Abstract
Background
A primary concern when targeting HIV-1 RNA by means of antisense related technologies is the accessibility of the targets. Using a library selection approach to define the most accessible sites for 20-mer oligonucleotides annealing within the highly structured 5'-UTR of the HIV-1 genome we have shown that there are at least four optimal targets available.
Results
The biological effect of antisense DNA and LNA oligonucleotides, DNA- and LNAzymes targeted to the four most accessible sites was tested for their abilities to block reverse transcription and dimerization of the HIV-1 RNA template in vitro, and to suppress HIV-1 production in cell culture. The neutralization of HIV-1 expression declined in the following order: antisense LNA > LNAzymes > DNAzymes and antisense DNA. The LNA modifications strongly enhanced the in vivo inhibitory activity of all the antisense constructs and some of the DNAzymes. Notably, two of the LNA modified antisense oligonucleotides inhibited HIV-1 production in cell culture very efficiently at concentration as low as 4 nM.
Conclusion
LNAs targeted to experimentally selected binding sites can function as very potent inhibitors of HIV-1 expression in cell culture and may potentially be developed as antiviral drug in patients.
Similar content being viewed by others
Background
Targeting specific mRNAs by annealing complementary oligonucleotides is a basic principle of several different gene silencing technologies. In the simplest form, antisense single stranded oligonucleotides (or derivatives hereof) are introduced into the cell to block gene expression by interfering with translation of the mRNA or by degrading the RNA in a DNA/RNA heteroduplex via an RNaseH dependent pathway. This antisense approach has been used for more than two decades to study gene function in the laboratory and in attempts to treat animal and human diseases [1–4]. However, the antisense technology has never fulfilled the initially anticipated break-through as a therapeutic tool. Poor intracellular delivery, in vivo instability of the single stranded oligonucleotide, chemical toxicity and lack of mRNA target accessibility are possibly obstacles for a lacking antisense effect. The latter problem is mainly due to the formation of stable RNA structures and assembly of the mRNA into nucleoprotein complexes rendering the target site inaccessible to base pairing [5, 6]. Furthermore, it has been estimated that only 2–5% of randomly chosen antisense oligonucleotides have any effect on gene expression [5, 7] and computer generated structure models are generally not sufficient for rational prediction of effective targets.
In a related approach, RNA- or DNA-based endonucleases (ribozymes and DNAzymes) are used to cleave complementary targets in mRNA. The most commonly used ribozyme, the hammerhead, has been used extensively in vitro and with more limited success in vivo (reviewed in [8, 9]). One of the main reasons is probably the notorious instability of unmodified RNA in vivo. DNAzymes do not appear to exist in nature, but have been selected in vitro from random DNA oligo pools. One of the most active DNAzymes, named 10–23, bears some structural resemblance to the hammerhead ribozyme [10–12] but, in spite of the higher in vivo stability of single stranded DNA compared to RNA, it also demonstrated only variable success in vivo [13]. In the reported examples of targeting nucleic acid enzymes to HIV-1 RNA, either relatively large concentrations and combination of catalytic molecules are required or an in vivo expression system is used [14–17]. Common to both the antisense and the enzymatic approach are that the knock down efficacy is restricted by the accessibility of the targets in the mRNA in vivo.
More recently, RNA interfering (RNAi) has been developed as a highly potent approach to knock down gene expression in mammalian cells with an unprecedented efficiency and specificity (Reviewed in [3]). The active molecule is a small interfering RNA (siRNA), a 20–23 nucleotides RNA duplex composed of two complementary strands, one of which is complementary to the mRNA target. Although it was initially suggested that the siRNA approach is less sensitive to RNA structure in the target, it was recently demonstrated that the efficiency of RNAi-mediated "knock down" can also be influenced by the RNA structure in HIV-1 [18–21].
To address the general problem of accessibility of mRNA we have previously developed a SELEX approach that selects for the most effectively binders from a 20-mer complete antisense library through repeated binding cycles [22]. The selection protocol was applied specifically to the 355-nucleotides 5'-terminal fragment of the HIV-1 RNA genome because: a) it contains several functionally important elements including the trans-activation response element (TAR), the 5' polyadenylation signal (Poly(A)), the primer binding site (PBS), the dimer initiation site (DIS), the major splice donor (SD) and the packaging signal (PSI) that precedes the Gag open reading frame (Fig. 1A; reviewed in [23, 24]); b) most of the region is positioned upstream of the major splice donor site and is therefore present in all viral mRNA species; c) this region is scanned by the ribosome prior to cap-dependent protein synthesis; d) it is the most conserved region of the HIV-1 genome, thus increasing the chance that all HIV-1 strains are inhibited and reducing the likelihood of escape mutants; and e) we have previously tested several siRNA targeted to this region and in all cases non or very low efficiencies were observed (unpublished data). This study revealed four sites that are particularly accessible to antisense binding and these targets are here subjected to further analysis.
Oligonucleotides and their respective targets in the 5' end of the HIV-1 RNA genome. (A) Secondary structural model of the HIV-1 leader RNA. The stem-loops are named according to assigned function (see text for details) and the sequence is numbered from the 5' end of the RNA transcript. (B) The targets for the various oligonucleotide constructs. The annealing sites for the oligonucleotides are indicated by a solid line and the cleavage sites of the DNA/LNAzymes are marked by arrows. (C) Sequences of the antisense oligonucleotides and DNAzymes containing the 10–23 catalytic motif [10] named according to their target sites shown in panel B. The selected target sequences for the antisense constructs include sequences downstream of the primer binding site (PBSD), the dimerization initiation site (DIS), the splice donor site (SD) and the Gag initiation codon (AUG). The nucleotides that are substituted with LNA residues in the LNA antisense gap-mers and LNAzymes constructs are circled. The target sequences of the "10–23" DNAzymes are indicated with grey letters.
Chemical modifications are often introduced at the ribose and/or phosphate group of the backbone to increase the stability of oligonucleotides for in vivo applications. In this report the antisense effect of DNA and DNAzyme was compared to oligos that are modified with locked nucleic acid (LNA) residues. This modification consist of a methylene bridge that connects the 2'oxygen with the 4'carbon of the furanose ring, This modification locks the structure into the C3'-endo configuration, which is ideal for recognition of RNA motifs, renders the nucleic acid inaccessible the nucleases and increases the melting temperature with the RNA target strands by 2–7°C per LNA residue [25, 26]. To enable efficient RNaseH cleavage of the target mRNA by the antisense oligo, it is important to avoid LNA residues in a stretch of at least 6 nucleotides, a design generally referred to as a gap-mer [27–29]. Moreover, in the design of DNAzymes with LNA modifications it has been reported that 2–3 modifications in each arm gives the optimal binding affinity versus binding kinetic [30, 31]. Here we tested DNA and LNA (gap-mer) antisense oligos, DNA- and LNAzymes directed towards four highly accessible targets in the HIV-1 leader. We found that the LNA antisense is the most potent inhibitor, neutralizing viral expression efficiently when applied in nanomolar concentrations. The LNAzymes had a moderate effect, whereas unmodified DNA/DNAzymes have no or very little effect.
Results
Construct design and LNA modification of targeting oligonucleotides
Four target sites were selected in the HIV-1 5'-UTR as potential target based on previous accessibility selection studies [22]: 1) a region immediately downstream from the primer binding site (PBSD, 203–222), 2) a region covering the DIS (DIS, 255–274), 3) a region encompassing the major splice donor site (SD, 278–297) and 4) a region covering the gag initiation site (AUG, 326–345; Fig. 1). Four different types of oligonucleotides with potential interfering properties were synthesized: DNA antisense, LNA antisense, "10–23" DNAzyme and "10–23" LNAzyme. All DNA- and LNA-antisense constructs contained 20 nucleotides that were complementary to the selected targets in the HIV-1 RNA, whereas the DNA- and LNAzymes contained two arms of 8–9 nucleotides complementary to the target (Fig. 1B and 1C). The LNA antisense oligonucleotides were designed as gap-mers with 5 LNA residues flanking a 10-mer phosphorothioate modified DNA body to enable RNase H cleavage. The incorporation of the LNA monomers was calculated to raise the Tm values by approximately 20 degrees.
Blocking reverse transcription with LNA oligonucleotides
Reverse transcription of the RNA genome into DNA is an essential step in the viral replication cycle, and antisense oligonucleotides may inhibit this step. We therefore tested the ability of the four LNA antisense oligonucleotides to inhibit reverse transcription in vitro (Fig. 2). The PBSD, SD, and AUG specific LNAs blocked reverse transcription from a downstream primer almost completely and precisely at the expected site (94–99%; Fig. 2, lanes 1, 2 and 4), whereas the LNADIS only showed a partial effect (Fig. 2, lane 3). Interestingly, the latter effect is not caused by insufficient binding of the LNA to DIS, since this LNA inhibits RNA dimerization almost completely (Fig. 3A). The extra band observed at a position corresponding to the PSI hairpin when adding LNADIS can be explained by partial sequence complementarity between the LNA and this region (Fig. 2 lane 3, marked by asterisk). When using the DNA versions of the same oligonucleotides only 60–70% inhibition of reverse transcription was observed for any of the selected sites ([22]; data not shown), clearly demonstrating the superior stability of RNA-LNA duplexes.
The effect of antisense LNA on reverse transcription of HIV-1 RNA. An equimolar amount (1pmol) of HIV RNA and LNA oligo was mixed and incubated prior to primer extension using a primer complementary to position 384–401. Antisense LNA oligonucleotides, included LNAAUG (lane 1), LNASD (lane 2), LNADIS (lane 3), LNAPBSD (lane 4), and the major sites of transcriptional termination are indicated to the right. Read through to the 5' end of the HIV-1 RNA is denoted by +1. A sequence latter obtained by dideoxysequencing of the HIV-1 RNA is included in lanes 6–9. The level of read through reverse transcription is calculated as read-through/(read through + paused) × 100% and indicated below.
The effect of antisense LNA versus RNA and DNA on dimerization of HIV-1 RNA. The ability of (A) antisense LNA oligonucleotides directed towards different targets or (B) antisense LNA, RNA or DNA oligonucleotide directed towards the DIS target, to inhibit the formation of the DIS dimer-complex during 30 min incubation were investigated. Monomeric and dimer bands are indicated to the left. (C) A similar experiment, but where the indicated antisense oligonucleotide was added after the dimers were allowed to pre-form for 30 mins and subsequently incubated for the indicated time, hence evaluating the efficiency of breaking a stable DIS dimer-complex as a function of time time. Diamonds = LNADIS; Bullet = RNADIS; Triangle = DNADIS.
Of all the LNAs tested only LNADIS blocked HIV-1 RNA dimerization and with an efficacy of 97% if added to the dimerization reaction prior to incubation (Fig. 3A). Similar levels of inhibition were observed for DNA and RNA oligos (Fig. 3B). However, if the antisense oligonucleotides were added after pre-dimerization of the HIV-1 RNA, the LNA modified antisense oligonucleotide was significantly more potent then RNA and DNA in dissociating the dimer (Fig. 3C).
Enzymatic cleavage of the HIV-1 leader sequence with DNAzymes and modified LNAzymes optimized for binding
We wanted to investigate whether the selected regions in the HIV-1 leader were accessible to enzymatic cleavage by DNAzymes. Nucleotide enzymes targeting the selected DIS and PBS sites were synthesized both as DNA (DNAzymeDIS and DNAzymePBSD, respectively; Fig. 1) and with two LNA modifications in each arm (LNAzymeDIS and LNAzymePBSD, respectively). The cleavage efficiency was assessed by incubating 5'-end radioactively labeled HIV-1 leader RNA with the DNAzymes or LNAzymes for different time points at 10 mM Mg2+ at an enzyme to substrate ratio of 20:1, 1:1 to 1:20. The HIV RNA cleavage products were separated by denaturing gel electrophoresis and quantified (Fig. 4). The DNAzymeDIS and DNAzymePBSD and their LNA modified counterpart oligonucleotides cleaved the HIV-1 RNA at the expected position, producing 5'-end labelled fragments of approximately 261 and 205 nucleotides, respectively.
In vitro cleavage of HIV-1 RNA by DNA- and LNAzymes. One hundred nmol 5' end labeled leader RNA (+1–355) was incubated with 5 nmol, 100 nmol or 2 pmol DNAzymes or LNAzymes for the indicated time. The DNAzymes targeted to the PBSD and DIS regions cleaved primarily at the expected site, yielding a 5'-end labeled fragment of 205 and 261 nucleotides, respectively (product; panel A and C). The same bands were obtained using the LNAzyme (panel B and C). The experiment was made in duplicates yielding essentially the same result and the cleavage efficiencies indicated below each autoradiogram were calculated as (cleaved RNA/cleaved RNA and uncleaved RNA) × 100% averaged over both experiments.
When incubating the HIV-1 RNA with an excess of enzyme (20:1) both DNAzymes showed significant levels of cleavage after 24 hours (Fig. 4A and 4C). At lower stoichiometric amounts (1:1 and 1:20) only DNAzymePBSD showed moderate cleavage in the PBS loop after 24 hours (Fig. 4C). Introduction of LNA in the arms of DNAzymeDIS strongly induced the efficacy to nearly 100% cleavage after 24 hours (20:1 excess) and to a moderate cleavage level at lower enzyme concentration (1:1 and 1:20) (Fig. 4B). In contrast, LNA modifications did not improve the activity of DNAzymePBSD (Fig. 4D). A small decline in its inhibitory activity was measured, indicating that the advantage of introducing LNA residues into a DNAzyme is not universal but rather depends on the nature of the target.
Blocking expression of HIV-1 in vivo
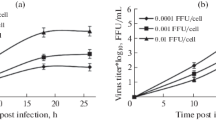
To evaluate the capacity of the antisense LNA to inhibit cellular HIV-1 expression the expression of the viral Gag derived CA-p24 protein was measured in the presence of 20 nM of the four different antisense and two mock LNAs (Fig. 5A). HEK 293-T cells were co-transfected with HIV-1 LAI genomic DNA plasmid, renilla luciferase plasmid and the LNAs. The CA-p24 production was strongly affected by all the HIV-1 specific LNAs, particularly by LNAPBSD and LNAAUG, which reduced protein production by 22- and 12-fold, respectively (Fig. 5A). In contrast, the internal luciferase control was only marginally affected (+/- 2-fold) by some of the LNAs (data no shown). The effect of the most potent LNAPBSD construct was investigated further at lower concentrations (Fig. 5B). Notably, CA-p24 expression was severely affected at concentrations as low as 4 nM (15-fold inhibition) and a complete block was apparent at 20 nM LNAPBS (Fig. 5B). This block was specific for HIV protein expression since the renilla luciferase signal was not affected at these concentrations of LNA (data no shown).
Inhibition of intracellular HIV-1 production in the presence of antisense LNA and LNAzymes. One hundred nanograms of HIV-1 genomic LAI plasmid was co-transfected with a renilla luciferase expression construct and the indicated amounts of antisense LNA or LNAzyme, and the HIV-1 production was measured by CA-p24 ELISA 72 hours later. (A) HIV-1 production in the presence 20 nM of the four different HIV-1 specific LNAs and 2 LNA controls (Mock 1 and Mock 2). (B) Measuring HIV-1 production in the presence of low range concentration of LNAPBSD (0.16–20 nM). The inhibition is calculated as the average value of two independent experiments where the relative CA-p24 expression is normalized for unspecific inhibition of renilla expression. (C) Comparing the inhibitory capacity of DNA versus LNA containing antisense or 10–23 enzymes targeted to the DIS and PBSD regions. The identity and the concentration of the oligonucleotide are indicated below. The assay was performed in duplicates.
To directly compare the inhibitory potential of the different strategies, different concentrations of oligo constructs (asDNA, asLNA, DNAzymes and LNAzymes) targeted to the PBS and DIS targets were tested for their ability to inhibit CA-p24 production in HIV-1 transfected cells (Fig. 5C). LNAPBS and LNADIS were clearly the most potent inhibitors, leading to almost complete knock down (below detection) at 20–100 nM and to a 3- and 18-fold inhibition, respectively, at 4 nM. In contrast, the DNA antisense oligos showed little effect. Notably, both the DNA- and LNAzymes led to a specific knock down. The LNAzymes were more effective than DNAzymes, giving a 100-fold knockdown at 100 nM (Fig. 5C). However, the LNAzymes also exhibited significant cell toxicity when applied at 100 nM concentration (data no shown).
Discussion
A major concern in the design of therapeutic antisense strategies against highly structured viral RNA genomes is the inaccessibility of the target sequence. To overcome this barrier we have chosen 4 targets in the HIV-1 genome that were previously selected as optimal annealing sites in vitro, and we tested them as targets for DNA and LNA antisense oligonucleotides, and DNA- and LNA-zymes. The antisense oligonucleotides were tested in vitro for their ability to interfere with reverse transcription and RNA dimerization and all inhibitors were assayed in vivo for their capacity to inhibit HIV-1 production in a cell culture assay.
Reverse transcription of viral RNA into double stranded DNA is an essential step in the retroviral replication cycle. A comparison of the antisense oligos for their ability to block this reaction revealed that all LNAs, except for LNADIS, caused a near complete block in reverse transcription. In addition to a significant level of read through, two pause sites were observed for LNADIS: one site mapped to the expected 5'end of the LNA, the other corresponded to the 3' nucleotide of the DIS loop (Fig. 1B). A likely interpretation for this observation is that a significant part of the LNADIS molecules anneal only to the exposed loop region, yet is unable to unzip the DIS stem in the HIV-1 RNA. This may explain the partial effect on reverse transcription. In contrast, RNA dimerization is nearly 100% blocked by LNADIS, suggesting that annealing of the LNA antisense to the single stranded loop of the DIS hairpin is very effective and sufficient to block dimerization.
The inhibitory potential of the antisense oligos in vivo was dramatically improved upon LNA incorporation. Especially the LNAs targeted to the PBSD, DIS and AUG regions were strong inhibitors of CA-p24 capsid protein expression. The advantage of LNA may in part rely on higher stability in the cells, but increased stability of the interaction between LNA and target most likely also plays an important role. The mechanism for the observed inhibition may involve numerous steps in the viral life cycle. All of the LNA gap-mers may degrade the mRNA via an RNase H dependent pathway and, if not degraded, block scanning of the ribosome during cap-dependent translation or the HIV-1 reverse transcriptase while copying the RNA template. In addition, the individual LNA may also have more specific actions that cannot directly be assessed in our single-round HIV-1 expression assay: the LNAPBSD may interfere with tRNA binding to the PBS and subsequent initiation of reverse transcription and the LNADIS may, in addition to dimerization, hinder effective packaging of the genome into viral particles. The LNASD covers the major splice donor site and may therefore interfere with splice site recognition but RNA packaging may also be disturbed based on the contribution of the region to this process [32–35]. Finally, the LNAAUG that covers the Gag initiation codon may interfere with the assembly of translational initiation complexes or disturb the long distance interaction recently reported between this region and upstream sequences [36, 37]. Hence, the multi-functional capacities of the LNAs applied in this study may be beneficial for their antiviral effect.
Of the four antisense targets we tested, LNAAUG was particular interesting since it overlaps with the previously described phosphorothioate modified 25-mer antisense oligonucleotide, GEM91 [38–40]. In cell culture experiments this oligo has been shown to inhibit HIV-1 replication for up to 20 days when applied at 1 μM [38, 40] and pharmacokinitical studies have been initiated in HIV-1 patients but abounded due to dose-limiting toxicity [41]. Considering that the general improved affect of LNAs compared to DNA it will be interesting to further develop the AUG directed LNA oligos as antiviral drugs for clinical use. In another study several antisense polyamide nucleotide analog (PNA) oligonucleotides targeted to the TAR stem-loop were tested, and one of the PNAs was found to have significant inhibitory potential on HIV-1 protein expression [42], however only at concentrations of more than 1 mM [43]. The efficacy of these constructs were recently improved by conjugating cell penetrating peptides to them [44].
In general the TAR-tat interaction have been objective for several antisense approaches using either LNA modified oligos [45, 46], LNA/DNA aptamers [47], mixmer of 2'-OMe and LNA modified oligos [48, 49] or PNA modified oligos [50–53]. These results do indeed indicate that the TAR region is a useful antisense target site and that various antisense oligos can inhibit the replication of HIV in different cell systems. The absence of binding sites in the TAR region in our screen suggest that this region is less accessible then the sites we have selected.
In another report, antisense gap-mer LNAs targeted to the DIS region have been tested [54]. One of these oligos resembles our LNADIS, but it is shifted a few nucleotides upstream and is two nucleotides shorter. A relatively modest 2-fold inhibitory effect was described, both in terms of in vitro dimerization and on HIV-1 expression in vivo in the presence of 160 nM oligonucleotide. This implies that small changes in target selection may have a dramatic effect, which is consistent with our in vitro binding studies [22]. However the results are not directly comparable since Elmén et al. used a subtype A HIV-1 strain that exhibits a different DIS loop sequence than the subtype B used in this report. The strong dependence on target availability creates a risk that adaptive mutations in the HIV-1 genome will render the antisense oligo less effective. It may therefore be favourable to combine the most effective LNA in future tests.
As an alternative to the antisense technology we tested the inhibitory capacity of one of the best-characterized DNAzymes, "10–23". The cleavage efficiency of this enzyme has previously been reported to be highly variable, which has limited its use [12, 31, 55–57]. The reason for this is believed to be the poor annealing of the DNAzymes to their targets. In this report, DNAzymes were targeted to the PBSD and DIS sites. The PBSD site was selected because it represents the most efficient target site for antisense molecules and hence may also be a good target for a DNAzyme and the DIS target due to it is partially inaccessible to at least one of the arms in the DNAzyme, allowing us to test the hypothesis that incorporation of LNA residues can enhance the cleavage efficiency ([30]; Fig. 1B). Indeed, we also observed that the activity of the DNAzyme directed towards the DIS target was strongly induced upon LNA incorporation, whereas the PBSD specific DNAzyme was approximately equally active in its modified and unmodified form. The LNAzymeDIS abrogated CA-p24 expression compared to the unmodified DNAzyme by at least 10-fold at 100 nM, which is consistent with a previous study on a cellular mRNA target [58]. In light of our in vitro data, this induction is most likely a result of increased cleavage of the target rather then being a stability issue. This interpretation is consistent with the much more modest effect observed when introducing LNA modifications in the more active DNAzymePBSD, both in vitro and in vivo. A potential disadvantage from introducing LNA residues in the arms of a DNAzyme is that the interaction between the LNAzyme and the target becomes too strong, which may reduce the turned over. This may explain why we never reached the point of multiple turnover using LNA modified enzymes, indicated by nearly complete digestion of target at sub-stoichiometric concentration of the enzyme.
As for antisense constructs, DNAzymes targeted against randomly selected sites are generally inactive. For instance, out of 8 DNAzymes targeting the HIV-1 TAR region, only 2 yielded detectable cleavage products and a relatively high concentration (1 μM) of inhibitor yielded only a 5–10 fold reduction in CA-p24 expression [56]. Both the pre-selected target sites tested here were cleaved by the DNAzymes reducing CA-p24 expression at a 10-fold lower concentration. The difficulties in rationally predicting efficient targets for nucleotide enzymes is also reflected by the observation that a DNAzyme directed against the natural tRNA primer binding site, which is generally assumed to be available for annealing, is unable to cleave the HIV-1 RNA (M.R.J. and J.K., unpublished observations).
siRNA targeted to the four highly accessible regions in the 5'UTR had almost no effect on HIV-1 gene expression (J.H., M.R.J. and J.K. unpublished observation). However, a direct comparison with the antisense approach is not meaningful since the selected target sequences are suboptimal using state of art design rules for siRNA [59]. Efficient knock down of HIV-1 expression by RNAi has been demonstrated using other targets [21, 60–62] and this approach is generally considered to be more potent than antisense. However, we find that the inhibitory effects observed with selected LNA antisense constructs in the low nanomolar range is able to match some of the best HIV-1 specific siRNAs reported in literature [63].
Conclusion
Four sites that were pre-selected as highly accessible regions in the HIV-1 leader were accessed as potential targets for various antisense based technologies and we conclude that antisense LNA targeted to specific sites in the PBS and the DIS regions were the most effective inhibitor of HIV-1 expression. LNA may have additional advantages for in vivo applications, such as more efficient cell uptake and increased stability. Moreover, the lower molecular weight and single stranded nature of LNA makes it potentially more inexpensive to synthesise in large quantities than double stranded siRNA. LNA therefore provides a serious alternative platform for development of therapeutics for human diseases.
Methods
Constructs
The plasmids pUC18-LAI and pUC18-LAI-1–444 contain +1–355 nucleotides and +1–444, respectively, of the HIV-1 genome sequence of the LAI isolate behind a T7 promoter and have been described earlier [64]. LNA oligonucleotides (Exiqon) were all designed as gap-mers with 5 LNA residues flanking a 10-mer phosphorothioate modified DNA body, except from MOCK-2 that were an 18-mer with only 4 LNA residues at each termini. Both MOCK-1 and MOCK-2 contain random sequences without extensive match to human or HIV-1 sequences. The LNAzymes (Exiqon) and DNAzymes (DNA Technology) were constructed as "10–23" enzymes [12] with an arm length of 9 nucleotides. RNA transcription was performed as described earlier [22].
Primer extension assay
The primer extension assay was performed using 1 pmol RNA spanning the first 444 nucleotides of the HIV-1 genome and 80 fmol 5'end-labeled RT primer (5'-CCTTAACCGAATTTTTTCCC-3') complementary to position 384–401. The template and the primer were annealed for 2 min at 90°C in a total volume of 6 μl annealing buffer (100 mM Tris-HCl, pH 7.5, 400 mM KCl) followed by 5 min at room temperature. Then 1 pmol LNA oligonucleotides were added and incubated for 20 min at 50°C. Reverse transcription and gel analysis was performed according to Damgaard et al. [65, 66].
Dimerization assay
One pmol [γ-P32] HIV-1 leader RNA and five pmol LNA oligonucleotide was incubated in 20 ul of water at 85°C for 5 min and then snap-cooled on ice for 5 minutes. The buffer was adjusted to dimerization conditions (50 mM Na-cacodylate; pH 7.5, 250 mM KCl, 5 mM MgCl2) and incubated at 37°C for 30 min in The sample was analyzed on a 6% native TBM gel (50 mM Tris-borate; pH 8.3, 5 mM MgCl2) and autoradiographed using phosphor image screens (Biorad).
In the kinetic assay, 1 pmol HIV-1 leader RNA was allowed to predimerize for 30 min at 37°C at dimerization conditions and then either 5 pmol LNA, DNA or RNA DIS oligonucleotide was added. After 0, 30, 120 and 240 minutes aliques were taken out and placed on ice before analyzed on a 6% native TBM gel and autoradiographed on phosphor image screens.
DNazyme and LNazyme cleavage assay
One hundred nmol 5' end labelled leader (+1–355) RNA was incubated at 85°C in 7 μl water for 5 min and cooled on ice for 5 min. The RNA were then mixed with 5 nmol, 100 nmol or 2 pmol DNAzymes or LNAzymes and incubated at 37°C for 1, 2, 4 or 24 hours in DNazyme buffer (10 mM MgCl2, 50 mM Tris-HCl; pH 8.0) and stopped with 100 mM EDTA. The samples were precipitated and analyzed on an 8% denaturing polyacrylamide gel run at 18 W and analyzed by phosphor imaging (Biorad).
HIV-1 production assay
HEK 293-T cells were seeded one day before transfection at 150.000 cells/ml/well in a 24-well plate. Transfection was performed at 40% confluency in duplicate using Lipofectamine-2000 (Invitrogen) in 400 μl medium without antibiotics. Per transfection, 100 ng of HIV-1 genomic LAI plasmid was diluted in 50 μl OPTIMEM and the respectively final concentration of the various oligonucleotides. Two μg Lipofectamine was added to 48 μl OPTIMEM and incubated for 5 min at RT. The diluted DNA and lipofectamine were combined to a final sample volume of 100 μl. This mixture was incubated for 20 min at 20°C before adding to cells. Six hours post-transfection 1 ml medium containing antibiotics replaced the original medium. Three days post-transfection 100 μl from the culture media was collected and inactivated by adding 10 μl 0.1% Empigen (final concentration) and heating at 65°C for 30 min. Production of HIV-1 CA-p24 was measured with p24 enzyme-linked immunosorbent assay (ELISA). As an internal control, 2.5 ng pRL was included and the Renilla luciferase expression levels were measured using the Dual-luciferase Reporter Assay System (Promega).
Abbreviations
- HIV:
-
Human Immunodeficiency Virus
- LNA:
-
Locked Nucleic Acid
- 5'-UTR:
-
5' Untranslated Region
- RT:
-
Reverse Transcriptase
- TAR:
-
Trans-activation response element
- PBS:
-
Primer binding site
- DIS:
-
Dimer initiation site
- SD:
-
Major splice donor
- PSI:
-
Packaging signal
- PBSD:
-
Primer binding site downstream
References
Kurreck J: Antisense technologies. Improvement through novel chemical modifications. Eur J Biochem. 2003, 270: 1628-1644. 10.1046/j.1432-1033.2003.03555.x.
Nielsen MH, Pedersen FS, Kjems J: Molecular strategies to inhibit HIV-1 replication. Retrovirology. 2005, 2: 10-10.1186/1742-4690-2-10.
Schubert S, Kurreck J: Oligonucleotide-based antiviral strategies. Handb Exp Pharmacol. 2006, 261-287.
Turner JJ, Fabani M, Arzumanov AA, Ivanova G, Gait MJ: Targeting the HIV-1 RNA leader sequence with synthetic oligonucleotides and siRNA: Chemistry and cell delivery. Biochim Biophys Acta. 2006, 1758: 290-300. 10.1016/j.bbamem.2005.10.013.
Sohail M, Southern EM: Selecting optimal antisense reagents. Adv Drug Deliv Rev. 2000, 44: 23-34. 10.1016/S0169-409X(00)00081-8.
Sohail M, Southern EM: Hybridization of antisense reagents to RNA. Curr Opin Mol Ther. 2000, 2: 264-271.
Sohail M, Hochegger H, Klotzbucher A, Guellec RL, Hunt T, Southern EM: Antisense oligonucleotides selected by hybridisation to scanning arrays are effective reagents in vivo. Nucleic Acids Res. 2001, 29: 2041-2051. 10.1093/nar/29.10.2041.
Schubert S, Kurreck J: Ribozyme- and deoxyribozyme-strategies for medical applications. Curr Drug Targets. 2004, 5: 667-681. 10.2174/1389450043345092.
Sioud M, Iversen PO: Ribozymes, DNAzymes and small interfering RNAs as therapeutics. Curr Drug Targets. 2005, 6: 647-653. 10.2174/1389450054863653.
Santoro SW, Joyce GF: A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci U S A. 1997, 94: 4262-4266. 10.1073/pnas.94.9.4262.
Santoro SW, Joyce GF: Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry. 1998, 37: 13330-13342. 10.1021/bi9812221.
Schubert S, Gul DC, Grunert HP, Zeichhardt H, Erdmann VA, Kurreck J: RNA cleaving '10-23' DNAzymes with enhanced stability and activity. Nucleic Acids Res. 2003, 31: 5982-5992. 10.1093/nar/gkg791.
Sun LQ, Cairns MJ, Gerlach WL, Witherington C, Wang L, King A: Suppression of smooth muscle cell proliferation by a c-myc RNA-cleaving deoxyribozyme. J Biol Chem. 1999, 274: 17236-17241. 10.1074/jbc.274.24.17236.
Sood V, Unwalla H, Gupta N, Chakraborti S, Banerjea AC: Potent knock down of HIV-1 replication by targeting HIV-1 Tat/Rev RNA sequences synergistically with catalytic RNA and DNA. Aids. 2007, 21: 31-40.
Barroso-DelJesus A, Puerta-Fernandez E, Tapia N, Romero-Lopez C, Sanchez-Luque FJ, Martinez MA, Berzal-Herranz A: Inhibition of HIV-1 replication by an improved hairpin ribozyme that includes an RNA decoy. RNA Biol. 2005, 2: 75-79.
Ikeda M, Habu Y, Miyano-Kurosaki N, Takaku H: Suppression of HIV-1 replication by a combination of endonucleolytic ribozymes (RNase P and tRNnase ZL). Nucleosides Nucleotides Nucleic Acids. 2006, 25: 427-437. 10.1080/01457630600684120.
Ramezani A, Ma XZ, Ameli M, Arora A, Joshi S: Assessment of an anti-HIV-1 combination gene therapy strategy using the antisense RNA and multimeric hammerhead ribozymes. Front Biosci. 2006, 11: 2940-2948. 10.2741/2023.
Westerhout EM, Vink M, Joost Haasnoot PC, Das AT, Berkhout B: A conditionally replicating HIV-based vector that stably expresses an antiviral shRNA against HIV-1 replication. Mol Ther. 2006
Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B: HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005, 33: 796-804. 10.1093/nar/gki220.
Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B: Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004, 78: 2601-2605. 10.1128/JVI.78.5.2601-2605.2004.
Berkhout B: RNA interference as an antiviral approach: targeting HIV-1. Curr Opin Mol Ther. 2004, 6: 141-145.
Jakobsen MR, Damgaard CK, Andersen ES, Podhajska A, Kjems J: A genomic selection strategy to identify accessible and dimerization blocking targets in the 5'-UTR of HIV-1 RNA. Nucleic Acids Res. 2004, 32: e67-10.1093/nar/gnh064.
Huthoff H, Berkhout B: Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry. 2002, 41: 10439-10445. 10.1021/bi025993n.
Berkhout B: Multiple biological roles associated with the repeat(R) region of the HIV-1 RNA genome. Adv in Pharmacology. 2000, 48: 29-73.
Wengel J, Petersen M, Nielsen KE, Jensen GA, Hakansson AE, Kumar R, Sorensen MD, Rajwanshi VK, Bryld T, Jacobsen JP: LNA (locked nucleic acid) and the diastereoisomeric alpha-L-LNA: conformational tuning and high-affinity recognition of DNA/RNA targets. Nucleosides Nucleotides Nucleic Acids. 2001, 20: 389-396. 10.1081/NCN-100002312.
Petersen M, Wengel J: LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003, 21: 74-81. 10.1016/S0167-7799(02)00038-0.
Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hokfelt T, Broberger C, Porreca F, Lai J, Ren K, Ossipov M, Koshkin A, Jakobsen N, Skouv J, Oerum H, Jacobsen MH, Wengel J: Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci U S A. 2000, 97: 5633-5638. 10.1073/pnas.97.10.5633.
Crinelli R, Bianchi M, Gentilini L, Magnani M: Design and characterization of decoy oligonucleotides containing locked nucleic acids. Nucleic Acids Res. 2002, 30: 2435-2443. 10.1093/nar/30.11.2435.
Kurreck J, Wyszko E, Gillen C, Erdmann VA: Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002, 30: 1911-1918. 10.1093/nar/30.9.1911.
Vester B, Hansen LH, Lundberg LB, Babu BR, Sorensen MD, Wengel J, Douthwaite S: Locked nucleoside analogues expand the potential of DNAzymes to cleave structured RNA targets. BMC Mol Biol. 2006, 7: 19-10.1186/1471-2199-7-19.
Vester B, Lundberg LB, Sorensen MD, Babu BR, Douthwaite S, Wengel J: Improved RNA cleavage by LNAzyme derivatives of DNAzymes. Biochem Soc Trans. 2004, 32: 37-40. 10.1042/BST0320037.
Amarasinghe GK, Zhou J, Miskimon M, Chancellor KJ, McDonald JA, Matthews AG, Miller RR, Rouse MD, Summers MF: Stem-loop SL4 of the HIV-1 psi RNA packaging signal exhibits weak affinity for the nucleocapsid protein. structural studies and implications for genome recognition. J Mol Biol. 2001, 314: 961-970. 10.1006/jmbi.2000.5182.
Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF: NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J Mol Biol. 2000, 301: 491-511. 10.1006/jmbi.2000.3979.
McBride MS, Panganiban AT: Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997, 71: 2050-2058.
McBride MS, Schwartz MD, Panganiban AT: Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997, 71: 4544-4554.
Berkhout B, Ooms M, Beerens N, Huthoff H, Southern E, Verhoef K: In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J Biol Chem. 2002, 277: 19967-19975. 10.1074/jbc.M200950200.
Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J: RNA interactions in the 5' region of the HIV-1 genome. J Mol Biol. 2004, 336: 369-379. 10.1016/j.jmb.2003.12.010.
Lisziewicz J, Sun D, Weichold FF, Thierry AR, Lusso P, Tang J, Gallo RC, Agrawal S: Antisense oligodeoxynucleotide phosphorothioate complementary to Gag mRNA blocks replication of human immunodeficiency virus type 1 in human peripheral blood cells. Proc Natl Acad Sci U S A. 1994, 91: 7942-7946. 10.1073/pnas.91.17.7942.
Yamaguchi K, Papp B, Zhang D, Ali AN, Agrawal S, Byrn RA: The multiple inhibitory mechanisms of GEM 91, a gag antisense phosphorothioate oligonucleotide, for human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1997, 13: 545-554.
Zhang R, Diasio RB, Lu Z, Liu T, Jiang Z, Galbraith WM, Agrawal S: Pharmacokinetics and tissue distribution in rats of an oligodeoxynucleotide phosphorothioate (GEM 91) developed as a therapeutic agent for human immunodeficiency virus type-1. Biochem Pharmacol. 1995, 49: 929-939. 10.1016/0006-2952(95)00010-W.
Sereni D, Tubiana R, Lascoux C, Katlama C, Taulera O, Bourque A, Cohen A, Dvorchik B, Martin RR, Tournerie C, Gouyette A, Schechter PJ: Pharmacokinetics and tolerability of intravenous trecovirsen (GEM 91), an antisense phosphorothioate oligonucleotide, in HIV-positive subjects. J Clin Pharmacol. 1999, 39: 47-54. 10.1177/00912709922007552.
Mayhood T, Kaushik N, Pandey PK, Kashanchi F, Deng L, Pandey VN: Inhibition of Tat-mediated transactivation of HIV-1 LTR transcription by polyamide nucleic acid targeted to TAR hairpin element. Biochemistry. 2000, 39: 11532-11539. 10.1021/bi000708q.
Kaushik N, Basu A, Pandey VN: Inhibition of HIV-1 replication by anti-trans-activation responsive polyamide nucleotide analog. Antiviral Res. 2002, 56: 13-27. 10.1016/S0166-3542(02)00024-4.
Tripathi S, Chaubey B, Ganguly S, Harris D, Casale RA, Pandey VN: Anti-HIV-1 activity of anti-TAR polyamide nucleic acid conjugated with various membrane transducing peptides. Nucleic Acids Res. 2005, 33: 4345-4356. 10.1093/nar/gki743.
Wengel J, Vester B, Lundberg LB, Douthwaite S, Sorensen MD, Babu BR, Gait MJ, Arzumanov A, Petersen M, Nielsen JT: LNA and alpha-L-LNA: towards therapeutic applications. Nucleosides Nucleotides Nucleic Acids. 2003, 22: 601-604. 10.1081/NCN-120021963.
Turner JJ, Arzumanov AA, Gait MJ: Synthesis, cellular uptake and HIV-1 Tat-dependent trans-activation inhibition activity of oligonucleotide analogues disulphide-conjugated to cell-penetrating peptides. Nucleic Acids Res. 2005, 33: 27-42. 10.1093/nar/gki142.
Darfeuille F, Reigadas S, Hansen JB, Orum H, Di Primo C, Toulme JJ: Aptamers targeted to an RNA hairpin show improved specificity compared to that of complementary oligonucleotides. Biochemistry. 2006, 45: 12076-12082. 10.1021/bi0606344.
Brown D, Arzumanov AA, Turner JJ, Stetsenko DA, Lever AM, Gait MJ: Antiviral activity of steric-block oligonucleotides targeting the HIV-1 trans-activation response and packaging signal stem-loop RNAs. Nucleosides Nucleotides Nucleic Acids. 2005, 24: 393-396. 10.1081/NCN-200059813.
Brown DE, Arzumanov A, Syed S, Gait MJ, Lever AM: Inhibition of HIV-1 replication by oligonucleotide analogues directed to the packaging signal and trans-activating response region. Antivir Chem Chemother. 2006, 17: 1-9.
Turner JJ, Jones S, Fabani MM, Ivanova G, Arzumanov AA, Gait MJ: RNA targeting with peptide conjugates of oligonucleotides, siRNA and PNA. Blood Cells Mol Dis. 2007, 38: 1-7. 10.1016/j.bcmd.2006.10.003.
Turner JJ, Ivanova GD, Verbeure B, Williams D, Arzumanov AA, Abes S, Lebleu B, Gait MJ: Cell-penetrating peptide conjugates of peptide nucleic acids (PNA) as inhibitors of HIV-1 Tat-dependent trans-activation in cells. Nucleic Acids Res. 2005, 33: 6837-6849. 10.1093/nar/gki991.
Riguet E, Tripathi S, Chaubey B, Desire J, Pandey VN, Decout JL: A peptide nucleic acid-neamine conjugate that targets and cleaves HIV-1 TAR RNA inhibits viral replication. J Med Chem. 2004, 47: 4806-4809. 10.1021/jm049642d.
Boulme F, Freund F, Moreau S, Nielsen PE, Gryaznov S, Toulme JJ, Litvak S: Modified (PNA, 2'-O-methyl and phosphoramidate) anti-TAR antisense oligonucleotides as strong and specific inhibitors of in vitro HIV-1 reverse transcription. Nucleic Acids Res. 1998, 26: 5492-5500. 10.1093/nar/26.23.5492.
Elmen J, Zhang HY, Zuber B, Ljungberg K, Wahren B, Wahlestedt C, Liang Z: Locked nucleic acid containing antisense oligonucleotides enhance inhibition of HIV-1 genome dimerization and inhibit virus replication. FEBS Lett. 2004, 578: 285-290. 10.1016/j.febslet.2004.11.015.
Wang DY, Lai BH, Feldman AR, Sen D: A general approach for the use of oligonucleotide effectors to regulate the catalysis of RNA-cleaving ribozymes and DNAzymes. Nucleic Acids Res. 2002, 30: 1735-1742. 10.1093/nar/30.8.1735.
Chakraborti S, Banerjea AC: Inhibition of HIV-1 gene expression by novel DNA enzymes targeted to cleave HIV-1 TAR RNA: potential effectiveness against all HIV-1 isolates. Mol Ther. 2003, 7: 817-826. 10.1016/S1525-0016(03)00096-0.
Chakraborti S, Banerjea AC: Identification of cleavage sites in the HIV-1 TAR RNA by 10-23 and 8-17 catalytic motif containing DNA enzymes. Biomacromolecules. 2003, 4: 568-571. 10.1021/bm025698i.
Fahmy RG, Khachigian LM: Locked nucleic acid modified DNA enzymes targeting early growth response-1 inhibit human vascular smooth muscle cell growth. Nucleic Acids Res. 2004, 32: 2281-2285. 10.1093/nar/gkh543.
Zamore PD, Aronin N: siRNAs knock down hepatitis. Nat Med. 2003, 9: 266-267. 10.1038/nm0303-266.
Hayafune M, Miyano-Kurosaki N, Park WS, Moori Y, Takaku H: Silencing of HIV-1 gene expression by two types of siRNA expression systems. Antivir Chem Chemother. 2006, 17: 241-249.
Nishitsuji H, Kohara M, Kannagi M, Masuda T: Effective suppression of human immunodeficiency virus type 1 through a combination of short- or long-hairpin RNAs targeting essential sequences for retroviral integration. J Virol. 2006, 80: 7658-7666. 10.1128/JVI.00078-06.
Konstantinova P, de Vries W, Haasnoot J, Ter Brake O, de Haan P, Berkhout B: Inhibition of human immunodeficiency virus type 1 by RNA interference using long-hairpin RNA. Gene Ther. 2006
Rossi JJ: RNAi as a treatment for HIV-1 infection. Biotechniques. 2006, Suppl: 25-29.
Andersen ES, Jeeninga RE, Damgaard CK, Berkhout B, Kjems J: Dimerization and template switching in the 5' untranslated region between various subtypes of human immunodeficiency virus type 1. J Virol. 2003, 77: 3020-3030. 10.1128/JVI.77.5.3020-3030.2003.
Andersen ES, Contera SA, Knudsen B, Damgaard CK, Besenbacher F, Kjems J: Role of the TAR hairpin in dimerization of HIV-1 RNA. J Biol Chem. 2004
Damgaard CK, Dyhr-Mikkelsen H, Kjems J: Mapping the RNA binding sites for human immunodeficiency virus type-1 gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 1998, 26: 3667-3676. 10.1093/nar/26.16.3667.
Acknowledgements
We are grateful to Rita Rosendahl for excellent technical assistance. We would also like to thank Exiqon for advice on LNA design. The work in the Kjems lab was supported in parts by grants from the Danish Natural Research Councils, The Danish AIDS Foundation and EU FP6 STREP "Hidden HIV". Studies in the Berkhout lab are supported by a ZonMw (vici grant) and NWO-CW (TOP grant). The interaction between the two labs was supported by NORFA.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
MRJ: Construct design, primer extension assay, enzymatic cleavage assay and manuscript preparation
JH: HIV-1 production assay
JW: LNA oligonucleotide and LNAzyme design
BB: Manuscript preparation
JK: Experimental design, manuscript preparation
All authors read and approved the final manuscript.
Martin R Jakobsen, Joost Haasnoot contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Jakobsen, M.R., Haasnoot, J., Wengel, J. et al. Efficient inhibition of HIV-1 expression by LNA modified antisense oligonucleotides and DNAzymes targeted to functionally selected binding sites. Retrovirology 4, 29 (2007). https://doi.org/10.1186/1742-4690-4-29
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1742-4690-4-29