Abstract
Background
Multiple Sclerosis (MS) is an autoimmune demyelinating disease that occurs more frequently in women than in men. Multiple Sclerosis Associated Retrovirus (MSRV) is a member of HERV-W, a multicopy human endogenous retroviral family repeatedly implicated in MS pathogenesis. MSRV envelope protein is elevated in the serum of MS patients and induces inflammation and demyelination but, in spite of this pathogenic potential, its exact genomic origin and mechanism of generation are unknown. A possible link between the HERV-W copy on chromosome Xq22.3, that contains an almost complete open reading frame, and the gender differential prevalence in MS has been suggested.
Results
MSRV transcription levels were higher in MS patients than in controls (U-Mann–Whitney; p = 0.004). Also, they were associated with the clinical forms (Spearman; p = 0.0003) and with the Multiple Sclerosis Severity Score (MSSS) (Spearman; p = 0.016). By mapping a 3 kb region in Xq22.3, including the HERV-W locus, we identified three polymorphisms: rs6622139 (T/C), rs6622140 (G/A) and rs1290413 (G/A). After genotyping 3127 individuals (1669 patients and 1458 controls) from two different Spanish cohorts, we found that in women rs6622139 T/C was associated with MS susceptibility: [χ2; p = 0.004; OR (95% CI) = 0.50 (0.31-0.81)] and severity, since CC women presented lower MSSS scores than CT (U-Mann–Whitney; p = 0.039) or TT patients (U-Mann–Whitney; p = 0.031). Concordantly with the susceptibility conferred in women, rs6622139*T was associated with higher MSRV expression (U-Mann–Whitney; p = 0.003).
Conclusions
Our present work supports the hypothesis of a direct involvement of HERV-W/MSRV in MS pathogenesis, identifying a genetic marker on chromosome X that could be one of the causes underlying the gender differences in MS.
Similar content being viewed by others
Background
Human endogenous retroviruses (HERVs) are believed to be remnants of ancient exogenous infections. They entered the germ line during primate evolution and as retroviruses were able to replicate and retrotranspose, increasing their copy number and spreading all over the genome. Indeed, approximately an 8% of the genome has a retroviral origin [1]. During evolution, most of the HERV proviruses have undergone extensive mutations and are unable to replicate. Nowadays, they reside in the genome of all human cells and are transmitted in Mendelian fashion.
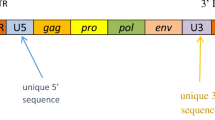
Retroviruses typically consist of an internal region containing gag, pro, pol, and env genes, flanked by two long terminal repeats (LTR). Some HERVs have retained open reading frames (ORFs) putatively encoding functional proteins. These proteins can display a physiological role, as Syncytin-1 which is involved in the formation of the syncytiotrophoblast during pregnancy [2]; but as viral products, they might show antigenic properties involving the immune system. Indeed, HERVs have been associated with several pathologies as cancer and autoimmune diseases. HERVs RNAs, proteins or virions have been found in different tumor tissues [3–7], in schizophrenia [8–10], rheumatoid arthritis [11] and especially in multiple sclerosis (MS) [12, 13].
Mainly two HERV families have been related to MS: HERV-W and HERV-H; and among them, the member most intensively studied is MSRV, a HERV-W-related retroelement [14].
Depending on the cohort studied, the percentage of MS serum or plasma samples positive to MSRV ranges from 50% [15] to 100% [16]. Enhanced expression of MSRV was also detected in MS plaques in independent post-mortem brain studies [17–19] and increased MSRV copy number in MS blood DNA was associated with a poorer MS prognosis [20, 21]. Besides, interferon-beta reduces MSRV load in MS patients with a good clinical response to the treatment [22], suggesting that it could be a biomarker to monitor disease progression and therapy outcome.
MSRV might be exerting its role in MS pathogenesis through the activation of innate immunity and subsequent release of pro-inflammatory cytokines [23] through Toll-like receptor (TLR-4) [24]. It acts as a superantigen producing polyclonal T-lymphocyte activation [25] and it triggers a T-cell mediated neuropathology in vivo[26].
In spite of its pathogenic potential, the exact genomic origin of MRSV is unknown. When MSRV was discovered, controversy arose regarding whether it was an endogenous or exogenous virus [14]; at present, it seems that MSRV is most likely transcribed from an HERV-W endogenous element [27]. The HERV-W family consists of approximately 650 elements dispersed through the human genome [28], but the only member showing a full-length proviral copy with an ORF is the locus ERVWE1 in the 7q21.2 chromosome, which encodes Syncytin-1 (Accession number: AF156963). In silico analyses have revealed that, in addition to the one previously described in chromosome 7, other putative complete retroviral units of HERV-W with gag, pro, pol and env might be replication competent [29]. Among all the HERV-W locations, the copy in chromosome Xq22.3 (ERVWE2) could be involved in MSRV expression due to several reasons. First of all, the env gene at this locus is the only known, besides ERVWE1, with an ORF only interrupted by a stop codon, and it has retained its coding capacity producing an almost complete N-truncated protein [30]in vitro. This sequence from chromosome X has important homologies with several reported MSRV clones derived from retrotranscription of RNA obtained from retroviral-like particles in MS patients. For example, the clone pV14 (AF331500) of MSRV env aligned with 98.5% of sequence identity and interestingly, the complete clone CL15 (AF127228) was 99.53% homologous to env + pol probably reflecting transcription of this region [27]. Moreover, a genetic origin in chromosome X could explain in part the higher prevalence of MS in women than in men (2:1 ratio).
In order to find polymorphisms associated with MS susceptibility that could be related with differential expression of MSRV in patients and controls, we mapped the HERV-W insertion in chromosome Xq22.3 and genotyped a total of 1669 patients and 1458 controls, from two different Spanish cohorts.
Results
Association of MSRV transcription levels with MS diagnosis and clinical evolution
To confirm the previously published association between MSRV RNA levels and MS, MSRV RNA expression was studied by RT-PCR in a randomly selected group of MS patients (n = 112; 55.6% women) and controls (n = 68; 53.3% women). The analysis was made with a set of primers and probe able to discriminate between syncytin-1 and MSRV env sequences [31]. MS patients presented higher MSRV RNA levels compared to controls (U-Mann–Whitney; p = 0.004) (Figure 1A). Also, we found a correlation with the clinical forms (Spearman’s rho = 0.28; p = 0.0003) and lower levels of MSRV expression were found in blood donors (BD) than in relapsing-remitting (RR) MS patients (U-Mann–Whitney; p = 0.005) or secondary-progressive (SP) patients (U-Mann–Whitney; p = 0.005) (Figure 1B).
Association of MSRV transcription levels with MS diagnosis and clinical evolution. A) MSRV relative expression* was higher in MS patients (n = 112) compared to controls (n = 68) (U-Mann–Whitney; p = 0.004). B) MSRV relative expression* correlated with clinical forms (Spearman’s rho; p = 3*10-4). MSRV relative expression* in BD (n = 68) vs. RR patients (n = 81) (U-Mann–Whitney; p = 0.005), RR vs. SP patients (n = 15) (U-Mann–Whitney; p = 0.108) and BD vs SP patients (U-Mann–Whitney; p = 0.005). C) Correlation of MSRV relative expression* and MSSS score in women (n = 60) (Spearman’s rho = 0.34; p = 0.017). Subjects were randomly selected. *MSRV Relative Expression = 2e-∆∆Ct = 2e-((Ct MSRV – Ct GUSB)sample-Mean (Ct MSRV – Ct GUSB)Blood Donors).
Regarding clinical parameters, a correlation between MSRV relative expression and Multiple Sclerosis Severity Score (MSSS) [32] (Spearman’s rho = 0.25; p = 0.016) was observed; also the number of relapses suffered in the last two years tended to correlate with MSRV expression (Spearman’s rho = 0.19; p = 0.06). When we stratified by sex, the correlation coefficient between the MSRV expression and MSSS increased in women (Spearman’s rho = 0.34; p = 0.017) (Figure 1C), while only a trend could be seen in men (Spearman’s rho = 0.22; p = 0.16).
Mapping of HERV-W copy in chromosome Xq22.3
First, we selected a region of 3 kb (106295193 – 106298196; Human GRCh37, March 2012) including the complete HERV-W locus. This sequence includes 3′ portions of the pol gene, the complete env gene, and the U3 and R regions of the 3′LTR (Figure 2). Then, we amplified the sequence through 18 overlapping amplicons. Each PCR product was analyzed by High Resolution Melting (HRM) curve analysis in 100 individuals, detecting 5 different mutations: T/C at 445 nt, A/T at 464 nt, T/C at 1158 nt, G/A at 1943 nt and G/A at 2611 nt from the beginning of the amplicon. Besides, an insertion of a G nucleotide was found at 1778 nt. Only 3 of these variants were detected in more than 5% of the individuals: T/C at 1158 nt position (SNP1: rs6622139), G/A at 1943 nt (SNP2: rs6622140) and G/A at 2611 nt (SNP3: rs1290413). The analysis in Haploview 4.0 showed that rs6622139 and rs6622140 were in perfect linkage disequilibrium (r2 = 1) and both map in the env open reading frame; therefore, only rs6622139 and the third polymorphism, rs1290413, were further studied in our cohort.
Association of rs6622139 and rs1290413 with MS susceptibility
A total of 3127 individuals from two different Spanish cohorts were genotyped for rs6622139 and rs1290413: 893 MS patients and 664 controls from the discovery cohort (Madrid) and 776 MS patients and 794 controls from the replication cohort (Andalusia). Clinical and demographic characteristics of patients and controls are described in Table 1.
The SNPs were analyzed separately in both sexes due to their location in chromosome X. Genotyping success rate was over 98% for both markers. Genotype distribution of the SNPs in controls and MS patients and p-values are shown in Table 2.
In the discovery cohort significant associations of rs6622139 and rs1290413 with MS were found in women. The homozygous mutant genotype of rs6622139 showed a protective effect, being more frequent in control than MS women [p = 0.02; OR (95% CI) =0.44 (0.20 – 0.96)] and the homozygous mutant genotype of rs1290413 also showed a nominal association with increased predisposition to MS in women [p = 0.05; OR (95% CI) = 1.81 (0.96-3.47)] (Table 2).
When data of a replication cohort were combined in order to increase the sample size and gain statistical power, the association of rs6622139*C in homozygosis was corroborated [p = 0.004; OR (95% CI) = 0.50 (0.31-0.81)] (Table 2).
Association of rs6622139 with MS disability
As rs6622139 was associated with MS susceptibility in women, we analyzed its role in clinical evolution. MSSS score was available from most patients from the Madrid cohort, thus the analysis was made including these subjects. Among MS women, carriers of the homozygous mutant genotype for rs6622139 (CC) (n = 9) presented lower MSSS scores [32] than either CT (n = 118) (U-Mann–Whitney; p = 0.039) or TT (n = 245) (U-Mann–Whitney; p = 0.031) carriers (Figure 3).
Association of rs6622139 with MSRV transcription levels
In agreement with the susceptibility conferred to women, rs6622139*T allele was associated with higher levels of MSRV RNA than rs6622139*C (U-Mann–Whitney; p = 0.003) in the women with available data analyzed (Figure 4A). Moreover, a dose-effect was evidenced from the association of rs6622139 genotypes and MSRV transcription levels (U-Mann–Whitney; CC vs. CT: p = 0.033 and CC vs. TT: p = 0.006) (Figure 4B). However, since the number of individuals with GG genotype in this group is very limited, any conclusion should be taken with caution.
Association of rs6622139 with MSRV transcription levels in women (MS patients and controls). A) rs6622139*T allele (n = 136) was associated to a higher MSRV relative expression* than rs6622139*C allele (n = 22) (U-Mann–Whitney; p = 0.003). B) MSRV relative expression* according to rs6622139 genotypes (U-Mann–Whitney; CC (n = 3) vs. CT (n = 16): p = 0.03 and CC vs. TT (n = 60): p = 0.006). *MSRV Relative Expression = 2e-∆∆Ct = 2e-((Ct MSRV – Ct GUSB)sample-Mean (Ct MSRV – Ct GUSB)Blood Donors).
Discussion
In the present study we confirm increased levels of MSRV transcripts in MS patients compared to controls and correlation between MSRV expression and clinical severity (secondary progressive condition and MSSS) supporting the role of this element in MS pathology. Besides, we performed a genetic screening of the HERV-W locus in chromosome Xq22.3, identifying a polymorphism that is associated with MS susceptibility and severity in women. Finally, we found significant differences between levels of MSRV expression comparing carriers of the rs6622139 genotypes. All these findings support the role of chromosome Xq22.3 HERV-W locus in MSRV expression and MS etiopathogenesis.
The polymorphism rs6622139 associated with the disease is in perfect linkage disequilibrium (r2 = 1) with rs6622140 and both map in the orf of the env sequence. Taking MSRV env sequence (AF33150) as reference, the locus ERVWE2 studied here presents a stop codon at position 39; however, this locus harbours an orf with a start codon (ATG) at position 68 that encodes for a N-terminally truncated 475 amino acid Env protein [30]. The N-truncated protein is expressed intracellularly ex vivo[30], while MSRV env, as a full-length retroviral envelope protein, is expected to be expressed at the cell surface. When the stop codon was reverted in vitro the resulting protein was expressed on the cell surface. Given the fundamental changes provoked by the elimination of the stop codon in ERVWE2, it has been speculated that MSRV env sequences could come from ERVWE2 variants lacking that stop codon [30]. After mapping ERVWE2 we did not find any mutation reversing the stop codon in our Spanish cohort.
Another hypothesis to explain the origin of the reported MSRV env sequences is that they might derive from recombination of transcripts from various HERV-W loci [27]. Apart from the full-length MSRV env, it has been also suggested that the N-truncated protein encoded by ERVW2 and other retroviral defective proteins may be expressed in vivo and exert functions in human physiology or pathology by themselves [30]. In fact, the primers used here and in other studies [31] that detect increased levels of HERV-W env (MSRV-type) RNA transcripts in MS patients compared to controls, would detect both the full-length MSRV env and the N-truncated env sequence encoded by ERVWE2, since they hybridize at the common C-terminal region. The SNPs rs6622139 (C/T) and rs6622140 (G/A) map at the residues 10 and 272, respectively, of the N-truncated protein (position 38 and 299 from the stop codon) and either the env protein is full-length or truncated, if ERVWE2 is involved in its origin, these polymorphisms would induce variations in its amino-acid sequence: rs6622139 (T/C) changes glycine by serine and rs6622140 (G/A) changes threonine by isoleucine. In both cases the substitution of an amino-acid with a hydroxyl (polar) group by an aliphatic one would be observed. This could affect the tridimensional configuration of the putative protein making epitopes more or less accessible to the immune cells, and therefore could alter its antigenic properties. On the other hand, changes in the nucleotide sequence of the RNA might affect its stability and thereby influence RNA levels.
The mechanism of generation of MSRV is unknown, but interestingly, complex interactions between herpesviruses, which have been historically studied for their involvement in MS pathogenesis, and HERVs have been brought to light. In vitro, Epstein-Barr virus (EBV) activates HERV-W/MSRV/syncytin-1 in cells derived from blood and brain [33] and antigens from other herpesviruses as HSV-1, HHV-6, and VZV are able to induce higher RT activity in peripheral lymphocytes from MS patients compared to controls [34].
It is well-known that MS is more prevalent in women than in men. Indeed, the female to male ratio for MS incidence currently ranges from 2:1 to 3:1 and varies by region [35]. Notably, over the past six decades, this ratio seems to have increased [36]. Beyond that, gender seems to influence other aspects of MS as age at onset [37], “parent-of-origin” effect on susceptibility [38] and risk for relatives of MS patients [38]. Although usually women do not have a poorer prognosis, higher levels of inflammatory lesions measured by MRI have been shown in female MS patients [39] and peripheral immune responses are more robust in females [40].
The relationship of MSRV with the gender differences in MS has already been suggested [20]. The DNA copy number of HERV-W (MSRV-type) is higher in MS patients than in controls, but it is specially increased in MS women. Indeed, a possible origin of MSRV in chromosome X is supported by the fact that the MSRV proviral load is higher in females than in men, not only among MS patients but also among controls [20]. Moreover, MSSS scores were higher among female patients with an elevated MSRV DNA load [20].
To our knowledge, this is the first demonstration of the genetic association with MS risk of the HERV-W env insert in chromosome X. The association supports the etiopathogenic role of this locus in MS risk, adding on previously published evidence.
The X chromosome has been proposed to constitute the common trait of the susceptibility to autoimmune diseases, other than to explain the female preponderance of these conditions [41]. A disease-promoting effect conferred by genes in the X chromosome has been suggested in females (since some genes escape X inactivation, resulting in double dosage in females) [41]. The critical role played by products of single genes located on the X chromosome or X-linked microRNAs in autoimmunity is evidenced also by the fact that most X-linked primary immune deficiencies carry significant autoimmune manifestations [41]. In this sense, the present data underline the crucial role of loci mapping to chromosome X in MS susceptibility.
Conclusions
Our present work reinforces the hypothesis of a direct involvement of HERV-W/MSRV in MS pathogenesis, identifying a genetic marker on chromosome X that might contribute to the gender differences in MS. This sex-related difference could be exploited to develop novel therapeutic approaches for MS. Recently, the safety and pharmacokinetic profiles of GNbAC1, a humanized monoclonal antibody which is directed against MSRV-Env, have been evaluated and appeared favorable in healthy male volunteers [42]. In light of the present results, the next Phase II development program for this innovative therapeutic approach should include female patients as well, as an enhanced effect would be expected in them and would provide a genetic rationale for the gender bias observed in MS.
Methods
Patients and controls
A total of 1669 patients and 1458 controls from two different regions in Spain were included: from Madrid 893 MS patients (66.4% women) and 664 ethnically and age matched healthy controls (55.9% women) and from Andalusia 776 MS patients (69.5% women) and 794 controls (64.6% women); all of them had European ancestors. MS diagnosis was established according to McDonald’s criteria [43]. All individuals participating in this study signed informed consent and the study was approved by the Ethics Committees of the respective hospitals.
None of the control subjects reported any first or second degree relatives with any immunological disease. Clinical and demographic characteristics are enclosed in Table 1.
Sequence data source
Previously published MSRV-env sequence (GenBank: AF33150) was used to map the HERV-W env element located on the negative strand of human chromosome Xq22.3. This sequence was aligned with the BLAST tool available at ensemble (http://www.ensembl.org) and enlarged 1500 nt at both 3′ and 5′ ends.
Primers design
Using PRIMER3 Input version (0.4.0) software (http://frodo.wi.mit.edu/), a pair of primers specific for the sequences adjacent to MSRV in chromosome X as well as 18 pairs of primers to perform High Resolution Melting curve analysis (HRM) [44] in overlapping amplicons (Additional file 1: Table S1) were designed.
PCR amplification of the HERV-W region at Xq22.3
The selected amplicon of 3004 bp (106295193 – 106298196; Human GRCh37, March 2012) including HERV-W locus in chromosome Xq22.3 was amplified by PCR using the above mentioned specific pair of primers (Additional file 1: Table S1). PCR reaction was performed on 40 ng of genomic DNA (35 cycles: 94ºC for 30s, 66ºC for 30 s and 68ºC for 180 s) in a final volume of 10ul of reaction mix containing the primers (0.4 uM), dNTPs (250uM), MgCl2 (2.5 mM) and 0.8 units of enzyme (BIO-X-ACT™ Long polymerase, Bioline). The amplification of the expected 3Kb fragment was confirmed by electrophoresis in 0.8% agarose gel.
High Resolution Melting curve analysis (HRM)
The PCR product of the specific Chr. X region diluted 1/80 on sterile water was used as template to perform HRM curve analysis on a 7900 Fast Real-Time PCR System (Applied Biosystems), according to manufacturer’s recommendations. Over 1800 melting curves from 18 different amplicons were analyzed in 100 DNA samples (50 MS patients and 50 healthy controls).
Sequencing analysis
At least three DNA samples from each group with altered melting curves compared with the average curve of wild-type subjects were sequenced. When a potential mutation or single nucleotide polymorphism (SNP) was detected, samples with the same melting curve were sequenced with BigDye Cycle sequencing kit (Applied Biosystems). Before sequencing, each HMR product was diluted 1/20 on sterile water.
SNPs analysis
The newly described SNPs were analyzed by TaqMan assays by design, from Applied Biosystems in an ABI 7900HT Fast Real-Time PCR system (Applied Biosystems), following the manufacturer’s recommendations. The genotyping success rate was higher than 98%.
MSRV expression analysis
Expression of MSRV was measured by quantitative real time RT-PCR with the relative method [45] which needs normalization with a housekeeping (HKG) gene. Before selecting Glucuronidase-B (GUS-B) as HKG gene, we analyzed the stability of its expression among MS patients and controls. The comparison of GUS-B Cts of both groups did not show significant differences (ANOVA, p = 0.98). Besides, efficiencies of both assays (MSRV and GUS-B) were tested by performing standard curves with serial dilutions of cDNA after retrotranscription of total human RNA treated with DNAse (Applied Biosystems); the efficiencies were 0.92 for GUS-B assay and 0.98 for MSRV assay.
RT positive controls and PCR controls were produced with the same batch of human total RNA (Applied Biosystems) as follows: part of the RNA diluted at 10 ng/ul was retrotranscribed into cDNA and then aliquoted to be used as “PCR control”; the rest of RNA was aliquoted to be used as “RT positive control” in each round of retrotranscription of our study. The cDNA was tested several times by real time PCR for MSRV and GUS-B and the means of the Cts were calculated. These values were used as references for the MSRV and GUS-B Cts of the “PCR controls” and the “RT positive controls” of each assay, since the same quantity of RNA was used in both cases. In case of failure, these two controls allowed us to detect at which step of the RT-PCR the problem occurred, i.e. if the retrotranscription step did not work correctly but the PCR did, the Ct of the PCR control would be similar to the reference value, but the Ct of the “RT positive control” would differ from them; on the contrary, if a problem occurred in the PCR step, both controls would differ from the reference value. PCR runs were accepted when both controls did not differ from the reference value more than 2% on Ct basis. This level of variation was used also to validate the duplicates of a sample [46].
From a group of randomly selected MS patients and blood donors RNA was extracted from PBMCs (Qiamp RNA Blood Mini kit. Qiagen) obtained by centrifugation of whole blood on CPT tubes (Becton Dickinson). The quality and concentration of RNA treated with DNAse (Applied Biosystems) were assessed by spectrophotometer. Then, sample volumes were adjusted to 10 ng/ul followed by two-step RT-PCR. Briefly, RNA was retrotranscribed with the First Strand cDNA Synthesis kit (Roche Diagnostics) with Oligo dT primers. In each round of retrotranscription, a positive control consisting of human total RNA with a known MSRV level of expression (RT positive control) and a “no RT” control of each sample, consisting of the same reaction mix without retrotranscriptase, were added. Then, a quantitative real time PCR was performed on the cDNA with a set of primers and a specific probe for MSRV[31] and for the HGK gene GUS-B (Applied Biosystems). Several controls were included in the PCR runs: 1) “No RT” control of each sample to detect possible DNA contamination; 2) RT positive control; 3) PCR control. Thus, the assays for the detection of GUS-B and MSRV env were considered acceptable in each sample when: 1) RT positive control and PCR control did not differ more than 2% on Ct basis; 2) Ct of HKG was lower than Mean + 2*S.D of HKG of all samples; 3) no amplification was detected in the “no RT” control; and 4) the duplicates of each sample had less than 2% of variability on Ct basis [46].
MSRV relative expression was measured by the 2e-ΔΔCt relative method [45]: MSRV Relative Expression = 2e-ΔΔCt = 2e-(ΔCt sample – MeanΔCtblood donors) = 2e-((Ct MSRV – Ct GUSB)sample-Mean(Ct MSRV – Ct GUSB)Blood Donors). The mean of the Δ Ct of all the Blood Donors was used as the calibrator in all the expression analysis.
Statistical analysis
Statistical analyses were performed using SPSS 15.0. Linkage disequilibrium values (D’) and Hardy-Weinberg equilibrium (H-W) were tested with Haploview 4.0 software. Chi-Square test (χ2) was used to compare allele and genotype frequencies. The Mantel–Haenszel (MH) test was used to perform the meta-analysis of allele and genotype frequencies of the two cohorts combined. MSRV relative expression was not normally distributed, therefore U-Mann Whitney test was used to compare MSRV relative expression between two groups. The non-parametric Spearman test was applied in order to evaluate the correlation between two variables. Statistically significant differences were considered when p-value < 0.05.
Abbreviations
- MS:
-
Multiple sclerosis
- MSRV:
-
Multiple sclerosis associated retrovirus
- HERVs:
-
Human endogenous retroviruses
- LTR:
-
Long terminal repeat
- ORF:
-
Open reading frame
- TLR:
-
Toll-like receptor
- BD:
-
Blood donor
- RR:
-
Relapsing-remitting
- SP:
-
Secondary progressive
- MSSS:
-
Multiple sclerosis severity score
- HRM:
-
High resolution melting
- CSF:
-
Cerebrospinal fluid
- EBV:
-
Epstein-barr virus
- HHV-6:
-
Human herpesvirus-6
- HKG:
-
Housekeeping gene.
References
Lower R, Lower J, Kurth R: The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996, 93: 5177-5184. 10.1073/pnas.93.11.5177.
Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, et al: An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000, 74: 3321-3329. 10.1128/JVI.74.7.3321-3329.2000.
Wang-Johanning F, Rycaj K, Plummer JB, Li M, Yin B, Frerich K, et al: Immunotherapeutic potential of anti-human endogenous retrovirus-k envelope protein antibodies in targeting breast tumors. J Natl Cancer Inst. 2012, 104: 189-210. 10.1093/jnci/djr540.
Liang Q, Xu Z, Xu R, Wu L, Zheng S: Expression patterns of non-coding spliced transcripts from human endogenous retrovirus HERV-H elements in colon cancer. PLoS One. 2012, 7: e29950-10.1371/journal.pone.0029950.
Goering W, Ribarska T, Schulz WA: Selective changes of retroelement expression in human prostate cancer. Carcinogenesis. 2011, 32: 1484-1492. 10.1093/carcin/bgr181.
Rakoff-Nahoum S, Kuebler PJ, Heymann JJ, Sheehy E, Ortiz M, Ogg S, et al: Detection of T lymphocytes specific for human endogenous retrovirus K (HERV-K) in patients with seminoma. AIDS Res Hum Retroviruses. 2006, 22: 52-56. 10.1089/aid.2006.22.52.
Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, et al: An endogenous retrovirus derived from human melanoma cells. Cancer Res. 2003, 63: 8735-8741.
Perron H, Hamdani N, Faucard R, Lajnef M, Jamain S, ban-Huard C, et al: Molecular characteristics of Human Endogenous Retrovirus type-W in schizophrenia and bipolar disorder. Transl Psychiatry. 2012, 2: e201-10.1038/tp.2012.125.
Yolken RH, Karlsson H, Yee F, Johnston-Wilson NL, Torrey EF: Endogenous retroviruses and schizophrenia. Brain Res Brain Res Rev. 2000, 31: 193-199. 10.1016/S0165-0173(99)00037-5.
Yao Y, Schroder J, Nellaker C, Bottmer C, Bachmann S, Yolken RH, et al: Elevated levels of human endogenous retrovirus-W transcripts in blood cells from patients with first episode schizophrenia. Genes Brain Behav. 2008, 7: 103-112.
Nakagawa K, Brusic V, McColl G, Harrison LC: Direct evidence for the expression of multiple endogenous retroviruses in the synovial compartment in rheumatoid arthritis. Arthritis Rheum. 1997, 40: 627-638. 10.1002/art.1780400407.
Perron H, Germi R, Bernard C, Garcia-Montojo M, Deluen C, Farinelli L, et al: Human endogenous retrovirus type W envelope expression in blood and brain cells provides new insights into multiple sclerosis disease. Mult Scler. 2012
Christensen T, Dissing SP, Riemann H, Hansen HJ, Munch M, Haahr S, et al: Molecular characterization of HERV-H variants associated with multiple sclerosis. Acta Neurol Scand. 2000, 101: 229-238.
Antony JM, Deslauriers AM, Bhat RK, Ellestad KK, Power C: Human endogenous retroviruses and multiple sclerosis: innocent bystanders or disease determinants?. Biochim Biophys Acta. 1812, 2011: 162-176.
Garson JA, Tuke PW, Giraud P, Paranhos-Baccala G, Perron H: Detection of virion-associated MSRV-RNA in serum of patients with multiple sclerosis. Lancet. 1998, 351: 33-10.1016/S0140-6736(98)24001-3.
Serra C, Sotgiu S, Mameli G, Pugliatti M, Rosati G, Dolei A: Multiple sclerosis and multiple sclerosis-associated retrovirus in Sardinia. Neurol Sci. 2001, 22: 171-173. 10.1007/s100720170019.
Perron H, Lazarini F, Ruprecht K, Pechoux-Longin C, Seilhean D, Sazdovitch V, et al: Human endogenous retrovirus (HERV)-W ENV and GAG proteins: physiological expression in human brain and pathophysiological modulation in multiple sclerosis lesions. J Neurovirol. 2005, 11: 23-33.
Mameli G, Astone V, Arru G, Marconi S, Lovato L, Serra C, et al: Brains and peripheral blood mononuclear cells of multiple sclerosis (MS) patients hyperexpress MS-associated retrovirus/HERV-W endogenous retrovirus, but not Human herpesvirus 6. J Gen Virol. 2007, 88: 264-274. 10.1099/vir.0.81890-0.
Antony JM, Izad M, Bar-Or A, Warren KG, Vodjgani M, Mallet F, et al: Quantitative analysis of human endogenous retrovirus-W env in neuroinflammatory diseases. AIDS Res Hum Retroviruses. 2006, 22: 1253-1259. 10.1089/aid.2006.22.1253.
Garcia-Montojo M, Dominguez-Mozo M, Arias-Leal A, Garcia-Martinez A, de Las Heras V, Casanova I, et al: The DNA copy number of human endogenous retrovirus-W (MSRV-type) is increased in multiple sclerosis patients and is influenced by gender and disease severity. PLoS One. 2013, 8 (1): Ref Type: Generic
Zawada M, Liwien I, Pernak M, Januszkiewicz-Lewandowska D, Nowicka-Kujawska K, Rembowska J, et al: MSRV pol sequence copy number as a potential marker of multiple sclerosis. Pol J Pharmacol. 2003, 55: 869-875.
Mameli G, Serra C, Astone V, Castellazzi M, Poddighe L, Fainardi E, et al: Inhibition of multiple-sclerosis-associated retrovirus as biomarker of interferon therapy. J Neurovirol. 2008, 14: 73-77. 10.1080/13550280701801107.
Saresella M, Rolland A, Marventano I, Cavarretta R, Caputo D, Marche P, et al: Multiple sclerosis-associated retroviral agent (MSRV)-stimulated cytokine production in patients with relapsing-remitting multiple sclerosis. Mult Scler. 2009, 15: 443-447. 10.1177/1352458508100840.
Rolland A, Jouvin-Marche E, Viret C, Faure M, Perron H, Marche PN: The envelope protein of a human endogenous retrovirus-W family activates innate immunity through CD14/TLR4 and promotes Th1-like responses. J Immunol. 2006, 176: 7636-7644.
Perron H, Jouvin-Marche E, Michel M, Ounanian-Paraz A, Camelo S, Dumon A, et al: Multiple sclerosis retrovirus particles and recombinant envelope trigger an abnormal immune response in vitro, by inducing polyclonal Vbeta16 T-lymphocyte activation. Virology. 2001, 287: 321-332. 10.1006/viro.2001.1045.
Firouzi R, Rolland A, Michel M, Jouvin-Marche E, Hauw JJ, Malcus-Vocanson C, et al: Multiple sclerosis-associated retrovirus particles cause T lymphocyte-dependent death with brain hemorrhage in humanized SCID mice model. J Neurovirol. 2003, 9: 79-93.
Laufer G, Mayer J, Mueller BF, Mueller-Lantzsch N, Ruprecht K: Analysis of transcribed human endogenous retrovirus W env loci clarifies the origin of multiple sclerosis-associated retrovirus env sequences. Retrovirology. 2009, 6: 37-10.1186/1742-4690-6-37.
Pavlicek A, Paces J, Elleder D, Hejnar J: Processed pseudogenes of human endogenous retroviruses generated by LINEs: their integration, stability, and distribution. Genome Res. 2002, 12: 391-399.
Voisset C, Bouton O, Bedin F, Duret L, Mandrand B, Mallet F, et al: Chromosomal distribution and coding capacity of the human endogenous retrovirus HERV-W family. AIDS Res Hum Retroviruses. 2000, 16: 731-740. 10.1089/088922200308738.
Roebke C, Wahl S, Laufer G, Stadelmann C, Sauter M, Mueller-Lantzsch N, et al: An N-terminally truncated envelope protein encoded by a human endogenous retrovirus W locus on chromosome Xq22.3. Retrovirology. 2010, 7: 69-10.1186/1742-4690-7-69.
Mameli G, Poddighe L, Astone V, Delogu G, Arru G, Sotgiu S, et al: Novel reliable real-time PCR for differential detection of MSRVenv and syncytin-1 in RNA and DNA from patients with multiple sclerosis. J Virol Methods. 2009, 161: 98-106. 10.1016/j.jviromet.2009.05.024.
Roxburgh RH, Seaman SR, Masterman T, Hensiek AE, Sawcer SJ, Vukusic S, et al: Multiple sclerosis severity score: using disability and disease duration to rate disease severity. Neurology. 2005, 64: 1144-1151. 10.1212/01.WNL.0000156155.19270.F8.
Mameli G, Poddighe L, Mei A, Uleri E, Sotgiu S, Serra C, et al: Expression and activation by Epstein Barr virus of human endogenous retroviruses-W in blood cells and astrocytes: inference for multiple sclerosis. PLoS One. 2012, 7: e44991-10.1371/journal.pone.0044991.
Brudek T, Luhdorf P, Christensen T, Hansen HJ, Moller-Larsen A: Activation of endogenous retrovirus reverse transcriptase in multiple sclerosis patient lymphocytes by inactivated HSV-1, HHV-6 and VZV. J Neuroimmunol. 2007, 187: 147-155. 10.1016/j.jneuroim.2007.04.003.
Koch-Henriksen N, Sorensen PS: The changing demographic pattern of multiple sclerosis epidemiology. Lancet Neurol. 2010, 9: 520-532. 10.1016/S1474-4422(10)70064-8.
Duquette P, Pleines J, Girard M, Charest L, Senecal-Quevillon M, Masse C: The increased susceptibility of women to multiple sclerosis. Can J Neurol Sci. 1992, 19: 466-471.
Ramagopalan SV, Yee IM, Dyment DA, Orton SM, Marrie RA, Sadovnick AD, et al: Parent-of-origin effect in multiple sclerosis: observations from interracial matings. Neurology. 2009, 73: 602-605. 10.1212/WNL.0b013e3181af33cf.
Ebers GC, Sadovnick AD, Dyment DA, Yee IM, Willer CJ, Risch N: Parent-of-origin effect in multiple sclerosis: observations in half-siblings. Lancet. 2004, 363: 1773-1774. 10.1016/S0140-6736(04)16304-6.
Pozzilli C, Tomassini V, Marinelli F, Paolillo A, Gasperini C, Bastianello S: ‘Gender gap’ in multiple sclerosis: magnetic resonance imaging evidence. Eur J Neurol. 2003, 10: 95-97. 10.1046/j.1468-1331.2003.00519.x.
Voskuhl RR, Gold SM: Sex-related factors in multiple sclerosis susceptibility and progression. Nat Rev Neurol. 2012, 8: 255-263. 10.1038/nrneurol.2012.43.
Bianchi I, Lleo A, Gershwin ME, Invernizzi P: The X chromosome and immune associated genes. J Autoimmun. 2012, 38: J187-J192. 10.1016/j.jaut.2011.11.012.
Curtin F, Lang AB, Perron H, Laumonier M, Vidal V, Porchet HC, Hartung HP: GNbAC1, a humanized monoclonal antibody against the envelope protein of multiple sclerosis associated endogenous retrovirus: a first in-humans randomized clinical study. Clin Ther. 2012, 34: 2268-2278. 10.1016/j.clinthera.2012.11.006.
McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, et al: Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001, 50: 121-127. 10.1002/ana.1032.
Rozen S, Skaletsky HJ: Primer3 on the WWW for general users and for biologist programmers. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Edited by: Krawetz S, Misener S. 2000, Totowa, NJ: Humana Press, 365-386.
Schmittgen TD, Livak KJ: Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008, 3: 1101-1108. 10.1038/nprot.2008.73.
Pfaffl MW: Quantification strategies in real-time PCR. A-Z of quantitative PCR. Edited by: Bustin SA. 2004, La Jolla, CA, USA: International University Line (IUL), 97-3
Acknowledgements
The authors thank M. C. Ramírez and M.J. Díez, who collected the specimens, and C. Martínez, for her skilful technical assistance.
Financial disclosure
M. García-Montojo, M. Dominguez-Mozo and A. Arias-Leal are recipient of contracts of “Fundación para la Investigación Biomédica-Hospital Clínico San Carlos”. R. Alvarez-Lafuente is recipient of a research contract of the Instituto de Salud Carlos III-Fondo Europeo de Desarrollo Regional (Feder) (CP07/00273). M.A. Garcia-Martinez is recipient of a technician contract from “REEM: Red Española de Esclerosis Múltiple” (RETICS-REEM RD07/0060; http://www.reem.es).
B. de la Hera is recipient of a PhD scholarship from ‘Fondo de Investigaciones Sanitarias’ (FI11/00560), J. Varade is recipient of a contract from “Ministerio de Economía y Competitividad“(PTA2011-6137-1) and E. Urcelay works for the “Fundación para la Investigación Biomédica-Hospital Clínico San Carlos”.
This work was supported by grants from: Instituto de Salud Carlos III-Fondo Investigaciones Sanitarias FIS (10/01985 and 09/02074), Fundación Genzyme, Fundación Alicia Koplowitz, Fundación Mutua Madrileña, and Fundación LAIR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MGM participated in the design of the study, carried out the expression analysis, performed statistical analysis and drafted the manuscript. BH carried out the HRM analysis, genotyping, sequencing and performed statistical analysis. JV participated in the design of the study, designed the primers and performed statistical analysis. AE and IC performed part of the molecular genetics studies. MDM, AAL and AGM prepared the samples for genetic and expression studies. IC, GI, ML and RA collected clinical data and samples from the patients. MF, AA FM carried out genotyping of samples. RA, FM, EU and RAL have been involved in the conception and design of the study and supervised the work. All the authors have revised critically the manuscript and approved the final version.
Marta García-Montojo, Belén de la Hera, Jezabel Varadé, Elena Urcelay and Roberto Alvarez-Lafuente contributed equally to this work.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
García-Montojo, M., de la Hera, B., Varadé, J. et al. HERV-W polymorphism in chromosome X is associated with multiple sclerosis risk and with differential expression of MSRV. Retrovirology 11, 2 (2014). https://doi.org/10.1186/1742-4690-11-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1742-4690-11-2