Abstract
Background
The phylogeography of the Y chromosome in Asia previously suggested that modern humans of African origin initially settled in mainland southern East Asia, and about 25,000–30,000 years ago, migrated northward, spreading throughout East Asia. However, the fragmented distribution of one East Asian specific Y chromosome lineage (D-M174), which is found at high frequencies only in Tibet, Japan and the Andaman Islands, is inconsistent with this scenario.
Results
In this study, we collected more than 5,000 male samples from 73 East Asian populations and reconstructed the phylogeography of the D-M174 lineage. Our results suggest that D-M174 represents an extremely ancient lineage of modern humans in East Asia, and a deep divergence was observed between northern and southern populations.
Conclusion
We proposed that D-M174 has a southern origin and its northward expansion occurred about 60,000 years ago, predating the northward migration of other major East Asian lineages. The Neolithic expansion of Han culture and the last glacial maximum are likely the key factors leading to the current relic distribution of D-M174 in East Asia. The Tibetan and Japanese populations are the admixture of two ancient populations represented by two major East Asian specific Y chromosome lineages, the O and D haplogroups.
Similar content being viewed by others
Introduction
The Y chromosome Alu polymorphism (YAP, also called M1) defines the deep-rooted haplogroup D/E of the global Y-chromosome phylogeny [1]. This D/E haplogroup is further branched into three sub-haplogroups DE*, D and E (Figure 1). The distribution of the D/E haplogroup is highly regional, and the three subgroups are geographically restricted to certain areas, therefore informative in tracing human prehistory (Table 1). The sub-haplogroup DE*, presumably the most ancient lineage of the D/E haplogroup was only found in Africans from Nigeria [2], supporting the "Out of Africa" hypothesis about modern human origin. The sub-haplogroup E (E-M40), defined by M40/SRY4064 and M96, was also suggested originated in Africa [3–6], and later dispersed to Middle East and Europe about 20,000 years ago [3, 4]. Interestingly, the sub-haplogroup D defined by M174 (D-M174) is East Asian specific with abundant appearance in Tibetan and Japanese (30–40%), but rare in most of other East Asian populations and populations from regions bordering East Asia (Central Asia, North Asia and Middle East) (usually less than 5%) [5–7]. Under D-M174, Japanese belongs to a separate sub-lineage defined by several mutations (e.g. M55, M57 and M64 etc.), which is different from those in Tibetans implicating relatively deep divergence between them [1]. The fragmented distribution of D-M174 in East Asia seems not consistent with the pattern of other East Asian specific lineages, i.e. O3-M122, O1-M119 and O2-M95 under haplogroup O [8, 9].
Besides Tibetans and Japanese, D-M174 is also prevalent in several southern ethnic populations in East Asia, including the Tibeto-Burman speaking populations from Yunnan province of southwestern China (14.0–72.3%), one Hmong-Mien population from Guangxi of southern China (30%) and one Daic population from Thailand (10%), which could be explained by fairly recent population admixture [9–11]. However, a recent study reported a high frequency of D-M174 in Andamanese (56.25%), people who live in the remote islands in the Indian Ocean and considered one of the earliest modern human settlers of African origin in Southeast Asia [12]. Another study by Underhill et al. suggested that the D-M174 lineage likely reached East Asia about 50,000 years ago [5]. This implies that the YAP lineage in East Asia could be indeed very ancient.
Our previous studies showed that the dominant and East Asian specific Y haplogroup O-M175 (44.3% on average) reflected possibly the earliest modern human expansion in East Asia [8, 9, 13]. Unlike the prevalence of O-M175 in most of the East Asian populations, populations with relatively high frequencies of D-M174 are mostly located at the peripheral regions of mainland East Asia with fragmented distribution [7, 9–11, 13–18], implying two possible explanations of human prehistory. Firstly, like O-M175, D-M174 may also be just one of the lineages going northward during the suggested Paleolithic migration of modern humans in East Asia [8, 9]. Subsequently, due to population substructure (the last glacial is likely a key factor) and recent expansion of Han culture [18], the distribution of D-M174 was fragmented into the current geographic pattern. The other possible scenario would suggest an independent earlier migration different from the one we proposed previously [8, 9]. To address this question, we conducted a systematic sampling and genetic analysis of more than 5,000 male individuals from 73 East Asian and Southeast Asian populations. Based on the Y chromosome SNP and STR data and the estimated ages of the major D-M174 lineages, we proposed that there was an independent Paleolithic northward migration of modern humans in East Asia, predating the previously suggested northward population movement [8, 9, 13, 19–21].
Methods
Samples
In this study, a total of 5,134 unrelated male samples were collected from 73 populations, covering the major geographic regions in East Asia and Southeast Asia (Table 2 and Figure 2). As shown in Figure 2, most populations were sampled from southern and southwestern China where about 80% Chinese ethnic populations live with inhabited histories longer than 3,000 years [22]. Samples from previous studies were also included, including 91 YAP+ samples (16 Japanese, 54 Tibetans, 3 Koreans, 1 Guam, 1 Cambodian, 4 Thais and 12 samples of China) from Su et al. [7, 9, 17] and 116 YAP+ samples (35 Africans, 13 Caucasians, 9 Indians, 26 Middle East and 33 Central Asians) from Wells et al. [23](Table 2).
Y Chromosome Markers and Genotyping
Initially all the samples were genotyped for three Y chromosome specific biallelic markers, including M1, M40 and M174. The samples belonging to sub-haplogroup D (D-M174) were subject to further typing of M15, M57 and p47 in order to designate the sub-clades. The sub-haplogroups were named by the characteristic mutations according to the Y-Chromosome-Consortium (2002). The PCR electrophoresis, PCR-RFLP and sequencing were used for genotyping [7]. The phylogenetic relationship of the Y biallelic markers is shown in Figure 1.
We typed the YAP locus in the 5,134 samples, of which 512 YAP+ were detected (9.97%), plus the previously published 207 Yap+ samples, and a total 719 Yap+ samples were then typed for the five biallelic markers (M174, M40, M15, M57 and p47) and eight STRs (DYS19/394, DYS388, DYS 389I, DYS389II, DYS 390, DYS 391, DYS 392 and DYS 393). Among the 719 YAP+, a total of 697 samples generated complete sets of allele counts for all the SNPs and STRs (data in the Additional files).
Data Analysis
For data analysis, we included the YAP+ samples from the published data on 90 Japanese and 44 Tibetans from Hammer et al. (2006), 19 Andamanese from Thangaraj et al. (2003) and 6 Nigerians from Weale et al. (2003). The divergence times between the sub-clades of D-M174 were estimated using the STR data following the SNP-STR coalescence method [4, 24, 25]. The average mutation rate of the Y-STRs tested is 0.00069 [26]. The network of the Y-STR haplotypes were constructed for each D-M174 sub-haplogroup using NETWORK4.2.0.1 http://www.fluxus-engineering.com, and then superimposed onto the established phylogeny of the D-M174 lineage (Figure 1). The average gene diversity of the populations was calculated with the use of the allele frequencies of the eight tested STR loci (Arlequin 3.0, http://lgb.unige.ch/arlequin/).
Results
Table 1 lists the reported YAP+ frequencies in worldwide populations (refer to table note for references). Africans have the highest frequency of YAP+, and all of them belong to the sub-haplogroup E-M40. In contrast, D-M174 is in general East Asian specific with sporadic occurrence in adjacent regions, i.e. Central Asia, Middle East and Northeast India. The average frequency of D-M174 in East Asians is 9.60% with high frequencies in Tibet (41.31%), Japan (35.08%) and Andaman Island (56.25%), but rare in other East Asian populations (< 5%). After genotyping the Y chromosome biallelic markers (M174, M40, M15, M57 and p47), the 719 Yap+ samples were assigned to 6 sub-haplogroups, i.e. DE*, E-M40, D*-M174, D1-M15, D2-M57 and D3-p47 (Figure 1). Further typing for eight STR loci of the 719 YAP+ generated the complete STR data sets for 697 samples. As shown in Table 2, consistent with previous reports [7, 9–11, 13, 16], the prevalence of D-M174 is mostly in western and southern China and Japan.
The distribution patterns of the four D-M174 sub-lineages (sub-haplogroups) (Figure 1) are different from each other. D1-M15 is widely distributed across East Asia including most of the Tibeto-Burman and Daic speaking populations (Table 2). D*-M174 and D3-p47 are mainly distributed in Tibeto-Burman populations with sporadic occurrence in the Daic populations. In surprise, we observed two DE* in the Tibetan samples, which was previously only observed in Africa (Nigerians), but not in other world populations. In contrast, D2-M57 only occurred in Japanese, an implication of the early divergence of this lineage from other D-M174 sub-haplogroups (Table 2). We identified four E-M40 individuals in the northwestern Han populations, a reflection of recent gene flow from Central Asia [23].
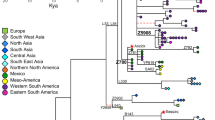
To reveal the detailed structure of the D-M174 lineage in East Asian populations, we conducted the network analysis combining the SNP and STR haplotype data (Figure 3). D*-M174 has a deep structure with no loops in the network. The D*-M174 lineage contains distinct STR haplotypes of Tibeto-Burman (mostly Tibetan), Daic and Andamanese respectively, and no haplotype sharing (i.e. shared by individuals from different geographic/linguistic populations) was observed, implying that D*-M174 is an ancient lineage. As the most common lineage, the network of D1-M15 is also deep-structured and a clear south vs. north divergence can be inferred though sporadic haplotype sharing exists. In contrast, D2-M57 is restricted to Japan and D3-p47 is prevalent in Tibet and its adjacent regions with sporadic appearance in Central Asian and Daic populations. The short-distanced and star-like network structure of these two sub-haplogroups indicates a long-term local existence and population expansion of the D-M174 lineage in these two geographically far away regions. The non-Tibetan Tibeto-Burman populations, e.g. Naxi, Pumi and Qiang only have a subset of the Tibetan haplotypes, again indicating a recent gene flow from Tibet as recorded in the literature [7, 22].
We next sought to estimate the age of the D-M174 sub-lineages. The coalescence analysis shows that the age of D*-M174, D3-p47 and D1-M15 are the oldest (66,392 ± 1,466, 52,103 ± 1,327 and 51,640 ± 2,563 years). The age of the Japanese specific lineage D2-M57 is the youngest (37,678 ± 2,216 years) (Table 3). Notably, these estimated ages are much older than that of O3-M122, the other East Asian specific haplogroup we reported before (25,000–30,000 years) [8].
Discussion
The biased distribution of D-M174 bears the different possible inferences of the human population prehistory in view of the origin and migratory pattern in East Asia. The hypothesis of northern origin of D-M174 is not supported by our data because D-M174 is rare in Central Asian populations (Table 1) and the few Central Asian D-M174s are all located at the peripheral positions of the Y-STR network (Figure 3). Our data also disapproves the notion of Indian origin since no D-M174 was detected in the 996 individuals across India. The sporadic occurrence of D-M174 in northeastern Indians (two D-M174s in 232 individuals tested) is because those populations are in fact Sino-Tibetan speaking populations (Table 1). The lack of gene flow between Tibet and India is likely due to the efficient geographic separation by the Himalayas [27].
On the other hand, the aborigines living on Andaman Island are genetically isolated in view of their Y chromosome haplotypes. Though phenotypically different from other Southeast Asian populations, the Andaman Islanders posses most of the major East Asian specific Y chromosome lineages including D-M174, O3-M122 and O2-M95, a strong indication of a relic Paleolithic population [28]. Also, the Daic and Hmong-Mien speaking populations are ancient southern populations in view of linguistic and archaeological evidences [22]. The network analysis indicates a clear divergence of D1-M15 between northern (Tibeto-Burman) and southern (Daic and Hmong) populations (Figure 3). Hence, the alternative explanation of northern origin of D-M174 is unlikely considering the absence of YAP+ in North Asia [29] and the sporadic appearance of D-M174 in Central Asia [23]. Consequently, the southern origin of D-M174 can be established, which is consistent with the proposed initial settlement of modern humans in mainland Southeast Asia and the migration pattern of other Y chromosome lineages [8, 9, 13].
There were studies arguing against the southern origin of East Asian, in which higher gene diversity was observed in northern populations compared with southern populations [14, 30]. As we discussed in our previous report, the data from Karafet et al. [14] suggested a false impression of the high diversity of northern populations without taking the recent admixture from Central Asia into consideration [8]. The study of Xue et al. [30] has the similar drawback though both Y-SNP and Y-STR data were used. In Xue et al (2006), the high gene diversity was claimed for Mongolian, Uygure and Manchurian, and all of them have recorded recent extensive admixture with Central Asian and Han Chinese populations [22]. In addition, the southern populations studied in Xue et al. (2006) were limited and the within population bottleneck effect caused by long-time geographic isolation might have a great impact on gene diversity estimation. When plenty southern populations were studied, we observed a higher diversity in those populations compared with the northern populations [8, 9].
The gene diversity based on the STR data in the southern populations is comparable with those in the northern populations. Tibetan has the highest diversity (0.525 ± 0.294), followed by Daic (0.484 ± 0.272), Japanese (0.419 ± 0.239) and Hmong Mien (0.347 ± 0.206). Gene diversity of other East Asian populations was not calculated due to small sample size. The higher diversity in Tibetan is largely due to the much larger effective population size of D-M174 in Tibetan compared with other populations. Tibetan and Japanese lived in two geographically far away regions and their D-M174 lineages belong to two different sub-haplogroups. These two sub-haplogroups all have a short-distanced and star-like network structure, which indicates a long-term local existence and recent population expansion (Figure 3). It should be noted that the estimation of gene diversity is subject to potential bias, e.g. the age difference of the D-M174 sub-lineages. The finding of two DE* in Tibet, which was only observed in Africa, supports the antiquity of D-M174 and suggests that the D-M174 lineage is among the earliest modern human settlers in East Asia. Additionally, the biased distribution of D-M174 and its ancient coalescent time suggests an independent Paleolithic migration of modern humans in East Asia.
Our previous data on the O3-M122 lineage suggested a prehistorically northward migration (about 25,000–30,000 years ago) of modern humans in East Asia, which explains the current phylogeography of the major East Asian specific Y chromosome lineages (O3-M122, O2-M95 and O1-M119) [8, 9, 13]. However, the data on D-M174 cannot be explained by the hypothesized migration pattern. Firstly, D-M174 is rare in the central part of eastern Asia, especially in Han Chinese populations. Though this could be explained by genetic drift, assuming D-M174 moving along with O3-M122 during the proposed northward migration, the prevalence of D-M174 in Tibet and Japan requires recurrent mutations or independent random enrichment of D-M174, which is unlikely. An independent earlier northward migration is more reasonable in explaining the current distribution of D-M174 in East Asia. We speculate that due to the later northward migration of O3-M122 and the Neolithic expansion of Han Chinese, the trace of the D-M174 migration in the heartland of East Asia was wiped out by the later and likely much larger migration of O3-M122. The current peripheral distribution pattern of D-M174 in East Asia is consistent with the proposed notion. Also, the age estimation supports that the northward migration of D-M174 may predate the migration of O3-M122.
The East African megadroughts (about 135–75 thousand years ago) during the early late-Pleistocene was suggested compelling modern humans out of Africa [31]. And the early modern human could occupy coastal areas and exploited the near-shore marine food resource by that time [32]. Then, modern humans was suggested expanding along the tropical coast, and the earliest modern human fossil found out of Africa was about 100,000 years ago [33]. The period of 80,000–10,000 years ago during the last glacial might have a huge impact on modern human migration, and the sea level had fallen 50–200 meters below present [34], which resulted in larger dry lands and possibility for human migration between currently separated lands by ocean, e.g. between Japan and the mainland.
Human fossil records and previous genetic data suggested that the earliest modern human settlement in East Asia likely occurred less than 60,000 years ago [8, 9, 13, 21, 35]. For example, the oldest Australian fossil (Lake Mungo 3) was dated about 45,000 ± 3,000 – 62,000 ± 6,000 years ago [36, 37], and the mitochondrial DNA and Y chromosome analysis of current Australian ethnic populations suggested the colonization of Australia about 50,000–70,000 years ago [38]. Our age estimation of the D-M174 lineages is consistent with this notion though there might be independent migrations of modern humans into East Asia and into Australia [38].
The estimated ages of the D-M174 lineages are older than those previously reported based on both Y chromosome and mtDNA variations in East Asia [8, 9, 21]. To see whether it is over-estimated, using the same method, we calculated the divergence time between DE* and E-M40. The estimated age is 27,176 years, which is much younger than the D-M174 lineage, but consistent with the previous estimation (27,800–37,000 years ago) [3]. Hence, the antiquity of D-M174 likely reflects the true prehistory of human populations in East Asia. The age estimation model developed by Zhivotovsky (2001) is not sensitive to effective population size and recent population expansion though the effect of population substructure cannot be totally ruled out. The antiquity of D-M174 was also supported by a previous study in which the origin of D-M174 was estimated more than 50,000 years ago [5].
The divergence time of haplogroup D is about 60,000 years ago, considering the wide though fragmented geographic distribution of D-M174, the proposed Paleolithic migration would be the first northward population movement of modern humans after their initial settlement in southern East Asia. As the last glacial occurred during 80,000–10,000 years ago, the northward migration of D-M174 is consistent with the proposed notion that modern humans might exploit the food of "Mammoth Steppe" [39]. Besides the later population expansion, the cold weather during the last glacial may also contribute to the current fragmented distribution of D-M174. Interestingly, a recent archaeological finding supported that modern humans explored the Tibetan plateau about 30,000–40,000 years ago, which is much earlier than previously suggested [40], but consistent with our hypothesis. The after-glacial sea level rise eventually led to the separation between Japan and the main continent, which explains the relic distribution of D-M174 in current Japanese populations. The archaeological data suggested that the initial colonization of modern humans in Japan occurred about 30,000 years ago [41, 42], consistent with our age estimation of D2-M57 (37,678 ± 2,216 years ago). Taken together, the current Tibetan and Japanese populations are probably the admixture of two ancient populations represented by D-M174 and O3-M122 respectively [7, 10, 16].
Conclusion
In summary, we demonstrated an ancient Paleolithic population migration in East Asia, predating the previously suggested northward population movement. The current fragmented distribution of D-M174 is likely due to the combination of later Neolithic population expansion and the last glacial.
References
Y-Chromosome-Consortium: A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002, 12 (2): 339-348. 10.1101/gr.217602.
Weale ME, Shah T, Jones AL, Greenhalgh J, Wilson JF, Nymadawa P, Zeitlin D, Connell BA, Bradman N, Thomas MG: Rare deep-rooting Y chromosome lineages in humans: lessons for phylogeography. Genetics. 2003, 165 (1): 229-234.
Cruciani F, La Fratta R, Trombetta B, Santolamazza P, Sellitto D, Colomb EB, Dugoujon JM, Crivellaro F, Benincasa T, Pascone R, et al: Tracing past human male movements in northern/eastern Africa and Western eurasia: new clues from y-chromosomal haplogroups e-m78 and j-m12. Mol Biol Evol. 2007, 24 (6): 1300-1311. 10.1093/molbev/msm049.
Semino O, Magri C, Benuzzi G, Lin AA, Al-Zahery N, Battaglia V, Maccioni L, Triantaphyllidis C, Shen P, Oefner PJ, et al: Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet. 2004, 74 (5): 1023-1034. 10.1086/386295.
Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley RA, Oefner PJ, Cavalli-Sforza LL: The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet. 2001, 65 (Pt 1): 43-62. 10.1046/j.1469-1809.2001.6510043.x.
Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, et al: Y chromosome sequence variation and the history of human populations. Nat Genet. 2000, 26 (3): 358-361. 10.1038/81685.
Su B, Xiao C, Deka R, Seielstad MT, Kangwanpong D, Xiao J, Lu D, Underhill P, Cavalli-Sforza L, Chakraborty R, et al: Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum Genet. 2000, 107 (6): 582-590. 10.1007/s004390000406.
Shi H, Dong YL, Wen B, Xiao CJ, Underhill PA, Shen PD, Chakraborty R, Jin L, Su B: Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122. Am J Hum Genet. 2005, 77 (3): 408-419. 10.1086/444436.
Su B, Xiao J, Underhill P, Deka R, Zhang W, Akey J, Huang W, Shen D, Lu D, Luo J, et al: Y-Chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am J Hum Genet. 1999, 65 (6): 1718-1724. 10.1086/302680.
Hammer MF, Karafet TM, Park H, Omoto K, Harihara S, Stoneking M, Horai S: Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes. J Hum Genet. 2006, 51 (1): 47-58. 10.1007/s10038-005-0322-0.
Wen B, Xie X, Gao S, Li H, Shi H, Song X, Qian T, Xiao C, Jin J, Su B, et al: Analyses of genetic structure of Tibeto-Burman populations reveals sex-biased admixture in southern Tibeto-Burmans. Am J Hum Genet. 2004, 74 (5): 856-865. 10.1086/386292.
Thangaraj K, Chaubey G, Kivisild T, Reddy AG, Singh VK, Rasalkar AA, Singh L: Reconstructing the origin of Andaman Islanders. Science. 2005, 308 (5724): 996-10.1126/science.1109987.
Jin L, Su B: Natives or immigrants: modern human origin in east Asia. Nat Rev Genet. 2000, 1 (2): 126-133. 10.1038/35038565.
Karafet T, Xu L, Du R, Wang W, Feng S, Wells RS, Redd AJ, Zegura SL, Hammer MF: Paternal population history of East Asia: sources, patterns, and microevolutionary processes. Am J Hum Genet. 2001, 69 (3): 615-628. 10.1086/323299.
Ke Y, Su B, Song X, Lu D, Chen L, Li H, Qi C, Marzuki S, Deka R, Underhill P, et al: African origin of modern humans in East Asia: a tale of 12,000 Y chromosomes. Science. 2001, 292 (5519): 1151-1153. 10.1126/science.1060011.
Qian Y, Qian B, Su B, Yu J, Ke Y, Chu Z, Shi L, Lu D, Chu J, Jin L: Multiple origins of Tibetan Y chromosomes. Hum Genet. 2000, 106 (4): 453-454. 10.1007/s004390000259.
Su B, Jin L, Underhill P, Martinson J, Saha N, McGarvey ST, Shriver MD, Chu J, Oefner P, Chakraborty R, et al: Polynesian origins: insights from the Y chromosome. Proc Natl Acad Sci USA. 2000, 97 (15): 8225-8228. 10.1073/pnas.97.15.8225.
Wen B, Li H, Lu D, Song X, Zhang F, He Y, Li F, Gao Y, Mao X, Zhang L, et al: Genetic evidence supports demic diffusion of Han culture. Nature. 2004, 431 (7006): 302-305. 10.1038/nature02878.
Ballinger SW, Schurr TG, Torroni A, Gan YY, Hodge JA, Hassan K, Chen KH, Wallace DC: Southeast Asian mitochondrial DNA analysis reveals genetic continuity of ancient mongoloid migrations. Genetics. 1992, 130 (1): 139-152.
Chu JY, Huang W, Kuang SQ, Wang JM, Xu JJ, Chu ZT, Yang ZQ, Lin KQ, Li P, Wu M, et al: Genetic relationship of populations in China. Proc Natl Acad Sci USA. 1998, 95 (20): 11763-11768. 10.1073/pnas.95.20.11763.
Yao YG, Kong QP, Bandelt HJ, Kivisild T, Zhang YP: Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet. 2002, 70 (3): 635-651. 10.1086/338999.
Wang ZH: History of nationalities in China. 1994, Beijin(in Chinese): China Social Science Press
Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, Su B, Pitchappan R, Shanmugalakshmi S, et al: The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA. 2001, 98 (18): 10244-10249. 10.1073/pnas.171305098.
Rootsi S, Magri C, Kivisild T, Benuzzi G, Help H, Bermisheva M, Kutuev I, Barac L, Pericic M, Balanovsky O, et al: Phylogeography of Y-chromosome haplogroup I reveals distinct domains of prehistoric gene flow in europe. Am J Hum Genet. 2004, 75 (1): 128-137. 10.1086/422196.
Zhivotovsky LA: Estimating divergence time with the use of microsatellite genetic distances: impacts of population growth and gene flow. Mol Biol Evol. 2001, 18 (5): 700-709.
Zhivotovsky LA, Underhill PA, Cinnioglu C, Kayser M, Morar B, Kivisild T, Scozzari R, Cruciani F, Destro-Bisol G, Spedini G, et al: The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet. 2004, 74 (1): 50-61. 10.1086/380911.
Cordaux R, Weiss G, Saha N, Stoneking M: The northeast Indian passageway: a barrier or corridor for human migrations?. Mol Biol Evol. 2004, 21 (8): 1525-1533. 10.1093/molbev/msh151.
Thangaraj K, Singh L, Reddy AG, Rao VR, Sehgal SC, Underhill PA, Pierson M, Frame IG, Hagelberg E: Genetic affinities of the Andaman Islanders, a vanishing human population. Curr Biol. 2003, 13 (2): 86-93. 10.1016/S0960-9822(02)01336-2.
Lell JT, Sukernik RI, Starikovskaya YB, Su B, Jin L, Schurr TG, Underhill PA, Wallace DC: The dual origin and Siberian affinities of Native American Y chromosomes. Am J Hum Genet. 2002, 70 (1): 192-206. 10.1086/338457.
Xue Y, Zerjal T, Bao W, Zhu S, Shu Q, Xu J, Du R, Fu S, Li P, Hurles ME, et al: Male demography in East Asia: a north-south contrast in human population expansion times. Genetics. 2006, 172 (4): 2431-2439. 10.1534/genetics.105.054270.
Scholz CA, Johnson TC, Cohen AS, King JW, Peck JA, Overpeck JT, Talbot MR, Brown ET, Kalindekafe L, Amoako PY, et al: East African megadroughts between 135 and 75 thousand years ago and bearing on early-modern human origins. Proc Natl Acad Sci USA. 2007, 104 (42): 16416-16421. 10.1073/pnas.0703874104.
Walter RC, Buffler RT, Bruggemann JH, Guillaume MM, Berhe SM, Negassi B, Libsekal Y, Cheng H, Edwards RL, von Cosel R, et al: Early human occupation of the Red Sea coast of Eritrea during the last interglacial. Nature. 2000, 405 (6782): 65-69. 10.1038/35011048.
Forster P, Matsumura S: Evolution. Did early humans go north or south?. Science. 2005, 308 (5724): 965-966. 10.1126/science.1113261.
Lambeck K, Chappell J: Sea level change through the last glacial cycle. Science. 2001, 292 (5517): 679-686. 10.1126/science.1059549.
Wu HC, Poirier FE, Wu XZ: Human Evolution in China: A Metric Description of the Fossils and a Review of the Sites. 1995, Oxford: Oxford Univ Press
Bowler JM, Johnston H, Olley JM, Prescott JR, Roberts RG, Shawcross W, Spooner NA: New ages for human occupation and climatic change at Lake Mungo, Australia. Nature. 2003, 421 (6925): 837-840. 10.1038/nature01383.
Thorne A, Grun R, Mortimer G, Spooner NA, Simpson JJ, McCulloch M, Taylor L, Curnoe D: Australia's oldest human remains: age of the Lake Mungo 3 skeleton. J Hum Evol. 1999, 36 (6): 591-612. 10.1006/jhev.1999.0305.
Hudjashov G, Kivisild T, Underhill PA, Endicott P, Sanchez JJ, Lin AA, Shen P, Oefner P, Renfrew C, Villems R, et al: Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis. Proc Natl Acad Sci USA. 2007, 104 (21): 8726-8730. 10.1073/pnas.0702928104.
Guthrie RD: Frozen fauna of the Mammoth Steppe. 1990, Chicago: University of Chicago Press
Yan YB, Huang WW, Zhang D: The modern human explored the northern Tibet plateau at the late Pleistocene. Chinese Science Bulletin (in Chinese). 2007, 52 (13): 1567-1571.
Ono A, Sato H, Tsutsumi T, Kudo Y: Radiocarbon dates and archaeology of the Late Pleistocene in the Japanese islands. Radiocarbon. 2002, 44: 477-494.
Takamiya H: Peopling of Western Japan, focusing on Kyushu, and Ryukyu Archipelago. Radiocarbon. 2002, 44: 495-502.
Cinnioglu C, King R, Kivisild T, Kalfoglu E, Atasoy S, Cavalleri GL, Lillie AS, Roseman CC, Lin AA, Prince K, et al: Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet. 2004, 114 (2): 127-148. 10.1007/s00439-003-1031-4.
Cruciani F, Santolamazza P, Shen P, Macaulay V, Moral P, Olckers A, Modiano D, Holmes S, Destro-Bisol G, Coia V, et al: A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet. 2002, 70 (5): 1197-1214. 10.1086/340257.
Hammer MF, Karafet T, Rasanayagam A, Wood ET, Altheide TK, Jenkins T, Griffiths RC, Templeton AR, Zegura SL: Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol Biol Evol. 1998, 15 (4): 427-441.
Jin HJ, Kwak KD, Hammer MF, Nakahori Y, Shinka T, Lee JW, Jin F, Jia X, Tyler-Smith C, Kim W: Y-chromosomal DNA haplogroups and their implications for the dual origins of the Koreans. Hum Genet. 2003, 114 (1): 27-35. 10.1007/s00439-003-1019-0.
Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, Metspalu E, Adojaan M, Tolk HV, Stepanov V, et al: The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet. 2003, 72 (2): 313-332. 10.1086/346068.
Ramana GV, Su B, Jin L, Singh L, Wang N, Underhill P, Chakraborty R: Y-chromosome SNP haplotypes suggest evidence of gene flow among caste, tribe, and the migrant Siddi populations of Andhra Pradesh, South India. Eur J Hum Genet. 2001, 9 (9): 695-700. 10.1038/sj.ejhg.5200708.
Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, et al: The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science. 2000, 290 (5494): 1155-1159. 10.1126/science.290.5494.1155.
Zerjal T, Wells RS, Yuldasheva N, Ruzibakiev R, Tyler-Smith C: A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia. Am J Hum Genet. 2002, 71 (3): 466-482. 10.1086/342096.
Acknowledgements
We are grateful to people who donated their DNA samples in this study. We thank Hui Zhang and Yi-Chuan Yu for their technical help. This study was supported by grants from the National 973 project of China (2006CB701506, 2007CB815705), the Chinese Academy of Sciences (KSCX1-YW-R-34), the National Natural Science Foundation of China (30700445, 30771181, 30370755, 30525028 and 30630013), and the Natural Science Foundation of Yunnan Province of China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
BS and LJ designed research. BS and HS analyzed data and wrote the paper. HS, HZ, YP, Y-LD and X-BQ performed research. FZ, LF-L, S-JT, R-LM, C-JX and SW provided some research samples.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Shi, H., Zhong, H., Peng, Y. et al. Y chromosome evidence of earliest modern human settlement in East Asia and multiple origins of Tibetan and Japanese populations . BMC Biol 6, 45 (2008). https://doi.org/10.1186/1741-7007-6-45
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1741-7007-6-45