Abstract
In the current era of antiviral drug therapy, combining multiple drugs is a primary approach for improving antiviral effects, reducing the doses of individual drugs, relieving the side effects of strong antiviral drugs, and preventing the emergence of drug-resistant viruses. Although a variety of new drugs have been developed for HIV, HCV and influenza virus, the optimal combinations of multiple drugs are incompletely understood. To optimize the benefits of multi-drugs combinations, we must investigate the interactions between the combined drugs and their target viruses. Mathematical models of viral infection dynamics provide an ideal tool for this purpose. Additionally, whether drug combinations computed by these models are synergistic can be assessed by two prominent drug combination theories, Loewe additivity and Bliss independence. By combining the mathematical modeling of virus dynamics with drug combination theories, we could show the principles by which drug combinations yield a synergistic effect. Here, we describe the theoretical aspects of multi-drugs therapy and discuss their application to antiviral research.
Similar content being viewed by others
Background
Several landmark mathematical modeling studies of anti-retroviral therapy were reported in 1995[1–4]. Thereafter, mathematical modeling has provided quantitative insights into antiviral drugs targeting HIV[5–13], HCV[14–18], HBV[19, 20] and influenza virus[21, 22]. These studies have elucidated the key steps of viral replication cycles and viral infection dynamics, such as the half-life of viruses and how viruses replicate in cells (reviewed in[23–27]). Although mathematical viral models could also search for drug combinations that improve antiviral effects and prevent the emergence of drug-resistant viruses, this potential of viral modeling has yet to be properly explored. Multi-drug administration is a standard treatment for HIV and HCV infection, and is also required in HBV and influenza. The established combination drug therapy for HIV, known as highly active anti-retroviral therapy (HAART), remains prohibitively expensive in developing countries. Additionally, HIV patients receiving long-term HAART may experience severe side effects such as lipodystrophy, hepatotoxicity, renal dysfunction, peripheral neuropathy and cardiovascular diseases (reviewed in[28]). By optimizing drug combinations, we could reduce the doses of individual drugs, thereby lowering the cost of treatment, reducing side effects, enhancing the antiviral effects and reducing the risk of drug-resistant viruses.
To accelerate the above benefits of drug combinations, we need to know whether or not a drug combination exerts a synergistic effect (reviewed in[29, 30]). A non-synergistic drug combination requires a larger than expected dose to achieve a regular antiviral effect. Although multi-drug therapy is standard practice, the complexity of the combined drugs’ responses is improperly understood, and rendered counter-intuitive by drug absorption, distribution, metabolism, and excretion through drug transporters[28]. To overcome these difficulties, comprehensive trials (or experiments) of various combinations and doses of drugs have been required. The combined effect has been investigated by empirical methods such as Loewe additivity[29, 31] and Bliss independence[29, 32]. Both theories have evolved from pharmacological research, and are used to classify combination effects as synergistic, additive or antagonistic[29, 33, 34]. These empirical frameworks have become widely accepted in pharmacology and have provided quantitative insight into drug combinations. However, because their underlying mechanics are unknown, the frameworks cannot provide reasons for a given combination effect (e.g. why is this drug combination additive, whereas that combination is antagonistic?). Showing the mechanisms of drug combination effects is expected to remarkably advance the development of multicomponent therapeutics. Thus, in addition to judging drug combination effects, knowledge of the mechanism is crucial to fundamental antiviral research.
Here we discuss two primary concepts of drug combination theory: Loewe additivity and Bliss independence. We apply these models to viral replication in a host cell, and discuss their utility for understanding two-drug interactions and viral dynamics. Finally, we discuss how drug combination theory might be applied to cancer chemotherapies and antiviral therapies. Combining mathematical models of viral replication with drug combination theory, we can establish an efficient framework for optimizing drug combinations.
Basic drug combination theory: Loewe additivity
Loewe additivity is regarded as a primary criterion for evaluating drug combination effects[29]. To conceptualize Loewe additivity, let us consider the simplest situation, in which a combination of drugs A and B has a synergistic, additive or antagonistic effect. If the effect is additive, the Loewe additivity is defined as[31]:
where and are the concentrations of drugs A and B respectively in the combined dose, and C A and C B are the respective concentrations of drugs A and B that produce the same effect as the drug combination. That is, the Loewe additivity specifies the concentration ratio of a single drug and its combination with another drug. Note that Loewe additivity assumes that two drugs target the same molecule or pathway. If two drugs do not mutually interact, they can be related through the combination index (CI) based on the mass action law derived by Chou and Talalay[35]:
where the right hand side of Eq. (2) is identical to the left side of Eq. (1). When CI < 1, the relationship is synergetic, when CI = 1, it is additive, and when CI > 1, it is antagonistic. For example, suppose that 4 μM of drug A and 5 μM of drug B exert the same effect as a combination of 1 μM of A and 2 μM of B. Substituting these concentrations into Eq. (2), we obtain CI = 0.65 (=1/4 + 2/5 < 1), implying that the drug combination is synergistic. Note that the CI does not express the extent of synergy or antagonism; it is merely a criterion that separates the two behaviors without quantifying them.
To evaluate a drug combination effect by the CI, we require dose–response curves of drugs A and B, from which we can determine and. The effect of a single drug E is modeled by the Hill function as follows:
where E max is the maximum effect, c is the drug concentration, h is the Hill coefficient that determines the steepness of the dose–response curve, and IC50 is the concentration at which E exerts 50% of its maximum effect. Substituting Eq. (3) into Eq. (1) and rearranging in the case of E max = 1, we obtain:
Numerically solving Eq. (4) for E, we can predict the additive effect at any drug concentrations and[35, 36]. The following section explains an example of applying Loewe additivity to estimate rational drug combinations and optimal doses.
Application of Loewe additivity for estimating rational drug combinations and optimal doses
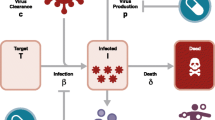
We here explain the usefulness of combining the Loewe additivity with mathematical modeling of hepatitis C virus (HCV) replication. The aim is to optimize the antiviral drug combination. Combining multiple drugs such as interferon-α and ribavirin is a standard therapy for enhancing the antiviral effects and reducing the risk of drug-resistant viruses. However, the conventional anti-HCV drugs fail to eradicate HCV genotype 1 in about 40% patients and may cause severe side effects. To overcome these problems, researchers have developed and administered direct acting antiviral (DAA) drugs that target viral-specific proteins such as viral proteases, as adjuncts to conventional anti-HCV drugs. To understand the principle behind optimal drug combinations and doses, we focused on the essential features of HCV replication and the mechanisms of the antiviral drugs (Figure 1A). Using previous mathematical models of HCV replication in a host cell[37–40], a modified mathematical model accounting for the mechanisms of drug actions would quantitatively simulate the anti-HCV effects at any dose of a drug combination. The effects of the drug combination at specific doses could then be evaluated by the Loewe additivity. In this way, we could estimate the synergistic dose point of a drug combination that enhances the anti-HCV effects (Figure 1B). The above analysis could predict the optimal drug combination that both reduces the total dose of each drug and enhances their anti-HCV effects.
Estimating the optimal drug combination from the mechanism of drug effects on viral replication. A) Conceptual diagram showing the effects of two anti-HCV drug effects on HCV replication. Drug A inhibits virus release, and drug B inhibits the production of viral polymerase. To calculate the effect of co-administered A and B on HCV production, the dynamics of each viral component during the replication are expressed by differential equations[37–40]. The presence of both drugs decreases the parameters, a production rate of viral proteins and a releasing rate of virions. B) Estimation of the optimal dose point in a drug combination. Each point (corresponding to a specific dose) denotes the difference between the effects of the combined drugs obtained by modeling the viral dynamics and the additive effects calculated by Loewe additivity. A positive (red) difference indicates that the drug combination surpasses the expected additive effect. At such synergistic dose points, the antiviral effect can be achieved by administering the drug in combination at doses below the single drug dose (the synergistic point).
The alternative drug combination theory: Bliss independence
Bliss independence is the other major criterion for clarifying drug combination effects[32]. Bliss independence defines the expectation of a combined drug effect, calculated by multiplying the probabilities of the individual drugs. It assumes that each drug independently targets a different stage of viral replication via a different mechanism, with no interaction between the drug actions. Bliss independence expresses the unaffected probability of a two-drug combination U AB as the product of the unaffected probabilities U A and U B of drugs A and B:
Intuitively, Eq. (5) implies that targets bypassed by drug A will be intercepted by drug B. The total probability of missed targets is obtained by multiplying the probabilities of the unaffected targets. Substituting U by 1 - P, where P denotes the probability of an intercepted target, Eq. (5) can be expressed as (1 - P AB ) = (1 - P A ) × (1 - P B ), or
If the combined effect of the single drug doses is consistent with Eq. (6), this drug combination is evaluated as additive according to the Bliss independence measure. For instance, suppose that drugs A and B exhibit 50% and 60% inhibition, respectively. The Bliss independence predicts that drugs A and B will exert a combined inhibitory effect of 80%, calculated as P AB = 0.5 + 0.6 - 0.5 × 0.6 = 0.8.
Unlike the Loewe additivity, the Bliss independence can evaluate the effect of a drug combination without requiring dose–response curves of the single drugs. If we attempt to assess a drug combination by the Loewe additivity, we must measure the effects at several single doses for constructing the dose–response curve. In contrast, the Bliss independence can assess the effects of a drug combination from a single point. Thus, the Bliss independence is useful if experimental data are limited. Additionally, in specific cases, the Bliss independence and Loewe additivity can be rendered equivalent using the Hill function Eq. (3)[41]. The following section explains an example of applying Bliss independence to quantifying the antiviral effects of intrinsic factors on HIV replication.
Quantifying the antiviral effects of APOBEC3G on HIV replication by Bliss independence
The Bliss independence measure can predict antiviral effects as well as evaluate drug combinations. To illustrate this idea, we estimate the anti-HIV effect of a well-known antiviral host factor by Bliss independence[42]. Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3G (APOBEC3G) suppresses HIV replication in cells by two main mechanisms: (1) inhibiting viral reverse transcriptase and (2) mutating cytidine (C) to uracil (U) in viral DNA by cytidine deaminase activity (Figure 2A). Although the anti-HIV effect of reverse transcriptase inhibition can be measured using a mutated APOBEC3G (with its C-to-U activity removed) (Figure 2A), the extent to which the mutation itself inhibits HIV replication cannot be experimentally determined. To quantify the anti-HIV effect of C-to-U mutation by APOBEC 3G, we can use Bliss independence to model the unaffected fractions of viral infectivity of wild-type (WT) and mutant APOBEC3G, denoted f WT and f CI respectively, in terms of the unaffected fractions of viral infectivity by reverse transcription inhibition and C-to-U mutation, f RT and f Mu , as follows:
Estimating the antiviral effects of C-to-U mutation by APOBEC3G on HIV production. A) Mechanism of APOBEC3G inhibition in HIV replication. B) Quantification of antiviral effects of mutant APOBEC3G on HIV infection. HIV production reduces with increasing expression of APOBEC3G (dots: experimental data, solid lines: theoretical predictions, purple: WT APOBEC3G, blue: mutated APOBEC3G). Red dotted line shows the expected anti-HIV effect of C-to-U mutation activity, determined by Bliss independence.
and
where x is the expression level of APOBEC3G, h RT and h Mu are Hill coefficients, and and are the expression levels required to achieve 50% inhibition by reverse transcription inhibition and C-to-U mutation, respectively. We estimated 4 parameters (h RT , h Mu ,, and) by fitting to experimental data of WT and mutated APOBEC3G (Figure 2B). Thus, the anti-HIV effect of C-to-U mutation by APOBEC3G can be quantitatively estimated by a Bliss independence-based modeling approach.
Potential applications of drug combination theory
The Loewe additivity and Bliss independence form the basis of sophisticated protocols for assessing drug combinations, especially in cancer-targeted chemotherapy[33, 34]. Research on biological network systems, as well as the biochemical characteristics of drugs, has also unraveled the mechanisms by which drug combinations produce synergistic effects (reviewed in[41, 43–45]). Such research is important because (1) it facilitates rational drug discovery and (2) it predicts unknown connectivities in biological networks. Regarding the first point, the effects of a developing drug combined with conventional drugs are maximized by investigating biological networks, and rational drug targets are decided. For example, using computational modeling of cancer signaling networks and animal experiments, Kirouac et al.[46] identified the optimal inhibitor combination for suppressing the growth of ERBB2-amplified breast cancer. They adopted Bliss independence as a synergy criterion. Supplementing this computational approach with experimental data is useful for developing theoretically effective novel drugs. Regarding point (2), the biological networks of signaling pathways constructed by various components can be predicted from the dose–response shapes of drug combinations (reviewed in[47]). Lehar et al.[48] used several drug combination criteria, including Loewe additivity and Bliss independence, to connect biological components from the results of cellular responses to chemical combinations. This approach assumes that the dose–response shape of a drug combination depends on how drug targets are connected in a biological system.
The above research potentially benefits not only cancer chemotherapy, but also antiviral therapy development. For instance, Owens et al.[49] showed how HCV replication responds to chemical combinations that inhibit some enzymes involved in the sterol biosynthesis pathway. Supplementing computational simulation with in vitro experiments, they identified a rational inhibitor combination as a novel drug candidate. Mathematical models of HCV dynamics during drug therapy have also been investigated at the intracellular level[18], the intercellular level[14–17, 50, 51], and on a scale encompassing both levels[51–54]. These studies have quantitatively elucidated the properties of anti-HCV drugs such as interferon and protease inhibitors. Furthermore, by inputting clinical data to a mathematical model of HCV dynamics, Rong et al.[55] derived a mechanism by which drug-resistant viruses can rapidly emerge during single-drug therapy. Applying drug combination theories to these studies, mathematical models of HCV dynamics could be used to optimize the anti-HCV drug dosage and combination, thereby enhancing the anti-viral effect and reducing the risk of drug-resistant viruses (Figure 3).
Concept of integrating drug combination theory and viral dynamics. The effect of a drug combination calculated by mathematical modeling of viral dynamics could be assessed by the Loewe additivity and Bliss independence. This approach could improve antiviral therapy by predicting the effect of a drug combination in vivo and in vitro, estimating the optimal drug dosage and combination, and discovering how the drugs interact.
Conclusion
We have reviewed two major drug combination theories, Loewe additivity and Bliss independence, and discussed how combining these theories with mathematical modeling of viral dynamics might assist antiviral drug therapy. In addition, we have proposed an efficient framework for optimizing drug combinations and quantifying the anti-viral effect. Based on previous studies of computational virology, the integration of drug combination theory and dynamic modeling is a new approach with great potential for showing viral responses to drug combinations, and accelerating novel antiviral drug discovery.
References
Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M: Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995, 373: 123-126. 10.1038/373123a0.
Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH: Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995, 373: 117-122. 10.1038/373117a0.
Nowak MA, Bonhoeffer S, Loveday C, Balfe P, Semple M, Kaye S, Tenant-Flowers M, Tedder R: HIV results in the frame. Results confirmed. Nature. 1995, 375: 193-
Coffin JM: HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995, 267: 483-489. 10.1126/science.7824947.
Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD: HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996, 271: 1582-1586. 10.1126/science.271.5255.1582.
Wein LM, Zenios SA, Nowak MA: Dynamic multidrug therapies for HIV: a control theoretic approach. J Theor Biol. 1997, 185: 15-29. 10.1006/jtbi.1996.0253.
Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD: Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997, 387: 188-191. 10.1038/387188a0.
Notermans DW, Goudsmit J, Danner SA, de Wolf F, Perelson AS, Mittler J: Rate of HIV-1 decline following antiretroviral therapy is related to viral load at baseline and drug regimen. AIDS. 1998, 12: 1483-1490. 10.1097/00002030-199812000-00010.
Wein LM, D’Amato RM, Perelson AS: Mathematical analysis of antiretroviral therapy aimed at HIV-1 eradication or maintenance of low viral loads. J Theor Biol. 1998, 192: 81-98. 10.1006/jtbi.1997.0622.
Di Mascio M, Markowitz M, Louie M, Hogan C, Hurley A, Chung C, Ho DD, Perelson AS: Viral blip dynamics during highly active antiretroviral therapy. J Virol. 2003, 77: 12165-12172. 10.1128/JVI.77.22.12165-12172.2003.
Sedaghat AR, Dinoso JB, Shen L, Wilke CO, Siliciano RF: Decay dynamics of HIV-1 depend on the inhibited stages of the viral life cycle. Proc Natl Acad Sci U S A. 2008, 105: 4832-4837. 10.1073/pnas.0711372105.
Shen L, Rabi SA, Sedaghat AR, Shan L, Lai J, Xing S, Siliciano RF: A critical subset model provides a conceptual basis for the high antiviral activity of major HIV drugs. Sci Transl Med. 2011, 3: 91ra63-
Rosenbloom DIS, Hill AL, Rabi SA, Siliciano RF, Nowak MA: Antiretroviral dynamics determines HIV evolution and predicts therapy outcome. Nat Med. 2012, 18: 1378-1385. 10.1038/nm.2892.
Zeuzem S, Schmidt JM, Lee JH, von Wagner M, Teuber G, Roth WK: Hepatitis C virus dynamics in vivo: effect of ribavirin and interferon alfa on viral turnover. Hepatology. 1998, 28: 245-252. 10.1002/hep.510280132.
Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS: Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998, 282: 103-107.
Dixit NM, Layden-Almer JE, Layden TJ, Perelson AS: Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature. 2004, 432: 922-924. 10.1038/nature03153.
Dahari H, Ribeiro RM, Perelson AS: Triphasic decline of hepatitis C virus RNA during antiviral therapy. Hepatology. 2007, 46: 16-21. 10.1002/hep.21657.
Dahari H, Sainz B, Perelson AS, Uprichard SL: Modeling subgenomic hepatitis C virus RNA kinetics during treatment with alpha interferon. J Virol. 2009, 83: 6383-6390. 10.1128/JVI.02612-08.
Lewin SR, Ribeiro RM, Walters T, Lau GK, Bowden S, Locarnini S, Perelson AS: Analysis of hepatitis B viral load decline under potent therapy: complex decay profiles observed. Hepatology. 2001, 34: 1012-1020. 10.1053/jhep.2001.28509.
Dahari H, Shudo E, Ribeiro RM, Perelson AS: Modeling complex decay profiles of hepatitis B virus during antiviral therapy. Hepatology. 2009, 49: 32-38. 10.1002/hep.22586.
Beauchemin CAA, McSharry JJ, Drusano GL, Nguyen JT, Went GT, Ribeiro RM, Perelson AS: Modeling amantadine treatment of influenza A virus in vitro. J Theor Biol. 2008, 254: 439-451. 10.1016/j.jtbi.2008.05.031.
Dobrovolny HM, Gieschke R, Davies BE, Jumbe NL, Beauchemin CAA: Neuraminidase inhibitors for treatment of human and avian strain influenza: A comparative modeling study. J Theor Biol. 2011, 269: 234-244. 10.1016/j.jtbi.2010.10.017.
Nowak M, May RM: Virus Dynamics: Mathematical Principles of Immunology and Virology. 2001, USA: Oxford University Press, 256-
Perelson AS: Modelling viral and immune system dynamics. Nat Rev Immunol. 2002, 2: 28-36. 10.1038/nri700.
Rong L, Perelson AS: Modeling HIV persistence, the latent reservoir, and viral blips. J Theor Biol. 2009, 260: 308-331. 10.1016/j.jtbi.2009.06.011.
Guedj J, Rong L, Dahari H, Perelson AS: A perspective on modelling hepatitis C virus infection. J Viral Hepat. 2010, 17: 825-833. 10.1111/j.1365-2893.2010.01348.x.
Chatterjee A, Guedj J, Perelson AS: Mathematical modelling of HCV infection: what can it teach us in the era of direct-acting antiviral agents?. Antivir Ther. 2012, 17: 1171-1182. 10.3851/IMP2428.
Hawkins T: Understanding and managing the adverse effects of antiretroviral therapy. Antiviral Res. 2010, 85: 201-209. 10.1016/j.antiviral.2009.10.016.
Greco WR, Bravo G, Parsons JC: The search for synergy: a critical review from a response surface perspective. Pharmacol Rev. 1995, 47: 331-385.
Tallarida RJ: Drug synergism: its detection and applications. J Pharmacol Exp Ther. 2001, 298: 865-872.
Loewe S: The problem of synergism and antagonism of combined drugs. Arzneimittelforschung. 1953, 3: 285-290.
Bliss CI: The toxicity of poisons applied jointly. Ann Appl Biol. 1939, 26: 585-615. 10.1111/j.1744-7348.1939.tb06990.x.
Chou TC: Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006, 58: 621-681. 10.1124/pr.58.3.10.
Chou TC: Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70: 440-446. 10.1158/0008-5472.CAN-09-1947.
Chou TC, Talalay P: Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984, 22: 27-55.
Martinez-Irujo JJ, Villahermosa ML, Alberdi E, Santiago E: A checkerboard method to evaluate interactions between drugs. Biochem Pharmacol. 1996, 51: 635-644. 10.1016/S0006-2952(95)02230-9.
Dahari H, Ribeiro RM, Rice CM, Perelson AS: Mathematical modeling of subgenomic hepatitis C virus replication in Huh-7 cells. J Virol. 2007, 81: 750-760. 10.1128/JVI.01304-06.
McLean AK, Luciani F, Tanaka MM: Trade-offs in resource allocation in the intracellular life-cycle of hepatitis C virus. J Theor Biol. 2010, 267: 565-572. 10.1016/j.jtbi.2010.09.031.
Nakabayashi J: A compartmentalization model of hepatitis C virus replication: an appropriate distribution of HCV RNA for the effective replication. J Theor Biol. 2012, 300: 110-117.
Binder M, Sulaimanov N, Clausznitzer D, Schulze M, Hüber CM, Lenz SM, Schlöder JP, Trippler M, Bartenschlager R, Lohmann V, Kaderali L: Replication vesicles are load- and choke-points in the hepatitis C virus lifecycle. PLoS Pathog. 2013, 9: e1003561-10.1371/journal.ppat.1003561.
Jonker DM, Visser SA, van der Graaf PH, Voskuyl RA, Danhof M: Towards a mechanism-based analysis of pharmacodynamic drug-drug interactions in vivo. Pharmacol Ther. 2005, 106: 1-10.1016/j.pharmthera.2004.10.014.
Kobayashi T, Koizumi Y, Takeuchi JS, Misawa N, Kimura Y, Morita S, Aihara K, Koyanagi Y, Iwami S, Sato K: Quantification of Deaminase Activity-Dependent and -Independent Restriction of HIV-1 Replication Mediated by APOBEC3F and APOBEC3G through Experimental-Mathematical Investigation. J Virol. 2014, 88: 5881-5887. 10.1128/JVI.00062-14.
Keith CT, Borisy AA, Stockwell BR: Multicomponent therapeutics for networked systems. Nat Rev Drug Discov. 2005, 4: 71-78. 10.1038/nrd1609.
Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK: Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol. 2006, 2: 458-466. 10.1038/nchembio817.
Zimmermann GR, Lehár J, Keith CT: Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov Today. 2007, 12: 34-42. 10.1016/j.drudis.2006.11.008.
Kirouac DC, Du JY, Lahdenranta J, Overland R, Yarar D, Paragas V, Pace E, McDonagh CF, Nielsen UB, Onsum MD: Computational modeling of ERBB2-amplified breast cancer identifies combined ErbB2/3 blockade as superior to the combination of MEK and AKT inhibitors. Sci Signal. 2013, 6: ra68-
Lehár J, Stockwell BR, Giaever G, Nislow C: Combination chemical genetics. Nat Chem Biol. 2008, 4: 674-681. 10.1038/nchembio.120.
Lehár J, Zimmermann GR, Krueger AS, Molnar RA, Ledell JT, Heilbut AM, Short GF, Giusti LC, Nolan GP, Magid OA, Lee MS, Borisy AA, Stockwell BR, Keith CT: Chemical combination effects predict connectivity in biological systems. Mol Syst Biol. 2007, 3: 80-
Owens CM, Mawhinney C, Grenier JM, Altmeyer R, Lee MS, Borisy AA, Lehár J, Johansen LM: Chemical combinations elucidate pathway interactions and regulation relevant to Hepatitis C replication. Mol Syst Biol. 2010, 6: 375-
Guedj J, Perelson AS: Second-phase hepatitis C virus RNA decline during telaprevir-based therapy increases with drug effectiveness: implications for treatment duration. Hepatology. 2011, 53: 1801-1808. 10.1002/hep.24272.
Guedj J, Neumann AU: Understanding hepatitis C viral dynamics with direct-acting antiviral agents due to the interplay between intracellular replication and cellular infection dynamics. J Theor Biol. 2010, 267: 330-340. 10.1016/j.jtbi.2010.08.036.
Guedj J, Dahari H, Rong L, Sansone ND, Nettles RE, Cotler SJ, Layden TJ, Uprichard SL, Perelson AS: Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc Natl Acad Sci U S A. 2013, 110: 3991-3996. 10.1073/pnas.1203110110.
Rong L, Guedj J, Dahari H, Coffield DJ, Levi M, Smith P, Perelson AS: Analysis of hepatitis C virus decline during treatment with the protease inhibitor danoprevir using a multiscale model. PLoS Comput Biol. 2013, 9: e1002959-10.1371/journal.pcbi.1002959.
Rong L, Perelson AS: Mathematical analysis of multiscale models for hepatitis C virus dynamics under therapy with direct-acting antiviral agents. Math Biosci. 2013, 245: 22-30. 10.1016/j.mbs.2013.04.012.
Rong L, Dahari H, Ribeiro RM, Perelson AS: Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci Transl Med. 2010, 2: 30ra32-
Acknowledgments
This research is partly supported by a Grants-in-Aid for Young Scientists B25800092 (to S.I.) from the Japan Society for the Promotion of Science (JSPS); the Kyushu University Interdisciplinary Programs in Education and Projects in Research Development (to S.I.); the Aihara Innovative Mathematical Modeling Project, JSPS through the “Funding Program for World-Leading Innovative R & D on Science and Technology (FIRST program)”, initiated by Council for Science and Technology Policy (to S.I.).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the study: YK SI. Wrote the paper: YK SI. Both authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Koizumi, Y., Iwami, S. Mathematical modeling of multi-drugs therapy: a challenge for determining the optimal combinations of antiviral drugs. Theor Biol Med Model 11, 41 (2014). https://doi.org/10.1186/1742-4682-11-41
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1742-4682-11-41