQ represents P. quasimodo.
Specimens from the locations of NWP, SEP, NEP, SWP and SWA have previously been named P. parvus, or P. indicus (e.g. [24, 74, 75]), although some have mentioned the morphological similarity of NWP and NEP to P. quasimodo[24, 76]. They are probably not connected to any described species and are thus referred to as Paracalanus sp. A (NWP), Paracalanus sp. B (SWP), Paracalanus sp. C (NEP), Paracalanus sp. D (SEP) and Paracalanus sp. E (SWA).
Phylogeography
Only Paracalanus indicus (PI) is truly widespread on an oceanic scale, sharing haplotypes between oceans. Gene flow seems to lack persistent geographic boundaries as has been seen in other copepod species (e.g. [77, 78]. In the past, at least two morphological species (P. parvus and P. indicus) have been identified from many regions around the globe [72], but the present study revealed that P. parvus is restricted probably to the Northeast Atlantic. Hence, the findings of P. parvus around the oceans have to be distributed to other MOTUs.
Seven MOTUs are found in temperate waters, each being restricted to one marine temperate ecoregion as assigned by [79], with the exception of SEA/NZ. Temperate environments in the southern hemisphere were established with the onset of the Antarctic convergence (~23 MYA (millions years ago); [80]. With the closure of the Indonesian Seaway (~13 MYA), temperate marine environments developed also in the Northwest Pacific. Sequence divergences between widespread tropical and geographically restricted temperate MOTUs for both the Atlantic and the Pacific were similare (Pacific: 14.0% (PI/PQ vs. NWP/NEP/SEP/SWP) and Atlantic: 14.5% (PT/PA vs. NEA/SEA/NWA). However, there are no direct estimates of mutation rates for copepods, but molecular clock calibrations for COI for other crustaceans resulted in mutation rates of approximately 1.4% per million years [81]. Thus, the temperate and tropical clades could have diverged somewhere around 10 – 11 MYA, which would be after the closure of the Indonesian Seaways and the establishment of temperate habitats. But taking into account the substitution saturation often seen in mitochondrial DNA such as COI, these divergence times could be underestimated.
Due to closure of the Central American Seaway (~4.6 MYA; [82], the Gulf Stream intensified creating favourable habitats in the North Atlantic. Low pairwise distances (7.9%) between the populations of Paracalanus sp. F (NWA, SEA/NZ) and P. parvus s.s. (NEA) could indicate that speciation processes in the Atlantic may have occurred more recently than in the Pacific which would coincide with the timing of paleoceanographic changes described above. Thus, all Paracalanus species possibly have a tropical ancestor, and temperate forms may have evolved concurrent with paleoceanographic changes that led to the establishment of temperate marine environments. Due to their high abundances and consequently a high number of adaptive mutations Paracalanus may have a great potential to rapidly adapt genetically as has been hypothesized for oceanic zooplankton in general [83].
In some regions it has also been noted that Paracalanus is abundant only during certain seasons [84]. Thus, it can be speculated that ecological factors (such as seasonality, food sources) of the temperate Paracalanus species are another important issue that enhance speciation and also function as boundaries to gene flow. From Acartia tonsa it is known that different salinity regimes in close vicinity coincide with genetic divergence [69], while sea water temperature seems to control lineages of Metridia lucens in the Southern Atlantic [85]. It has also been suspected that planktonic taxa may drift anywhere but successfully reproduce only in their favourite environment [6]. Based on this assumption that sympatric or parapatric speciation processes may play a more important role in pelagic evolution than vicariant or allopatric models, which is supported by other findings [63]. However, the linages of P. tropicus may be subject to allopatric speciation as they are separated by ocean (PA in the Atlantic, PT in the Indo-Pacific). The genetic isolation between oceans is also found in other copepods species, e.g. Clausocalanus lividus and has been explained by the rising of the Isthmus of Panama [77].
SEA/NZ (Paracalanus sp. F) includes specimens from the Southeast Atlantic and New Zealand. Their presence in Southwest Pacific waters may be explained by two possibilities. The first hypothesis is that these specimens are transported regularly, possibly by the Antarctic Circumpolar Current, to southern New Zealand. Due to high gene flow they have not yet separated from the Southeast Atlantic specimens. This theory is supported by our observation that the sequenced specimens from New Zealand in this study are morphologically similar to the description of Bradford [34]. The second hypothesis is that individuals are transported to New Zealand in ballast water tanks of commercial ships. P. parvus s.l. is common in many coastal waters and often found in ballast water tanks (e.g. [86, 87]). It has also been suggested that cosmopolitan distribution of many coastal species may be partly attributed to ballast water transport [88]. However, the present results indicate that both the circumglobal distribution of temperate coastal species and the influence on species distribution may be questionable in case of Paracalanus.
Furthermore, the presented biogeography of the Paracalanus species can be biased due to the low specimen number as has been shown for other marine organisms [4, 88]. Small sample sizes and restricted geographic sampling suggested that the circumpolar crinoid species Promachocrinus kerguelensis as a complex of several cryptic species, some geographically limited and others widespread [4]. A later study included circum-Antarctic samples and revealed that all of the lineages were circumpolar [89]. This shows that there is a need of adequate geographic sampling. The present study includes samples from many locations but still there are large geographic gaps, which could either hide more cryptic species or change the present biogeography. The East Pacific is only covered by a few locations (coastal waters of Oregon and Chile). The same accounts for the Southwest Atlantic and the Indian Ocean.
Conclusions
The circumglobal distribution of many marine planktonic copepod species is currently under debate (e.g. Paracalanus parvus, Acartia tonsa, Oithona similis, Paracalanus parvus). For the Paracalanus parvus species complex the present study has provided clear evidence for cryptic and pseudocryptic speciation, revealing 10 to 12 putative species with differing biogeographic distribution. All species delimitation methods were largely congruent, which indicates that the species diversity was effectively assessed. One major insight was that Paracalanus parvus s.s. was only identified from samples the northeastern Atlantic, and not panmictic.
COI has proven to be a good indicator of specimen identification in Paracalanus and thus the present data set can serve as a database for future identification of Paracalanus specimens from other locations.
In conclusion, the Paracalanus parvus species complex can serve as a role model to investigate cryptic speciation in other widely distributed marine copepod species complexes and may help to better understand speciation processes within the pelagic marine environment in the future.
References
McManus GB, Katz LA: Molecular and morphological methods for identifying plankton: what makes a successful marriage?. J Plankton Res. 2009, 31: 1119-1129. 10.1093/plankt/fbp061.
Goetze E: Cryptic speciation on the high seas; global phylogenetics of the copepod family Eucalanidae. Proc R Soc B. 2003, 270: 2321-2331. 10.1098/rspb.2003.2505.
Jörger KM, Norenburg JL, Wilson NG, Schrödl M: Barcoding against a paradox? Combined molecular species delineations reveal multiple cryptic lineages in elusive meiofaunal sea slugs. BMC Evol Biol. 2012, 12: 245-10.1186/1471-2148-12-245.
Wilson NG, Hunter RL, Lockhart SJ, Halanych KM: Multiple lineages and absence of panmixia in the “circumpolar” crinoid Promachocrinus kerguelensis from the Atlantic sector of Antarctica. Mar Biol. 2007, 152: 895-904. 10.1007/s00227-007-0742-9.
Knowlton N: Sibling Species in the Sea. Annu Rev Ecol Syst. 1993, 24: 189-216. 10.1146/annurev.es.24.110193.001201.
Norris RD: Pelagic species diversity, biogeography, and evolution. Paleobiology. 2000, 26: 236-258. 10.1666/0094-8373(2000)26[236:PSDBAE]2.0.CO;2.
Sáez AG, Probert I, Geisen M, Quinn P, Young JR, Medlin LK: Pseudo-cryptic speciation in coccolithophores. Proc Natl Acad Sci U S A. 2003, 100: 7163-7168. 10.1073/pnas.1132069100.
Cowen RK, Sponaugle S: Larval Dispersal and Marine Population Connectivity. Annu Rev Mar Sci. 2009, 1: 443-466. 10.1146/annurev.marine.010908.163757.
Knowlton N: Molecular genetic analyses of species boundaries in the sea. Hydrobiologia. 2000, 420: 73-90. 10.1023/A:1003933603879.
Bucklin A, Ortman BD, Jennings RM, Nigro LM, Sweetman CJ, Copley NJ, Sutton T, Wiebe PH: A “Rosetta Stone” for metazoan zooplankton: DNA barcode analysis of species diversity of the Sargasso Sea (Northwest Atlantic Ocean). Deep-Sea Res II. 2010, 57: 2234-2247. 10.1016/j.dsr2.2010.09.025.
Miyamoto H, Machida RJ, Nishida S: Global phylogeography of the deep-sea pelagic chaetognath Eukrohnia hamata. Prog Oceanogr. 2012, 104: 99-109.
Peijnenburg KTCA, Breeuwer JAJ, Pierrot-Bults AC, Menken SBJ: Phylogeography of the planktonic chaetognath Sagitta setosa reveals isolation in European Seas. Evolution. 2004, 58: 1472-1487.
Vogler AP, Monaghan MT: Recent advances in DNA taxonomy. J Zool Syst Evol Res. 2006, 45: 1-10.
Wiens JJ: Species delimitation: new approaches for discovering diversity. Syst Biol. 2007, 56: 875-878. 10.1080/10635150701748506.
Jones M, Ghoorah A, Blaxter ML: jMOTU and Taxonerator: Turning DNA barcode sequences into annotated operational taxonomic units. PLoS ONE. 2011, 6: e19259-10.1371/journal.pone.0019259.
Pons J, Barraclough T, Gomez-Zurita J, Cardoso A, Duran D, Hazell S, Kamoun S, Sumlin W, Vogler AP: Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol. 2006, 55: 595-609. 10.1080/10635150600852011.
Puillandre N, Lambert A, Brouillet S, Achaz G: ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol Ecol. 1864–1877, 2012: 21-
Rosenberg NA: Statistical tests for taxonomic distinctiveness from observations of monophyly. Evolution. 2007, 61: 317-323. 10.1111/j.1558-5646.2007.00023.x.
Cornils A, Schulz J, Schmitt P, Lanuru M, Richter C, Schnack-Schiel SB: Mesozooplankton distribution in the Spermonde Archipelago (Indonesia, Sulawesi) with special reference to the Calanoida (Copepoda). Deep-Sea Res II. 2076–2088, 2010: 57-
Escribano R, Hidalgo P: Spatial distribution of copepods in the north of the Humboldt Current region off Chile during coastal upwelling. J Mar Biol Ass UK. 2000, 80: 283-290. 10.1017/S002531549900185X.
Hwang J-S, Souissi S, Tseng L-C, Seuront L, Schmitt FG, Fang L-S, Peng S-H, Cheng-Han WU, Hsiao S-H, Twan W-H: A 5-year study of the influence of the northeast and southwest monsoons on copepod assemblages in the boundary coastal waters between the East China Sea and the Taiwan Strait. J Plankton Res. 2006, 28: 943-958. 10.1093/plankt/fbl031.
Kang HK: Effects of suspended sediments on reproductive responses of Paracalanus sp. (Copepoda: Calanoida) in the laboratory. J Plankton Res. 2012, 34: 626-635. 10.1093/plankt/fbs033.
Paffenhöfer G-A: Does Paracalanus feed with a leaky sieve?. Limnol Oceangr. 1984, 29: 155-160. 10.4319/lo.1984.29.1.0155.
Suzuki K, Nakamura Y, Hiromi J: Feeding by the small calanoid copepod Paracalanus sp. on heterotrophic dinoflagellates and ciliates. Aquat Microb Ecol. 1999, 17: 99-103.
Shen CJ, Lee FS: The estuarine Copepoda of Chiekong and Zaikong Rivers, Kwangtung Province, China. Acta Zool Sin. 1963, 15: 571-596.
Vervoort W: Report on some Copepoda collected during the Melanesia Expedition of the Osaka Museum of Natural History. Publ Seto Mar Biol Lab. 1962, 10: 393-470.
Floyd R, Abebe E, Papert A, Blaxter ML: Molecular barcodes for soil nematode identification. Mol Ecol. 2002, 11: 839-850. 10.1046/j.1365-294X.2002.01485.x.
Andronov VN: Paracalanus tropicus sp. n. (Copepoda, Paracalanidae) from South-East Atlantic. Zool Zh. 1977, 56: 154-156.
Bowman TE: The distribution of calanoid copepods off the southeastern United States between Cape Hatteras and southern Florida. Smithson Contrib Zool. 1971, 96: 1-58.
Bradford-Grieve JM, Markhaseva EL, Rocha C, Abiahy B: Copepoda. South Atlantic Zooplankton. Volume 2. Edited by: Boltovskoy D. 1999, Leiden: Backhuys Publishers, 869-1098.
Blanco-Bercial L, Bradford-Grieve JM, Bucklin A: Molecular phylogeny of the Calanoida (Crustacea: Copepoda). Mol Phylogenet Evol. 2011, 59: 103-113. 10.1016/j.ympev.2011.01.008.
Cornils A, Blanco-Bercial L: Phylogeny of the Paracalanidae Giesbrecht, 1888 (Crustacea: Copepoda: Calanoida). Mol Phylogenet Evol. 2013, 69: 861-872. 10.1016/j.ympev.2013.06.018.
Jungbluth MJ, Lenz PH: Copepod diversity in a subtropical bay based on a fragment of the mitochondrial COI gene. J Plankton Res. 2013, 35: 630-643. 10.1093/plankt/fbt015.
Bradford JM: Paracalanus indicus Wolfenden and Corycaeus aucklandicus Kraemer, two neritic pelagic copepods from New Zealand. J R Soc NZ. 1978, 8: 133-141. 10.1080/03036758.1978.10429386.
Merritt TJ, Shi L, Chase MC, Rex MA, Etter RJ, Quattro JM: Universal cytochrome b primers facilitate intraspecific studies in molluscan taxa. Mol Mar Biol Biotechnol. 1998, 7: 7-11.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R: DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994, 3: 294-299.
Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P: Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994, 87: 651-701.
Maddison W, Maddison D: Mesquite: a modular system for evolutionary analysis. Version 2.75. [http://mesquiteproject.org]
Buhay JE: “COI-like” sequences are becoming problematic in molecular systematic and DNA barcoding studies. J Crustac Biol. 2009, 29: 96-110. 10.1651/08-3020.1.
Song H, Buhay JE, Whiting MF, Crandall KA: Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc Natl Acad Sci U S A. 2008, 105: 13486-13491. 10.1073/pnas.0803076105.
Calvignac S, Konecny L, Malard F, Douady CJ: Preventing the pollution of mitochondrial datasets with nuclear mitochondrial paralogs (numts). Mitochondrion. 2011, 11: 246-254. 10.1016/j.mito.2010.10.004.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S: MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011, 28: 2731-2739. 10.1093/molbev/msr121.
Stamatakis A: RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006, 22: 2688-2690. 10.1093/bioinformatics/btl446.
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP: MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012, 61: 539-542. 10.1093/sysbio/sys029.
Pattengale ND, Alipour M, Bininda-Emonds OR, Moret BM, Stamatakis A: How many bootstrap replicates are necessary?. J Comput Biol. 2009, 17: 337-354.
Guindon S, Gascuel O: A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003, 52: 696-704. 10.1080/10635150390235520.
Posada D: jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008, 25: 1253-1256. 10.1093/molbev/msn083.
Clement MJ, Posada D, Crandall KA: TCS: A computer program to estimate gene genealogies. Mol Ecol. 2000, 9: 1657-1659. 10.1046/j.1365-294x.2000.01020.x.
Edzard T, Fujisawa T, Barraclough TG: SPLITS: SPecies’ LImits by Threshold Statistics. R package version 1.0-18/r45. [http://r-forge.r-project.org/projects/splits/]
Fujisawa T, Barraclough TG: Delimiting species using single-locus data and the generalized mixed yule coalescent approach: a revised method and evaluation on simulated data sets. Syst Biol. 2013, 62: 707-724. 10.1093/sysbio/syt033.
Drummond AJ, Rambaut A: BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007, 7: 214-10.1186/1471-2148-7-214.
Hebert PDN, Ratnasingham S, de Waard JR: Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc B. 2003, 270: S96-S99. 10.1098/rsbl.2003.0025.
Collins RA, Cruickshank RH: The seven deadly sins of DNA barcoding. Mol Ecol Resour. 2013, 13: 969-975.
Brown SDJ, Collins RA, Boyer S, Lefort M-C, Malumbres-Olarte J, Vink CJ, Cruickshank RH: Spider: An R package for the analysis of species identity and evolution, with particular reference to DNA barcoding. Mol Ecol Resour. 2012, 12: 562-565. 10.1111/j.1755-0998.2011.03108.x.
Laakmann S, Gerdts G, Erler R, Knebelsberger T, Martínez Arbizu P, Raupach MJ: Comparison of molecular species identification for North Sea calanoid copepods (Crustacea) using proteome fingerprints and DNA sequences. Mol Ecol Resour. 2013, 13: 862-876. 10.1111/1755-0998.12139.
Bucklin A, Guarneri M, Hill RS, Bentley A, Kaartvedt S: Taxonomic and systematic assessment of planktonic copepods using mitochondrial COI sequence variation and competitive, species-specific PCR. Hydrobiologia. 1999, 401: 239-254.
Hill R, Allen LD, Bucklin A: Multiplexed species-specific PCR protocol to discriminate four N. Atlantic Calanus species, with an mtCOI gene tree for ten Calanus species. Mar Biol. 2001, 139: 279-287. 10.1007/s002270100548.
Bucklin A, Frost BW: Morphological and molecular phylogenetic analysis of evolutionary lineages within Clausocalanus (Copepoda: Calanoida). J Crustac Biol. 2009, 29: 111-120. 10.1651/07-2879.1.
De Queiroz K: Species concepts and species delimitation. Syst Biol. 2007, 56: 879-886. 10.1080/10635150701701083.
Hare MP: Prospects for nuclear gene phylogeography. Trends Ecol Evol. 2001, 16: 700-706. 10.1016/S0169-5347(01)02326-6.
Petit RJ, Excoffier L: Gene flow and species delimitation. Trends Ecol Evol. 2009, 24: 386-393. 10.1016/j.tree.2009.02.011.
Carrillo B-G, Miller CB, Wiebe PH: Failure of interbreeding between Atlantic and Pacific populations of the marine calanoid copepod Acartia clausi Giesbrecht. Limnol Oceangr. 1974, 19: 452-458. 10.4319/lo.1974.19.3.0452.
Lee CE: Global phylogeography of a cryptic copepod species complex and reproductive isolation between genetically proximate “populations”. Evolution. 2014–2027, 2000: 54-
Böttger-Schnack R, Machida RJ: Comparison of morphological and molecular traits for species identification and taxonomic grouping of oncaeid copepods. Hydrobiologia. 2010, 666: 111-125.
Marrone F, Brutto Lo S, Hundsdoerfer AK, Arculeo M: Overlooked cryptic endemism in copepods: Systematics and natural history of the calanoid subgenus Occidodiaptomus Borutzky 1991 (Copepoda, Calanoida, Diaptomidae). Mol Phylogenet Evol. 2013, 66: 190-202. 10.1016/j.ympev.2012.09.016.
Chen G, Hare MP: Cryptic diversity and comparative phylogeography of the estuarine copepod Acartia tonsa on the US Atlantic coast. Mol Ecol. 2011, 20: 2425-2441. 10.1111/j.1365-294X.2011.05079.x.
Thum RA, Harrison RG: Deep genetic divergences among morphologically similar and parapatric Skistodiaptomus (Copepoda: Calanoida: Diaptomidae) challenge the hypothesis of Pleistocene speciation. Biol J Linnean Soc. 2009, 96: 150-165.
Caudill CC, Bucklin A: Molecular phylogeography and evolutionary history of the estuarine copepod, Acartia tonsa, on the Northwest Atlantic Coast. Hydrobiologia. 2004, 511: 91-102.
Chen G, Hare MP: Cryptic ecological diversification of a planktonic estuarine copepod, Acartia tonsa. Mol Ecol. 2008, 17: 1451-1468. 10.1111/j.1365-294X.2007.03657.x.
Lee CE, Frost BW: Morphological stasis in the Eurytemora affinis species complex (Copepoda: Temoridae). Hydrobiologia. 2002, 480: 111-128. 10.1023/A:1021293203512.
Claus CFW: Die frei lebenden Copepoden: Mit besonderer Berücksichtigung der Fauna Deutschlands, der Nordsee und des Mittelmeeres. 1863, Leipzig: W. Engelmann
Razouls C, de Bovée F, Kouwenberg J, Desreumaux N: Diversity and Geographic Distribution of Marine Planktonic Copepods. [http://copepodes.obs-banyuls.fr/en/]
Wolfenden RN: Notes on the collection of Copepoda. The Fauna and Geography of the Maldive and Laccadive Archipelagoes. Edited by: Gardiner JS. 1905, Cambridge: University Press, 989-1040.
Hidalgo P, Escribano R, Vergara O, Jorquera E, Donoso K, Mendoza P: Patterns of copepod diversity in the Chilean coastal upwelling system. Deep-Sea Res II. 2089–2097, 2010: 57-
McKinnon AD, Duggan S, De'ath G: Mesozooplankton dynamics in nearshore waters of the Great Barrier Reef. Est Coast Shelf Sci. 2005, 63: 497-511. 10.1016/j.ecss.2004.12.011.
Kimmerer WJ: Distribution patterns of zooplankton in Tomales Bay, California. Estuaries. 1993, 16: 264-272. 10.2307/1352499.
Blanco-Bercial L, Álvarez-Marqués F, Bucklin A: Comparative phylogeography and connectivity of sibling species of the marine copepod Clausocalanus (Calanoida). J Exp Mar Biol Ecol. 2011, 404: 108-115. 10.1016/j.jembe.2011.05.011.
Goetze E, Bradford-Grieve J: Genetic and morphological description of Eucalanus spinifer T. Scott, 1894 (Calanoida: Eucalanidae), a circumglobal sister species of the copepod E. hyalinus s.s. (Claus, 1866). Prog Oceanogr. 2005, 65: 55-87. 10.1016/j.pocean.2005.02.015.
Spalding MD, Fox HE, Allen GR, Davidson N: Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. Bioscience. 2007, 57: 573-583. 10.1641/B570707.
Knowlton N, Weigt LA: New dates and new rates for divergence across the Isthmus of Panama. Proc R Soc Lond B. 1998, 265: 2257-2263. 10.1098/rspb.1998.0568.
White BN: Vicariance biogeography of the open-ocean Pacific. Prog Oceanogr. 1994, 34: 257-284. 10.1016/0079-6611(94)90012-4.
Haug GH, Tiedemann R: Effect of the formation of the Isthmus of Panama on Atlantic Ocean thermohaline circulation. Nature. 1998, 393: 673-676. 10.1038/31447.
Peijnenburg KTCA, Goetze E: High evolutionary potential of marine zooplankton. Ecol Evol. 2013, 3: 2765-2781. 10.1002/ece3.644.
Peterson WT, Keister JE: Interannual variability in copepod community composition at a coastal station in the northern California Current: a multivariate approach. Deep-Sea Res II. 2003, 50: 2499-2517. 10.1016/S0967-0645(03)00130-9.
Stupnikova AN, Molodtsova TN, Mugue NS, Neretina TV: Genetic variability of the Metridia lucens complex (Copepoda) in the Southern Ocean. J Mar Syst. 2013, 128: 175-184.
DiBacco C, Humphrey DB, Nasmith LE, Levings CD: Ballast water transport of non-indigenous zooplankton to Canadian ports. ICES J Mar Sci. 2012, 69: 483-491. 10.1093/icesjms/fsr133.
Levings CD, Cordell JR, Ong S, Piercey GE: The origin and identity of invertebrate organisms being transported to Canada's Pacific coast by ballast water. Can J Fish Aquat Sci. 2004, 61: 1-11. 10.1139/f03-135.
Hallegraeff GM, Bolch CJ: Transport of diatom and dinoflagellate resting spores in ships' ballast water: implications for plankton biogeography and aquaculture. J Plankton Res. 1992, 14: 1067-1084. 10.1093/plankt/14.8.1067.
Hemery LG, Eléaume M, Roussel V, Améziane N, Gallut C, Steinke D, Cruaud C, Couloux A, Wilson NG: Comprehensive sampling reveals circumpolarity and sympatry in seven mitochondrial lineages of the Southern Ocean crinoid species Promachocrinus kerguelensis (Echinodermata). Mol Ecol. 2012, 21: 2502-2518. 10.1111/j.1365-294X.2012.05512.x.
Acknowledgements
This study would not have been possible without the help of colleagues who provided samples from various regions of the oceans: Holger Auel, Leocadio Blanco-Bercial, Ruth Böttger-Schnack, Ann Bucklin, Ruben Escribano, Janet Grieve, Monica Hoffmeyer, Carin Jantzen, Meriem Khelifi Touhami, Rainer Kiko, Dave McKinnon, Benjamin Kürten, Jürgen Laudien, Chaolun Li, Ilka Peeken, William T. Peterson, Gertraud Schmidt, Sigrid Schnack-Schiel, Maximilian Schulz, Hans Verheye, Lidia Yebra. Many thanks also to Joy Smith for the english revision of the manuscript. This manuscript has also been improved by two anonymous reviewers. We are grateful for the support of the Alfred P. Sloan Foundation. This study is a contribution from the Census of Marine Zooplankton (CMarZ), a Census of Marine Life project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AC and CH designed the general approach of this study. AC performed the analysis and wrote the manuscript. Both authors contributed in editing the manuscript.
Electronic supplementary material
12983_2013_308_MOESM2_ESM.docx
Additional file 2:Morphological observations made prior to DNA extraction (A), and from paratype specimens (B) for each MOTU (molecular operational taxonomic unit). Identification characters were selected according to Bradford [34], (all specimens except NWP in ethanol). For NWP only specimens in formalin were available. Abbreviations (see also Figure 1): n (Number of specimens); TL (Total length), SD (Standard deviation); P:U (Prosome:Urosome ratio); A1 (antennules); GS (genital segment); CR (Caudal rami); U3 (Urosome segment 3); U4 (Urosome segment 4); AS (Anal segment); P2, P3, P4 (swimming legs 2, 3, 4); Exp3 (Exopod segment 3); B1 (Coxa). (DOCX 135 KB)
12983_2013_308_MOESM3_ESM.docx
Additional file 3:Number of Cytb sequences (n), haplotypes (H) and contig sizes. MOTU: Molecular operational taxonomic unit. (DOCX 38 KB)
12983_2013_308_MOESM4_ESM.zip
Additional file 4:Results of Maximum Likelihood/Bayesian Inference analyses (ML/BI), jMOTU analyses with 3% and 5% thresholds (determined with the function localMinima of the R package spider), ABGD, GMYC and Rosenberg) with resulting putative species.(ZIP 131 KB)
12983_2013_308_MOESM5_ESM.pdf
Additional file 5:Color heatmap representing uncorrected p-distances (Cyt b ) among the haplotypes of the Paracalanus parvus species complex.(PDF 121 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Cornils, A., Held, C. Evidence of cryptic and pseudocryptic speciation in the Paracalanus parvus species complex (Crustacea, Copepoda, Calanoida). Front Zool 11, 19 (2014). https://doi.org/10.1186/1742-9994-11-19
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1742-9994-11-19