Abstract
Background
The bacterial family Enterobacteriaceae gave rise to a variety of symbiotic forms, from the loosely associated commensals, often designated as secondary (S) symbionts, to obligate mutualists, called primary (P) symbionts. Determination of the evolutionary processes behind this phenomenon has long been hampered by the unreliability of phylogenetic reconstructions within this group of bacteria. The main reasons have been the absence of sufficient data, the highly derived nature of the symbiont genomes and lack of appropriate phylogenetic methods. Due to the extremely aberrant nature of their DNA, the symbiotic lineages within Enterobacteriaceae form long branches and tend to cluster as a monophyletic group. This state of phylogenetic uncertainty is now improving with an increasing number of complete bacterial genomes and development of new methods. In this study, we address the monophyly versus polyphyly of enterobacterial symbionts by exploring a multigene matrix within a complex phylogenetic framework.
Results
We assembled the richest taxon sampling of Enterobacteriaceae to date (50 taxa, 69 orthologous genes with no missing data) and analyzed both nucleic and amino acid data sets using several probabilistic methods. We particularly focused on the long-branch attraction-reducing methods, such as a nucleotide and amino acid data recoding and exclusion (including our new approach and slow-fast analysis), taxa exclusion and usage of complex evolutionary models, such as nonhomogeneous model and models accounting for site-specific features of protein evolution (CAT and CAT+GTR). Our data strongly suggest independent origins of four symbiotic clusters; the first is formed by Hamiltonella and Regiella (S-symbionts) placed as a sister clade to Yersinia, the second comprises Arsenophonus and Riesia (S- and P-symbionts) as a sister clade to Proteus, the third Sodalis, Baumannia, Blochmannia and Wigglesworthia (S- and P-symbionts) as a sister or paraphyletic clade to the Pectobacterium and Dickeya clade and, finally, Buchnera species and Ishikawaella (P-symbionts) clustering with the Erwinia and Pantoea clade.
Conclusions
The results of this study confirm the efficiency of several artifact-reducing methods and strongly point towards the polyphyly of P-symbionts within Enterobacteriaceae. Interestingly, the model species of symbiotic bacteria research, Buchnera and Wigglesworthia, originated from closely related, but different, ancestors. The possible origins of intracellular symbiotic bacteria from gut-associated or pathogenic bacteria are suggested, as well as the role of facultative secondary symbionts as a source of bacteria that can gradually become obligate maternally transferred symbionts.
Similar content being viewed by others
Background
One of the most fundamental evolutionary questions concerning insect-bacteria symbiosis is the origin and phylogenetic relationships of various symbiotic lineages. This knowledge is necessary for understanding the dynamics and mechanisms of symbiosis establishment and maintenance within the host. For instance, close relationships between symbionts and pathogenic bacteria suggests a transition from pathogenicity to symbiosis; polyphyly of the symbionts within a single host group is evidence of their multiple independent origins and close relationships among symbionts of different biology indicate high ecological flexibility within a given symbiotic group [1–6]. These implications are particularly important within Enterobacteriaceae, the group containing a broad spectrum of symbiotic lineages and forms described from various groups of insects. Their biology varies from loosely associated facultative symbionts (often called Secondary (S) symbionts) to obligatory mutualists of a highly derived nature, called Primary (P) symbionts [7–9]. However, the concept of the P- and S-symbionts and the associated terminology are a major oversimplification and they become inadequate for the description of the ever increasing complexity of the symbiotic system within Enterobacteriaceae. This complexity is manifested by such phenomena as the presence of multiple symbionts in a single host [10], occurrence of intermediate symbiotic forms and the replacement of symbionts within a host [11–14] or close phylogenetic relationships between typical S- and P-symbionts revealing their high ecological versatility [15]. A good example of such a complex system is provided by the occurrence of multiple obligate symbionts within Auchenorrhyncha [10], universally harboring Sulcia muelleri (Bacteroidetes) [16] with either Hodgkinia cicadicola (α-Proteobacteria) in cicadas, Zinderia insecticola (β-Proteobacteria) in spittlebugs or Baumannia cicadellinicola (γ-Proteobacteria) in sharpshooters. All of these latter symbionts are obligate and have been cospeciating with their hosts for millions of years [17–21]. A close phylogenetic relationship between typical S- and P-symbionts has been so far demonstrated in two well defined and often studied groups, the enterobacterial genera Arsenophonus and Sodalis [5, 22, 23]. The general capability of S-symbionts to supplement the metabolic functions of P-symbionts or even replace them was demonstrated experimentally by replacement of Buchnera with Serratia in aphids [24].
It is obvious that all these fascinating processes can only be studied on a reliable phylogenetic background [9, 25–28]. Unfortunately, under current conditions, the phylogeny within Enterobacteriaceae and the placement of various symbiotic lineages are very unstable. Particularly, the P-symbionts present an extremely difficult challenge to phylogenetic computation due to their strongly modified genomes [9]. There are several root problems that are responsible for this dissatisfactory state. Traditionally, 16S rDNA was frequently used as an exclusive molecular marker for the description of a new symbiont. Many lineages are thus represented only by this gene, which has been shown within Enterobacteriaceae to be inadequate for inferring a reliable phylogeny [29]. In addition, it is notoriously known that the phylogenetic information of symbiotic bacteria is often seriously distorted due to the conditions associated with the symbiotic lifestyle. The effect of strong bottlenecks accompanied by reduced purifying selection and the overall degeneration of symbiotic genomes have been thoroughly discussed in many studies [30–33]. As a result of these degenerative processes, symbiotic lineages may experience parallel changes of their DNAs and these convergences produce the main source of phylogenetic artifacts. Among the most important features are biased nucleotide composition favoring adenine-thymine bases and rapid sequence evolution. While the compositional bias leads to the introduction of homoplasies at both nucleotide and amino acid levels, the accelerated evolution is a well known source of the long-branch attraction phenomenon [34, 35]. Due to these circumstances, symbionts almost always appear as long branches in phylogenetic trees and tend to cluster together [36].
Various methodological approaches have been tested to overcome these difficulties (Additional file 1). They are based mainly on the concatenation of a large number of genes through the whole genome [37–39], the supertree and the consensus approach [37], exclusion of amino acids (FYMINK: phenylalanine, tyrosine, methionine, isoleucine, asparagine and lysine) most affected by nucleotide bias [37], modifications of sequence evolution models [11, 12, 36, 40] and use of the genome structure as a source of phylogenetic data [41]. Phylogenomic studies based on large concatenated sets frequently imply monophyly of the typical P-symbionts (Additional file 1). However, due to the limited number of available genomes, these studies are usually based on inadequate taxon sampling. For example, secondary symbionts and plant pathogens that were shown to break the P-symbiont monophyly in the analysis using a nonhomogeneous model [40] could not be included into these phylogenomic studies. It is important to note that P-symbionts are probably only distantly related to the Escherichia/Salmonella/Yersinia clade. Therefore, the monophyly of P-symbionts derived from such a phylogenomic dataset is logically inevitable, but does not carry any evolutionary information.
The non-monophyletic nature of P-symbionts has been recently suggested in several studies. Perhaps the most inspiring is a study based on a nonhomogeneous model that separates P-symbionts into two independent lineages [40]. As an alternative, a paraphyletic arrangement of these symbionts in respect to several free-living taxa has been revealed from gene-order analysis based on break-point and inversion distances [41]. Most recently, Williams et al. [42] performed a 'telescoping' multiprotein phylogenomic analysis of 104 γ-Proteobacterial genomes. The phylogeny of Enterobacteriaceae endosymbionts was difficult to resolve, although it appeared that there were independent origins of at least the Sodalis and Buchnera lineages.
Thus, there is now a spectrum of hypotheses on the phylogeny of insect symbionts, ranging from complete polyphyly with multiple independent origins to complete monophyly with one common origin. In this study, we take advantage of current progress in computational methods to investigate phylogenetic relationships among the symbiotic lineages. One of the promising recent methodological advances is the introduction of a site-heterogeneous non-parametric mixture CAT model that allows for site-specific features of protein evolution [43]. This model was shown to solve the long-branch attraction (LBA) artifacts and outperform the previous models [44–47]. Similarly, the slow-fast method based on removal of the fastest evolving sites was shown to reduce phylogenetic artifacts [48–54], as well as purine/pyrimidine (RY) data recoding [55–58] or amino acid data recoding [59, 60]. We used these methods as the core of a complex approach and tried to investigate series of methods, models and parameters to detect common trends in changes of the topologies. To do this, we applied two parallel approaches, one based on the application of recently developed algorithms and the other on the removal or recoding of the positions most affected by rapid sequence evolution and/or compositional (AT) bias. In addition, we paid particular attention to the sampling and used as much of a complete set of both symbiotic and free-living lineages as possible. This approach is particularly important to avoid interpretation uncertainties due to the absence of phylogenetically important lineages.
Results
The complete methodological design of this study and the resulting topologies are depicted in Figure 1. All matrices, alignments and phylogenetic trees are available in the TreeBASE database http://purl.org/phylo/treebase/phylows/study/TB2:S11451, as supplementary material, or on the webpage http://users.prf.jcu.cz/husnif00.
Study design. General design of the study summarizing all analyses and results. Individual topologies show the gradient of acquired results; method names are written above and below the arrows. Notice an increasing number of independent origins of symbionts and decreasing number of phylogenetic artifacts along the continuum towards the 'derived' methods. 1+2: third codon positions excluded; AT/GC(BI/4-11): AT/GC datasets 4-11 analyzed by BI; BI: Bayesian inference; Dayhoff6/Dayhoff4/HP: amino acid recoded matrices; ML: maximum likelihood; nhPhyML: ML under nonhomogeneous model; MP: maximum parsimony; RY: purine/pyrimidine recoded matrix; SF: slow-fasted datasets.
Standard maximum likelihood and Bayesian inference
The single gene maximum likelihood (ML) analyses of both nucleic and amino acid data provided an array of mutually exclusive topologies. The majority consensus based on amino acid data (Additional file 2a) groups almost all symbionts into polytomy with only two pairs of sister symbiotic species being resolved (Buchnera and Blochmannia). Phylogenetic trees inferred by ML and Bayesian inference (BI) from the nucleic acid concatenated data using the General Time Reversible model with an estimated proportion of invariable sites (I) and heterogeneity of evolutionary rates modeled by the four substitution rate categories of the gamma (Γ) distribution with the gamma shape parameter (alpha) estimated from the data (GTR+I+Γ) were apparently affected by phylogenetic artifacts, as demonstrated by placement of Riesia and Wigglesworthia within the Buchnera cluster with high posterior probabilities in the BI tree (Figure 2) and the attraction of two outgroup species (Haemophilus and Pasteurella) in the ML tree with high bootstrap support (Additional file 2b). Similar topologies were also retrieved from the amino acid concatenate by ML and BI using the LG+I+Γ, WAG+I+Γ and GTR+I+Γ models (Figure 3). Nevertheless, in contrast to the nucleotide-derived results, the monophyly of the Buchnera clade was not disrupted and Hamiltonella and Regiella were unambiguously separated from the other symbionts and clustered with Yersinia.
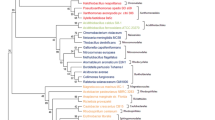
MrBayes phylogram - 69 genes, nucleotide matrix. Phylogenetic tree inferred from the concatenated nucleotide matrix using BI under the GTR+I+Γ model. Asterisks designate nodes with posterior probabilities equal to 1.0, values next to species names represent GC content calculated from the 69-gene dataset, genomic GC content can be found in Additional file 4. BI: Bayesian inference.
MrBayes phylogram - 69 genes, amino acid matrix. Phylogram inferred from the concatenated amino acid matrix using BI under the WAG+I+Γ model. Values at nodes represent posterior probabilities (WAG+I+Γ model, GTR+I+Γ protein model) and bootstrap supports from ML analysis (LG+I+Γ model). Asterisks designate nodes with posterior probabilities or bootstrap supports equal to 1.0, dashes designate values lower than 0.5 or 50, values next to species names represent GC content calculated from the 69-gene dataset, genomic GC content can be found in Additional file 4. BI: Bayesian inference. ML: maximum likelihood.
PhyloBayes, non-homogenous PhyML and modified matrices
The phylogenetic trees acquired under the CAT+GTR PhyloBayes model from 14 and 55 concatenated genes (Figure 4 and Additional file 2p) split symbiotic bacteria into four and three independent lineages, respectively. First, Arsenophonus nasoniae is a sister species to Proteus mirabilis; second, Hamiltonella and Regiella form a sister clade to Yersinia pestis; third, the Sodalis, Baumannia, Blochmannia, Wigglesworthia, Riesia and Buchnera clade form a sister clade to Dickeya/Pectobacterium. The position of Ishikawaella differs between the two datasets. In the 14-gene dataset, Ishikawaella forms a sister clade to Pantoea (Figure 4) and in the 55-gene dataset, it is attracted to the P-symbiont cluster (Additional file 2p).
PhyloBayes phylogram - 14 genes, amino acid matrix. Phylogram derived from concatenation of 14 genes (AceE, ArgS, AspS, EngA, GidA, GlyS, InfB, PheT, Pgi, Pnp, RpoB, RpoC, TrmE and YidC) using PhyloBayes under the CAT+GTR model. Asterisks designate nodes with posterior probabilities equal to 1.0, values next to species names represent GC content calculated from the 69-gene dataset, genomic GC content can be found in Additional file 4.
A topology with four independent symbiotic clades resulted from the trees derived from dayhoff6 and dayhoff4 recoded amino acid data sets analyzed by CAT and CAT+GTR models (Figure 5, Additional file 2r, q) and partially with the hp (hydrophobic-polar) recoded dataset (Additional file 2c) - which was on the other hand affected by the substantial loss of phylogenetic information. The first clade is Buchnera+Ishikawaella as a sister clade to the Erwinia/Pantoea clade, the second clade is Riesia+Arsenophonus as a sister clade to Proteus, the third clade is Hamiltonella+Regiella as a sister clade to Yersinia, and the last clade is composed of Sodalis, Baumannia, Blochmannia and Wigglesworthia.
PhyloBayes cladogram - 69 genes, Dayhoff6 amino acid recoded matrix. Cladogram inferred from amino acid matrix recoded with Dayhoff6 scheme using PhyloBayes with the CAT and CAT+GTR model. Because of the length of symbiotic branches, phylogram is presented only as a preview (original phylogram can be found in Additional trees on our website). Values at nodes represent posterior probabilities from CAT and CAT+GTR analyses, respectively (asterisks designate nodes with posterior probabilities equal to 1.0). Values next to species names represent GC content calculated from the 69-gene dataset, genomic GC content can be found in Additional file 4.
The analyses testing each symbiont independently, using a CAT+GTR model on the dayhoff6 recoded datasets, resulted in topologies supporting multiple origins of endosymbiosis (Additional file 2s). Arsenophonus clusters with Proteus; Hamiltonella clusters with Yersinia; Regiella clusters with Yersinia; and Sodalis, Blochmannia, Baumannia, Riesia and Wigglesworthia grouped into polytomy with the basal enterobacterial clades. Most importantly, the Buchnera clade clusters as a sister clade to the Erwinia clade and Ishikawaella is placed in polytomy with the Pantoea and Erwinia clade.
The non-homogenous (nh) PhyML nucleotide analyses with two different starting trees resulted in two different topologies (Figure 6 and Additional file 2d, e, f). When compared by the approximately unbiased (AU) test, the topology with four independent origins of symbiotic bacteria prevailed (P = 1) over the topology with monophyly of P-symbionts, which therefore corresponds to a local minimum due to a tree search failure (complete matrix: P = 2 × 10-67; matrix without the third positions: P = 9 × 10-87). The only incongruence in topologies based on the complete matrix (Additional file 2d) and the matrix without the third positions (Figure 6) was the placement of the Sodalis+Baumannia+Blochmannia+Wigglesworthia clade as a sister clade to the Edwardsiella or Dickeya/Pectobacterium clades.
Matrices obtained by removing positions according to the AT/GC contents produced trees covering the whole continuum illustrated in Figure 1. The most severe restrictions, that is, removal of all positions that contain both AT and GC categories or relaxing for up to three taxa (see BI trees in Additional file 2g, h, i, j), yielded topologies compatible with the results of the CAT model applied on the recoded amino acid data and of the nhPhyML analysis. Further relaxing the restriction rule led to a variety of trees along the Figure 1 continuum, with a less clear relation between the used parameter and the resulting topology (Additional file 3).
Compared to the ML analysis of all nucleotide positions, the analysis of first plus second positions reduced the obvious artifact of outgroup attraction (Additional file 2k). Nevertheless, it also sorted symbionts according to their branch length. Analysis of the RY recoded nucleotide matrix produced a tree compatible with the results of the CAT+GTR model (Additional file 2l). Analysis of the RY recoded nucleotide matrix without the third positions resulted in a topology with a Sodalis+Baumannia+Blochmannia cluster (as a sister to the Pectobacterium/Dickeya clade) separated from the rest of the P-symbionts, which clustered with the Erwinia/Pantoea clade (Additional file 2m). Slow-fast analyses with gradual reduction of saturated positions did not produce the polyphyly of P-symbionts (Additional file 3; only the first five trees presented, subsequent trees are identical to the fifth tree). However, this analysis shows an increasing effect of LBA artifacts associated with the increasing number of remaining saturated positions, especially Riesia attraction and swapping of symbiotic branches according to their length.
Discussion
Performance of the methods: convergence towards non-monophyly
The results obtained in this study strongly indicate that the frequently retrieved monophyly of P-symbionts is an artifact caused by their highly modified genomes. None of the most widely used methods, that is, ML and BI with different models used on nucleic (GTR+I+Γ) and amino acid (GTR/LG/WAG+I+Γ) data, were capable of resolving deep phylogenetic relationships and correct placement of the symbiotic taxa. This conclusion is evidenced by obvious artifacts, such as the inclusion of Riesia into the P-symbiotic lineage or the even more conspicuous distorted placing of Wigglesworthia within the Buchnera cluster. The arrangement of such trees suggests that these methods sort the symbionts according to their branch lengths and/or AT contents and attach the whole symbiotic cluster to the longest branch available. While the difficulty with placement of the most aberrant taxa, such as Riesia, Wigglesworthia and Buchnera (Cinara cedri) was also observed when using the mixture model accounting for site specific characteristics of protein evolution (Figure 4; Additional files 2p and 5), these artifacts disappeared after amino acid data recoding followed by CAT and CAT+GTR model analysis and the application of a nonhomogeneous model.
Additional support for the non-monophyly view stems from the second, parallel approach based on the restricted matrices. While our newly developed method shares the basic principles with the slow-fast and recoding methods, such as the removal of the positions that are likely to distort the phylogenetic relationships due to their aberrant evolution, it differs in the criteria of their removal and thus produces different input data. It is therefore significant that this method led independently to the same picture, the non-monophyly of the P-symbionts with clustering identical to the above analyses: Ishikawaella+Buchnera and Sodalis+Baumannia+Blochmannia+Wigglesworthia. The removal of the heteropecillous sites was recently shown to have similar effectiveness as our new method [61], which further supports the results. Moreover, this topology was obtained even under the maximum parsimony (MP) criterion (Additional file 3), which is known to be extremely sensitive to LBA [34]. On the other hand, although slow-fast analysis is generally considered a powerful tool for resolving relationships among taxa with different rates of evolution, we show in our data that the mere exclusion of the fast evolving sites is not sufficient when using empirical models and should be followed by analysis using some of the complex models, such as the CAT-like models. In addition, since this method usually requires an a priori definition of monophyletic groups, it should be used and interpreted with caution. Similar to the slow-fast method, RY recoding and exclusion of third codon positions were not sufficient for resolving deep symbiont phylogeny. However, all these methods can remove at least some of the artifacts and provide insight for further analyses.
Summarizing the topologies obtained in this study (Figure 1), a convergence can be detected towards a particular non-monophyletic arrangement of P-symbionts, as revealed under the most 'derived' methods. This result strongly supports the view of multiple origins of insect endosymbionts, as first revealed by the nonhomogeneous model of sequence evolution [40], and is partially congruent with the analyses of gene order [41] and phylogenomics of Gammaproteobacteria [42]. It is also important to note that, apart from multiple symbiont clustering, the arrangement of the non-symbiotic taxa corresponds to most of the phylogenomic analyses using Escherichia/Salmonella/Yersinia taxon sampling [37–39].
Biological significance of P-symbionts non-monophyly
Considering that most of the 'artifact-resistant' analyses point towards the non-monophyly of enterobacterial P-symbionts, the questions of how many symbiotic lineages are represented by the known symbiotic diversity and what are their closest free-living relatives now becomes of particular importance. It is not clear whether the split of the original P-symbiotic cluster into two lineages is definite or these two groups will be further divided after yet more sensitive methods and more complete data are available. At the moment there are still several clusters composed exclusively of derived symbiotic forms. In principle, three different processes may be responsible for the occurrence of such clusters: first, horizontal transmission of established symbiotic forms among host species; second, inadequate sampling with missing free-living relatives; or third, phylogenetic artifacts. All of these factors are likely to play a role in the current topological patterns. Being the main issues of this study, the role of methodological artifacts has been discussed above. Horizontal transmission, as the basis of non-artificial symbiotic clusters, is likely to take part at least in some cases. Perhaps the most convincing example is the Wolbachia cluster [62]: while within Enterobacteriaceae it may apply to Arsenophonus, Sodalis and possibly some other S-symbionts.
Recognition of the third cause, the incomplete sampling, and identification of the closest free-living relatives, now becomes a crucial step in future research. It is often assumed that symbionts originate from bacteria common to the environment typical for a given insect group. For example, cicadas spend most of their life cycle underground and feed primarily on plant roots. Consequently, their α-Proteobacterial symbiont Hodgkinia cicadicola originated within Rhizobiales [19]. A similar ecological background can be noticed in yet different hosts, the ixodid and argasid ticks. Several reports have shown that some of the tick-transmitted pathogens are related to their symbiotic fauna [63–65]. Many of the insect taxa associated with symbiotic Enterobacteriaceae are phytophagous, and plant pathogens thus fit well into this hypothesis as hypothetical ancestors of various insect symbionts lineages. The presence of a type III secretion system, which is used in pathogenic bacteria for host cell invasion, in secondary symbionts [66–69] and its remnant in the primary symbiont of Sitophilus spp. weevils [70] could further support the theory of pathogenic ancestors of insect symbionts. It can only be speculated that these bacteria first became S-symbiont type and were horizontally transferred to various other insect species. Within some of the infected species, facultative symbionts eventually became obligatory primary symbionts. An identical situation can be observed in symbiotic clades with numerous species, such as Wolbachia [71, 72], Sodalis [23, 73, 74] or Arsenophonus [5].
In our study, we gave particular attention to the sampling of free-living Enterobacteriaceae to provide as complete a background for the symbiotic lineages as possible under the current state of knowledge (that is, the availability of the genomic data). The most consistent picture derived from the presented analyses places the four main symbiotic clusters into the following positions. First, for the Buchnera cluster, its previously suggested relationship to Erwinia was confirmed. Erwinia, as a genus of mostly plant pathogenic bacteria, has been previously suggested to represent an ancestral organism, which upon ingestion by aphids at least 180 million years ago [75] turned into an intracellular symbiotic bacterium [76]. However, it is not known whether it was primarily pathogenic to aphids, similar to Erwinia aphidicola [77], or a gut associated symbiotic bacterium as in pentatomid stinkbugs [78], thrips [79, 80] or Tephritidae flies [81–83]. Ishikawaella capsulata, an extracellular gut symbiont of plataspid stinkbugs [84], was the only symbiotic bacterium that clustered in our 'derived' analyses with the Buchnera clade. However, several single-gene studies indicate that this group contains some additional symbiotic lineages for which sequenced genome data is not currently available. These are, in particular, the extracellular symbionts of acanthosomatid stinkbugs [85], parastrachid stinkbugs [86], scutellerid stinkbugs [87, 88] and some of the symbionts in pentatomid stinkbugs [78].
The second clade, represented in our analysis by Sodalis+Baumannia+Blochmannia+Wigglesworthia, is likely to encompass many other P- and S-symbionts [89–92]. The possible single origin of these symbionts has to be further tested, however the interspersion of both forms, together with basal position of Sodalis, seem to support a transition from a secondary to primary symbiotic lifestyle [15]. In our analysis, the whole clade was placed between pathogenic bacteria of plants and animals, the Edwardsiella and Pectobacterium/Dickeya clades, or as a sister to the latter group. Recently, another symbiotic bacterium (called BEV, Euscelidius variegatus host) was shown to be a sister species to Pectobacterium [93].
Two additional independent origins of insect symbionts are represented by the Arsenophonus/Riesia clade and Hamiltonella+Regiella. Both of these clades clustered in our analyses in the positions indicated by previous studies, that is, as related to Proteus and Yersinia, respectively [5, 67, 93–97].
While the position of individual symbiotic lineages is remarkably consistent across our 'artifact-resistant' analyses and are well compatible with some of the previous studies, the topology can only provide a rough picture of the relationships within Enterobacteriaceae. To get a more precise and phylogenetically meaningful background for an evolutionary interpretation, the sample of free-living bacteria as a possible source of symbiotic lineages has to be much improved. An illuminating example is provided by the bacterium Biostraticola tofi, described from water biofilms. When analyzed using 16S rDNA, this bacterium seemed to be closely related to Sodalis [98]. Its position as a sister group to the Sodalis/Baumannia/Blochmannia/Wigglesworthia clade was also retrieved in our single-gene analysis (groEL, data not shown). If confirmed by more precise multigene approach, Biostraticola would represent the closest bacterium to the large symbiotic cluster.
Conclusions
The topologies obtained by several independent approaches strongly support the non-monophyletic view of enterobacterial P-symbionts. Particularly, they show that at least three independent origins led to highly specialized symbiotic forms, the first giving rise to Sodalis, Baumannia, Blochmannia and Wigglesworthia (S- and P-symbionts), the second to Buchnera and Ishikawella and the last to Riesia and Arsenophonus (S- and P-symbionts). This separation of symbiotic clusters poses an interesting question as to whether the presented disbandment of the P-symbiotic cluster is definite or if it will continue after yet more complete data are available and more realistic evolutionary models [99–101] are applied. One obvious drawback of the current state is that many additional symbiotic lineages already known within Enterobacteriaceae cannot be at the moment included into serious phylogenetic analyses due to the lack of sufficient molecular data and will have to be revisited once complete genomic data are available. These bacteria include symbionts of mealybugs [89, 102], psyllids [90, 103], lice [2, 91], weevils [11, 12, 92], reed beetles [104, 105], true bugs [78, 84–88, 106, 107] and symbionts of leeches [108, 109]. Similarly, the importance of free-living bacteria and variety of S-symbionts as possible ancestors of P-symbionts should not be underestimated when assembling datasets for phylogenetic analyses. The shift from polymerase chain reaction-based gene-centered sequencing towards high-throughput next-generation sequencing may soon provide sufficient data for more complete analyses of the Enterobacteriaceae phylogeny.
Methods
Matrices and multiple sequence alignments
The genes used in this study were extracted from 50 complete genome sequences of γ-Proteobacteria available in GenBank (Additional file 4), including 14 endosymbiotic Enterobacteriaceae. We did not include Carsonella ruddii [110] since this psyllid symbiotic bacterium does not appear to be a member of the Enterobacteriaceae clade [90, 111] and is only attracted there by the AT rich taxa. After removal of the AT rich lineages from the analysis, Carsonella ruddii clusters with the genus Pseudomonas [42]. Also, we did not include Serratia symbiotica [95] because its genome only became available after completion of our datasets. However, the phylogenetic position of this symbiotic bacterium within Serratia genus is robust and was confirmed in several studies [6, 14, 112].
To minimize the introduction of a false phylogenetic signal, we compared the genomes of all symbiotic bacteria and selected only single-copy genes present in all of the included symbiotic and free-living taxa. Such strict gene exclusion was also necessary regarding the usage of computationally demanding methods; it was one of our goals to produce a taxonomically representative data set of efficient size with no missing data. Altogether, 69 orthologous genes, mostly involved in translation, ribosomal structure and biogenesis (Additional file 4) were selected according to the Clusters of Orthologous Groups of proteins (COGs) [113, 114]. Single-gene nucleotide data sets were downloaded via their COG numbers from a freely available database (MicrobesOnline [115]).
All protein coding sequences were translated into amino acids in SeaView version 4 [116], aligned by the MAFFT version 6 L-INS-i algorithm [117] and toggled back to the nucleotide sequences. Ambiguously aligned positions (codons) were excluded by Gblocks v0.91b [118, 119] with the following parameters: minimum number of sequences for a conserved position: 26; minimum number of sequences for a flanking position: 43; maximum number of contiguous nonconserved positions: 8; minimum length of a block: 10; allowed gap positions: with half. The resulting trimmed alignments were checked and manually corrected in BioEdit v7.0.5 [120]. Alignments were concatenated in SeaView. The 69 gene concatenate resulted in an alignment of 63, 462 nucleic acid positions with 42, 481 parsimony-informative and 48, 527 variable sites and 21, 154 amino acid positions with 12, 735 parsimony-informative and 15, 986 variable sites.
Phylogenetic analyses
We used two different approaches to deal with the distortions caused by the highly modified nature of symbiotic genomes, which are the main source of the phylogenetic artifacts in phylogenetic analyses.
First, we applied complex models of molecular evolution. Using PhyloBayes 3.2f [121], we applied non-parametric site heterogeneous CAT and CAT+GTR models [43]. For all PhyloBayes analyses, we ran two chains with an automatic stopping option set to end the chain when all discrepancies were lower than 0.3 and all effective sizes were larger than 100. Under the CAT and CAT+GTR models, the four independent PhyloBayes runs were stuck in a local maximum (maxdiff = 1) even after 25, 000 and 10, 000 cycles, respectively, and we were not able to reach Markov Chain Monte Carlo (MCMC) convergence. Therefore, we present these trees only as supplementary material (although they mostly point toward multiple origins of symbiosis; Additional file 5) and we ran the CAT+GTR analyses with the reduced dataset based on 14 genes with the number of parsimony-informative amino acid positions higher than 300 (AceE, ArgS, AspS, EngA, GidA, GlyS, InfB, PheT, Pgi, Pnp, RpoB, RpoC, TrmE and YidC). To check for compatibility of these arbitrary selected 14 genes with the rest of the data, we also analyzed, in a separate analysis, the remaining 55-gene dataset under the CAT+GTR model. Using nhPhyML [122], we applied a nonhomogeneous nonstationary model of sequence evolution [123, 124], which can deal with artifacts caused by compositional heterogeneity [40, 125, 126]. We used two different starting trees (Additional file 2n) and ran the analyses with and without the third codon positions. The resulting trees were evaluated by an AU test in CONSEL [127].
The second approach relies on the selective restriction of the data matrix. We used four previously established methods of data weighting and/or exclusion (see Background): RY data recoding, amino acid data recoding, exclusion of third codon positions and slow-fast analysis, and developed one additional method: since transition from G/C to A/T at many positions is a common homoplasy of symbiotic genomes, we removed from the matrix all positions containing both the G/C and A/T states. All substitutions considered in the subsequent analyses thus included exclusively transversions within the A/T or G/C categories. To analyze an effect of this restriction on the reduction of the data, we prepared 11 matrices with a partially relaxed rule (removing all positions with AT+GC, allowing for one taxon exception, two taxa exception, and so on, up until a 10 taxa exception). Since this method has never been tested, we analyzed the restricted matrices by the BI, ML (parameters as for standard analyses) and MP using PAUP* 4.0b10 with the tree bisection and reconnection algorithm [128]. Four other types of data weighting and/or exclusion were used to increase the phylogenetic signal to noise ratio and determine the robustness of our results. First, the third codon positions were removed in SeaView. Second, RY recoding was performed on all and first plus second positions. Third, saturated positions were excluded from the concatenated data sets by SlowFaster [129]. To assign substitutional rates to individual positions, unambiguously monophyletic groups were chosen on a polytomic tree (Additional file 2o), positions with the highest rates were gradually excluded and 21 restricted matrices were produced. These weighted data sets were analyzed by ML. Fourth, amino acid data recoding was performed in PhyloBayes with hp (A, C, F, G, I, L, M, V, W) (D, E, H, K, N, P, Q, R, S, T, Y), dayhoff4 (A, G, P, S, T) (D, E, N, Q) (H, K, R) (F, Y, W, I, L, M, V) (C = ?) and dayhoff6 (A, G, P, S, T) (D, E, N, Q) (H, K, R) (F, Y, W) (I, L, M, V) (C) recoding schemes. In addition, we have prepared 10 dayhoff6 recoded matrices to test individual symbiotic lineages without the presence of other symbionts. Amino acid recoded matrices were analyzed using the CAT and CAT+GTR models, which are more immune to phylogenetic artifacts than one-matrix models.
To allow for comparison of the results with previously published studies, as well as to separate the effect of newly used models and methods from changes due to the extended sampling, we also used standard procedures of phylogenetic inference, ML and BI. The following programs, algorithms and parameters were used in the ML and BI analyses. ML was applied to single-gene and concatenated alignments of both nucleotides and amino acids using PhyML v3.0 [130] with the subtree pruning and regrafting tree search algorithm. BI was performed in MrBayes 3.1.2 [131] with one to five million generations and tree sampling every 100 generations. Exploration of MCMC convergence and burn-in determination was performed in AWTY and Tracer v1.5 [132, 133]. Evolutionary substitution models for proteins were selected by ProtTest 2.4 [134] and for DNA by jModelTest 0.1.1 [135] according to the Akaike Information Criterion. For DNA sequences, the GTR+I+Γ model was used [136–138]. Transition and transversion models [139] were used with I+Γ under ML for the first two AT/GC datasets. LG+I+Γ [140], WAG+I+Γ [141] and GTR+I+Γ models were used for amino acid data. A cross-validation method implemented in PhyloBayes [121, 142] was used to estimate the fit of CAT-like models. For both datasets, the 14 selected genes as well as the complete 69 genes set, the cross-validation was performed according to the PhyloBayes manual in 10 replicates each with 1, 100 cycles. The CAT-Poisson model had significantly better fit to the data than the GTR model (Δl 157.37 ± 56.9379 for the 14-gene matrix and Δl 3923.9 ± 1963.5 for the 69-gene matrix); of the CAT-like models, the CAT+GTR model was found to be significantly better than the CAT-Poisson model (Δl 536.71 ± 32.8341 for the 14-gene matrix and Δl 1633.4 ± 123.482 for the 69-gene matrix) in all 10 replicates.
References
Gottlieb Y, Ghanim M, Gueguen G, Kontsedalov S, Vavre F, Fleury F, Zchori-Fein E: Inherited intracellular ecosystem: symbiotic bacteria share bacteriocytes in whiteflies. FASEB J. 2008, 22 (7): 2591-2599.
Hypša V, Křížek J: Molecular evidence for polyphyletic origin of the primary symbionts of sucking lice (Phthiraptera, Anoplura). Microb Ecol. 2007, 54 (2): 242-251.
Bordenstein SR, Paraskevopoulos C, Hotopp JC, Sapountzis P, Lo N, Bandi C, Tettelin H, Werren JH, Bourtzis K: Parasitism and mutualism in Wolbachia: what the phylogenomic trees can and cannot say. Mol Biol Evol. 2009, 26 (1): 231-241.
Caspi-Fluger A, Zchori-Fein E: Do plants and insects share the same symbionts?. Isr J Plant Sci. 2010, 58 (2): 113-119.
Nováková E, Hypša V, Moran NA: Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol. 2009, 9: 143-
Moran NA, Russell JA, Koga R, Fukatsu T: Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl Environ Microbiol. 2005, 71 (6): 3302-3310.
Baumann P: Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005, 59: 155-189.
Douglas AE: Mycetocyte symbiosis in insects. Biol Rev Camb Philos Soc. 1989, 64 (4): 409-434.
Moran NA, McCutcheon JP, Nakabachi A: Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008, 42: 165-190.
Takiya DM, Tran PL, Dietrich CH, Moran NA: Co-cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol Ecol. 2006, 15 (13): 4175-4191.
Lefevre C, Charles H, Vallier A, Delobel B, Farrell B, Heddi A: Endosymbiont phylogenesis in the Dryophthoridae weevils: evidence for bacterial replacement. Mol Biol Evol. 2004, 21 (6): 965-973.
Conord C, Despres L, Vallier A, Balmand S, Miquel C, Zundel S, Lemperiere G, Heddi A: Long-term evolutionary stability of bacterial endosymbiosis in Curculionoidea: additional evidence of symbiont replacement in the Dryophthoridae family. Mol Biol Evol. 2008, 25 (5): 859-868.
Perez-Brocal V, Gil R, Ramos S, Lamelas A, Postigo M, Michelena JM, Silva FJ, Moya A, Latorre A: A small microbial genome: the end of a long symbiotic relationship?. Science. 2006, 314 (5797): 312-313.
Lamelas A, Perez-Brocal V, Gomez-Valero L, Gosalbes MJ, Moya A, Latorre A: Evolution of the secondary symbiont "Candidatus Serratia symbiotica" in aphid species of the subfamily Lachninae. Appl Environ Microbiol. 2008, 74 (13): 4236-4240.
Wernegreen JJ, Kauppinen SN, Brady SG, Ward PS: One nutritional symbiosis begat another: phylogenetic evidence that the ant tribe Camponotini acquired Blochmannia by tending sap-feeding insects. BMC Evol Biol. 2009, 9 (1): 292-
Moran NA, Tran P, Gerardo NM: Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol. 2005, 71 (12): 8802-8810.
Wu D, Daugherty SC, Van Aken SE, Pai GH, Watkins KL, Khouri H, Tallon LJ, Zaborsky JM, Dunbar HE, Tran PL, Moran NA, Eisen JA: Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 2006, 4 (6): e188-
McCutcheon JP, Moran NA: Functional convergence in reduced genomes of bacterial symbionts spanning 200 million years of evolution. Genome Biol Evol. 2010, 2: 708-718.
McCutcheon JP, McDonald BR, Moran NA: Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 2009, 106 (36): 15394-15399.
McCutcheon JP, McDonald BR, Moran NA: Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. Plos Genet. 2009, 5 (7): e1000565-
McCutcheon JP, Moran NA: Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 2007, 104 (49): 19392-19397.
Snyder AK, McMillen CM, Wallenhorst P, Rio RV: The phylogeny of Sodalis-like symbionts as reconstructed using surface-encoding loci. FEMS Microbiol Lett. 2011, 317 (2): 143-151.
Fukatsu T, Koga R, Smith WA, Tanaka K, Nikoh N, Sasaki-Fukatsu K, Yoshizawa K, Dale C, Clayton DH: Bacterial endosymbiont of the slender pigeon louse, Columbicola columbae, allied to endosymbionts of grain weevils and tsetse flies. Appl Environ Microbiol. 2007, 73 (20): 6660-6668.
Koga R, Tsuchida T, Fukatsu T: Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc R Soc Lond B. 2003, 270 (1533): 2543-2550.
Oliver KM, Degnan PH, Burke GR, Moran NA: Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol. 2010, 55: 247-266.
Moya A, Pereto J, Gil R, Latorre A: Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet. 2008, 9 (3): 218-229.
Gosalbes MJ, Latorre A, Lamelas A, Moya A: Genomics of intracellular symbionts in insects. Int J Med Microbiol. 2010, 300 (5): 271-278.
Toft C, Andersson SGE: Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet. 2010, 11 (7): 465-475.
Naum M, Brown EW, Mason-Gamer RJ: Is 16S rDNA a reliable phylogenetic marker to characterize relationships below the family level in the Enterobacteriaceae?. J Mol Evol. 2008, 66 (6): 630-642.
Mira A, Moran NA: Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microb Ecol. 2002, 44 (2): 137-143.
Wernegreen JJ, Moran NA: Evidence for genetic drift in endosymbionts (Buchnera): analyses of protein-coding genes. Mol Biol Evol. 1999, 16 (1): 83-97.
Lambert JD, Moran NA: Deleterious mutations destabilize ribosomal RNA in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1998, 95 (8): 4458-4462.
Allen JM, Light JE, Perotti MA, Braig HR, Reed DL: Mutational meltdown in primary endosymbionts: selection limits Muller's ratchet. Plos One. 2009, 4 (3): e4969-
Bergsten J: A review of long-branch attraction. Cladistics. 2005, 21 (2): 163-193.
Ruano-Rubio V, Fares MA: Artifactual phylogenies caused by correlated distribution of substitution rates among sites and lineages: the good, the bad, and the ugly. Syst Biol. 2007, 56 (1): 68-82.
Charles H, Heddi A, Rahbe Y: A putative insect intracellular endosymbiont stem clade, within the Enterobacteriaceae, infered from phylogenetic analysis based on a heterogeneous model of DNA evolution. C R Acad Sci Ser III Sci Vie. 2001, 324 (5): 489-494.
Comas I, Moya A, Gonzalez-Candelas F: From phylogenetics to phylogenomics: the evolutionary relationships of insect endosymbiotic γ-Proteobacteria as a test case. Syst Biol. 2007, 56 (1): 1-16.
Lerat E, Daubin V, Moran NA: From gene trees to organismal phylogeny in prokaryotes: the case of the γ-Proteobacteria. PLoS Biol. 2003, 1 (1): e19-
Canback B, Tamas I, Andersson SG: A phylogenomic study of endosymbiotic bacteria. Mol Biol Evol. 2004, 21 (6): 1110-1122.
Herbeck JT, Degnan PH, Wernegreen JJ: Nonhomogeneous model of sequence evolution indicates independent origins of primary endosymbionts within the Enterobacteriales (γ-Proteobacteria). Mol Biol Evol. 2005, 22 (3): 520-532.
Belda E, Moya A, Silva FJ: Genome rearrangement distances and gene order phylogeny in γ-Proteobacteria. Mol Biol Evol. 2005, 22 (6): 1456-1467.
Williams KP, Gillespie JJ, Sobral BW, Nordberg EK, Snyder EE, Shallom JM, Dickerman AW: Phylogeny of Gammaproteobacteria. J Bacteriol. 2010, 192 (9): 2305-2314.
Lartillot N, Philippe H: A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004, 21 (6): 1095-1109.
Lartillot N, Brinkmann H, Philippe H: Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 2007, 7: S4-
Lartillot N, Philippe H: Improvement of molecular phylogenetic inference and the phylogeny of Bilateria. Philos Trans R Soc Lond B Biol Sci. 2008, 363 (1496): 1463-1472.
Nesnidal MP, Helmkampf M, Bruchhaus I, Hausdorf B: Compositional heterogeneity and phylogenomic inference of metazoan relationships. Mol Biol Evol. 2010, 27 (9): 2095-2104.
Philippe H, Brinkman FS, Martinez P, Riutort M, Baguna J: Acoel flatworms are not platyhelminthes: evidence from phylogenomics. Plos One. 2007, 2 (8): e717-
Brinkmann H, Philippe H: Archaea sister group of Bacteria? Indications from tree reconstruction artifacts in ancient phylogenies. Mol Biol Evol. 1999, 16 (6): 817-825.
Brochier C, Philippe H: A non-hyperthermophilic ancestor for bacteria. Nature. 2002, 417 (6886): 244-
Hampl V, Čepička I, Flegr J, Tachezy J, Kulda J: Critical analysis of the topology and rooting of the parabasalian 16S rRNA tree. Mol Phylogenet Evol. 2004, 32 (3): 711-723.
Bapteste E, Susko E, Leigh J, Ruiz-Trillo I, Bucknam J, Doolittle WF: Alternative methods for concatenation of core genes indicate a lack of resolution in deep nodes of the prokaryotic phylogeny. Mol Biol Evol. 2008, 25 (1): 83-91.
Philippe H, Lopez P, Brinkmann H, Budin K, Germot A, Laurent J, Moreira D, Muller M, Le Guyader H: Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proc R Soc Lond B. 2000, 267 (1449): 1213-1221.
Hirt RP, Logsdon JM, Healy B, Dorey MW, Doolittle WF, Embley TM: Microsporidia are related to Fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc Natl Acad Sci USA. 1999, 96 (2): 580-585.
Philippe H, Lartillot N, Brinkmann H: Multigene analyses of bilaterian animals corroborate the monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Mol Biol Evol. 2005, 22 (5): 1246-1253.
Phillips MJ, Delsuc F, Penny D: Genome-scale phylogeny and the detection of systematic biases. Mol Biol Evol. 2004, 21 (7): 1455-1458.
Delsuc F, Phillips MJ, Penny D: Comment on "Hexapod origins: monophyletic or paraphyletic?". Science. 2003, 301 (5639): 1482-
Phillips MJ, Penny D: The root of the mammalian tree inferred from whole mitochondrial genomes. Mol Phylogenet Evol. 2003, 28 (2): 171-185.
Gibson A, Gowri-Shankar V, Higgs PG, Rattray M: A comprehensive analysis of mammalian mitochondrial genome base composition and improved phylogenetic methods. Mol Biol Evol. 2005, 22 (2): 251-264.
Hrdý I, Hirt RP, Doležal P, Bardoňová L, Foster PG, Tachezy J, Embley TM: Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004, 432 (7017): 618-622.
Embley TM, van der Giezen M, Horner DS, Dyal PL, Foster P: Mitochondria and hydrogenosomes are two forms of the same fundamental organelle. Philos Trans R Soc Lond B Biol Sci. 2003, 358 (1429): 191-201. discussion 201-203
Philippe H, Roure B: Site-specific time heterogeneity of the substitution process and its impact on phylogenetic inference. BMC Evol Biol. 2011, 11: 17-
Raychoudhury R, Baldo L, Oliveira DC, Werren JH: Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution. 2009, 63 (1): 165-183.
Perlman SJ, Hunter MS, Zchori-Fein E: The emerging diversity of Rickettsia. Proc R Soc B. 2006, 273 (1598): 2097-2106.
Noda H, Munderloh UG, Kurtti TJ: Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl Environ Microbiol. 1997, 63 (10): 3926-3932.
Sassera D, Beninati T, Bandi C, Bouman EAP, Sacchi L, Fabbi M, Lo N: 'Candidatus Midichloria mitochondrii', an endosymbiont of the tick Ixodes ricinus with a unique intramitochondrial lifestyle. Int J Syst Evol Microbiol. 2006, 56: 2535-2540.
Degnan PH, Leonardo TE, Cass BN, Hurwitz B, Stern D, Gibbs RA, Richards S, Moran NA: Dynamics of genome evolution in facultative symbionts of aphids. Environ Microbiol. 2009, 12 (8): 2060-2069.
Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA: Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc Natl Acad Sci USA. 2009, 106 (22): 9063-9068.
Toh H, Weiss BL, Perkin SA, Yamashita A, Oshima K, Hattori M, Aksoy S: Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006, 16 (2): 149-156.
Wilkes TE, Darby AC, Choi JH, Colbourne JK, Werren JH, Hurst GD: The draft genome sequence of Arsenophonus nasoniae, son-killer bacterium of Nasonia vitripennis, reveals genes associated with virulence and symbiosis. Insect Mol Biol. 2010, 19 (Sp. Iss. SI Suppl. 1): 59-73.
Dale C, Plague GR, Wang B, Ochman H, Moran NA: Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc Natl Acad Sci USA. 2002, 99 (19): 12397-12402.
Hosokawa T, Koga R, Kikuchi Y, Meng XY, Fukatsu T: Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Natl Acad Sci USA. 2010, 107 (2): 769-774.
Dedeine F, Vavre F, Fleury F, Loppin B, Hochberg ME, Bouletreau M: Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proc Natl Acad Sci USA. 2001, 98 (11): 6247-6252.
Heddi A, Charles H, Khatchadourian C, Bonnot G, Nardon P: Molecular characterization of the principal symbiotic bacteria of the weevil Sitophilus oryzae: a peculiar G + C content of an endocytobiotic DNA. J Mol Evol. 1998, 47 (1): 52-61.
Nováková E, Hypša V: A new Sodalis lineage from bloodsucking fly Craterina melbae (Diptera, Hippoboscoidea) originated independently of the tsetse flies symbiont Sodalis glossinidius. FEMS Microbiol Lett. 2007, 269 (1): 131-135.
Moran N, Munson M, Baumann P, Ishikawa H: A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc R Soc Lond B. 1993, 253 (1337): 167-171.
Harada H, Oyaizu H, Ishikawa H: A consideration about the origin of aphid intracellular symbiont in connection with gut bacterial flora. J Gen Appl Microbiol. 1996, 42 (1): 17-26.
Harada H, Oyaizu H, Kosako Y, Ishikawa H: Erwinia aphidicola, a new species isolated from pea aphid, Acyrthosiphon pisum. J Gen Appl Microbiol. 1997, 43 (6): 349-354.
Prado SS, Almeida RP: Phylogenetic placement of pentatomid stink bug gut symbionts. Curr Microbiol. 2009, 58 (1): 64-69.
Chanbusarakum L, Ullman D: Characterization of bacterial symbionts in Frankliniella occidentalis (Pergande), Western flower thrips. J Invertebr Pathol. 2008, 99 (3): 318-325.
De Vries EJ, Van der Wurff AWG, Jacobs G, Breeuwer JAJ: Onion thrips, Thrips tabaci, have gut bacteria that are closely related to the symbionts of the western flower thrips, Frankliniella occidentalis. J Insect Sci. 2008, 8: 1-11.
Mazzon L, Martinez-Sanudo I, Simonato M, Squartini A, Savio C, Girolami V: Phylogenetic relationships between flies of the Tephritinae subfamily (Diptera, Tephritidae) and their symbiotic bacteria. Mol Phylogenet Evol. 2010, 56 (1): 312-326.
Mazzon L, Piscedda A, Simonato M, Martinez-Sanudo I, Squartini A, Girolami V: Presence of specific symbiotic bacteria in flies of the subfamily Tephritinae (Diptera Tephritidae) and their phylogenetic relationships: proposal of 'Candidatus Stammerula tephritidis'. Int J Syst Evol Microbiol. 2008, 58 (Pt 6): 1277-1287.
Capuzzo C, Firrao G, Mazzon L, Squartini A, Girolami V: 'Candidatus Erwinia dacicola', a coevolved symbiotic bacterium of the olive fly Bactrocera oleae (Gmelin). Int J Syst Evol Microbiol. 2005, 55 (Pt 4): 1641-1647.
Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T: Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 2006, 4 (10): e337-
Kikuchi Y, Hosokawa T, Nikoh N, Meng XY, Kamagata Y, Fukatsu T: Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 2009, 7: 2-
Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Hironaka M, Fukatsu T: Phylogenetic position and peculiar genetic traits of a midgut bacterial symbiont of the stinkbug Parastrachia japonensis. Appl Environ Microbiol. 2010, 76 (13): 4130-4135.
Kaiwa N, Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Kimura N, Ito M, Fukatsu T: Primary gut symbiont and secondary Sodalis-allied symbiont in the scutellerid stinkbug Cantao ocellatus. Appl Environ Microbiol. 2010, 76 (11): 3486-3494.
Kaiwa N, Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Kimura N, Ito M, Fukatsu T: Bacterial symbionts of the giant jewel stinkbug Eucorysses grandis (Hemiptera: Scutelleridae). Zool Sci. 2011, 28 (3): 169-174.
Thao ML, Gullan PJ, Baumann P: Secondary (γ-Proteobacteria) endosymbionts infect the primary (β-Proteobacteria) endosymbionts of mealybugs multiple times and coevolve with their hosts. Appl Environ Microbiol. 2002, 68 (7): 3190-3197.
Spaulding AW, von Dohlen CD: Phylogenetic characterization and molecular evolution of bacterial endosymbionts in psyllids (Hemiptera: Sternorrhyncha). Mol Biol Evol. 1998, 15 (11): 1506-1513.
Fukatsu T, Hosokawa T, Koga R, Nikoh N, Kato T, Hayama S, Takefushi H, Tanaka I: Intestinal endocellular symbiotic bacterium of the macaque louse Pedicinus obtusus: Distinct endosymbiont origins in anthropoid primate lice and the old world monkey louse. Appl Environ Microbiol. 2009, 75 (11): 3796-3799.
Toju H, Hosokawa T, Koga R, Nikoh N, Meng XY, Kimura N, Fukatsu T: "Candidatus Curculioniphilus buchneri", a novel clade of bacterial endocellular symbionts from weevils of the genus Curculio. Appl Environ Microbiol. 2009, 76 (1): 275-282.
Degnan PH, Bittleston LS, Hansen AK, Sabree ZL, Moran NA, Almeida RP: Origin and examination of a leafhopper facultative endosymbiont. Curr Microbiol. 2011, 62 (5): 1565-1572.
Allen JM, Reed DL, Perotti MA, Braig HR: Evolutionary relationships of "Candidatus Riesia spp., " endosymbiotic Enterobacteriaceae living within hematophagous primate lice. Appl Environ Microbiol. 2007, 73 (5): 1659-1664.
Burke GR, Moran NA: Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol. 2011, 3: 195-208.
Gherna RL, Werren JH, Weisburg W, Cote R, Woese CR, Mandelco L, Brenner DJ: Arsenophonus nasoniae gen. nov, sp. nov, the causative agent of the son-killer trait in the parasitic wasp Nasonia vitripennis. Int J Syst Bacteriol. 1991, 41 (4): 563-565.
Dale C, Beeton M, Harbison C, Jones T, Pontes M: Isolation, pure culture, and characterization of "Candidatus Arsenophonus arthropodicus, " an intracellular secondary endosymbiont from the hippoboscid louse fly Pseudolynchia canariensis. Appl Environ Microbiol. 2006, 72 (4): 2997-3004.
Verbarg S, Fruhling A, Cousin S, Brambilla E, Gronow S, Lunsdorf H, Stackebrandt E: Biostraticola tofi gen. nov., spec. nov., a novel member of the family Enterobacteriaceae. Curr Microbiol. 2008, 56 (6): 603-608.
Blanquart S, Lartillot N: A site- and time-heterogeneous model of amino acid replacement. Mol Biol Evol. 2008, 25 (5): 842-858.
Blanquart S, Lartillot N: A Bayesian compound stochastic process for modeling nonstationary and nonhomogeneous sequence evolution. Mol Biol Evol. 2006, 23 (11): 2058-2071.
Foster PG: Modeling compositional heterogeneity. Syst Biol. 2004, 53 (3): 485-495.
Kono M, Koga R, Shimada M, Fukatsu T: Infection dynamics of coexisting Beta- and Gammaproteobacteria in the nested endosymbiotic system of mealybugs. Appl Environ Microbiol. 2008, 74 (13): 4175-4184.
Thao ML, Clark MA, Baumann L, Brennan EB, Moran NA, Baumann P: Secondary endosymbionts of psyllids have been acquired multiple times. Curr Microbiol. 2000, 41 (4): 300-304.
Kolsch G, Matz-Grund C, Pedersen BV: Ultrastructural and molecular characterization of endosymbionts of the reed beetle genus Macroplea (Chrysomelidae, Donaciinae), and proposal of "Candidatus Macropleicola appendiculatae" and "Candidatus Macropleicola muticae". Can J Microbiol. 2009, 55 (11): 1250-1260.
Kolsch G, Pedersen BV: Can the tight co-speciation between reed beetles (Col., Chrysomelidae, Donaciinae) and their bacterial endosymbionts, which provide cocoon material, clarify the deeper phylogeny of the hosts?. Mol Phylogenet Evol. 2010, 54 (3): 810-821.
Kuchler SM, Dettner K, Kehl S: Molecular characterization and localization of the obligate endosymbiotic bacterium in the birch catkin bug Kleidocerys resedae (Heteroptera Lygaeidae, Ischnorhynchinae). FEMS Microbiol Ecol. 2010, 73 (2): 408-418.
Kuechler SM, Dettner K, Kehl S: Characterization of an obligate intracellular bacterium in the midgut epithelium of the bulrush bug Chilacis typhae (Heteroptera, Lygaeidae, Artheneinae). Appl Environ Microbiol. 2011, 77 (9): 2869-2876.
Kikuchi Y, Fukatsu T: Endosymbiotic bacteria in the esophageal organ of glossiphoniid leeches. Appl Environ Microbiol. 2002, 68 (9): 4637-4641.
Perkins SL, Budinoff RB, Siddall ME: New Gammaproteobacteria associated with blood-feeding leeches and a broad phylogenetic analysis of leech endosymbionts. Appl Environ Microbiol. 2005, 71 (9): 5219-5224.
Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M: The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006, 314 (5797): 267-
Thao ML, Baumann P: Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl Environ Microbiol. 2004, 70 (6): 3401-3406.
Burke GR, Normark BB, Favret C, Moran NA: Evolution and diversity of facultative symbionts from the aphid subfamily Lachninae. Appl Environ Microbiol. 2009, 75 (16): 5328-5335.
Tatusov RL, Koonin EV, Lipman DJ: A genomic perspective on protein families. Science. 1997, 278 (5338): 631-637.
Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA: The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003, 4: 41-
Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, Dubchak IL, Alm EJ, Arkin AP: MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 2010, D396-D400. 38 Database
Gouy M, Guindon S, Gascuel O: SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010, 27 (2): 221-224.
Katoh K, Toh H: Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008, 9 (4): 286-298.
Talavera G, Castresana J: Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007, 56 (4): 564-577.
Castresana J: Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000, 17 (4): 540-552.
Hall TA: BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999, 41: 95-98.
Lartillot N, Lepage T, Blanquart S: PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009, 25 (17): 2286-2288.
Boussau B, Gouy M: Efficient likelihood computations with nonreversible models of evolution. Syst Biol. 2006, 55 (5): 756-768.
Galtier N, Gouy M: Inferring pattern and process: maximum-likelihood implementation of a nonhomogeneous model of DNA sequence evolution for phylogenetic analysis. Mol Biol Evol. 1998, 15 (7): 871-879.
Galtier N, Gouy M: Inferring phylogenies from DNA sequences of unequal base compositions. Proc Natl Acad Sci USA. 1995, 92 (24): 11317-11321.
Tarrio R, Rodriguez-Trelles F, Ayala FJ: Shared nucleotide composition biases among species and their impact on phylogenetic reconstructions of the Drosophilidae. Mol Biol Evol. 2001, 18 (8): 1464-1473.
Galtier N, Tourasse N, Gouy M: A nonhyperthermophilic common ancestor to extant life forms. Science. 1999, 283 (5399): 220-221.
Shimodaira H, Hasegawa M: CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001, 17 (12): 1246-1247.
Swofford DL: PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. 2002, Sunderland, Massachusetts: Sinauer Associates
Kostka M, Uzlíková M, Čepička I, Flegr J: SlowFaster, a user-friendly program for slow-fast analysis and its application on phylogeny of Blastocystis. BMC Bioinformatics. 2008, 9: 341-
Guindon S, Gascuel O: A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003, 52 (5): 696-704.
Ronquist F, Huelsenbeck JP: MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003, 19 (12): 1572-1574.
Nylander JA, Wilgenbusch JC, Warren DL, Swofford DL: AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008, 24 (4): 581-583.
Rambaut A, Drummond AJ: Tracer v1.5. 2009, [http://tree.bio.ed.ac.uk/software/tracer/]
Abascal F, Zardoya R, Posada D: ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005, 21 (9): 2104-2105.
Posada D: jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008, 25 (7): 1253-1256.
Tavaré S: Some probabilistic and statistical problems in the analysis of DNA sequences. Lect Math Life Sci. 1986, 17: 57-86.
Lanave C, Preparata G, Saccone C, Serio G: A new method for calculating evolutionary substitution rates. J Mol Evol. 1984, 20 (1): 86-93.
Yang Z: Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994, 39 (3): 306-314.
Posada D: Using MODELTEST and PAUP* to select a model of nucleotide substitution. Curr Protoc Bioinformatics. 2003, 6.5.1-6.5.14.
Le SQ, Gascuel O: An improved general amino acid replacement matrix. Mol Biol Evol. 2008, 25 (7): 1307-1320.
Whelan S, Goldman N: A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001, 18 (5): 691-699.
Smyth P: Model selection for probabilistic clustering using cross-validated likelihood. Stat Comput. 2000, 10 (1): 63-72.
Acknowledgements
This work was supported by the Grant Agency of Academy of Sciences of the Czech Republic (grant number P505/10/1401 to VH); the Student Grant Agency of the Faculty of Science, University of South Bohemia (grant number SGA2009002 to FH); and the Ministry of Education, Youth, and Sports of the Czech Republic (grant numbers LC06073 and MSM 60076605801 to VH). Access to the MetaCentrum computing facilities provided under the 'Projects of Large Infrastructure for Research, Development, and Innovations' program LM2010005 funded by the Ministry of Education, Youth, and Sports of the Czech Republic is highly appreciated.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
FH carried out the sequence alignments and phylogenetic analyses, and participated in the study design, evolutionary interpretation of the results and preparation of the manuscript. TCH compiled and analyzed the AT/GC reduced matrices. VH conceived of the study and participated in its design, evolutionary interpretation of the results and preparation of the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12915_2011_516_MOESM3_ESM.RAR
Additional file 3: All phylogenetic trees derived from AT-GC and SF datasets. A rar file of all phylogenetic trees obtained under BI, ML and MP from 11 AT/GC datasets, and under ML from five slow-fasted datasets. Trees are in phylip and nexus formats and can be viewed, for example, in TreeView http://taxonomy.zoology.gla.ac.uk/rod/treeview.html or Mesquite http://mesquiteproject.org/mesquite/mesquite.html. (RAR 34 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Husník, F., Chrudimský, T. & Hypša, V. Multiple origins of endosymbiosis within the Enterobacteriaceae (γ-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol 9, 87 (2011). https://doi.org/10.1186/1741-7007-9-87
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1741-7007-9-87