Abstract
Cytokines play a very important role in nearly all aspects of inflammation and immunity. The term 'interleukin' (IL) has been used to describe a group of cytokines with complex immunomodulatory functions -- including cell proliferation, maturation, migration and adhesion. These cytokines also play an important role in immune cell differentiation and activation. Determining the exact function of a particular cytokine is complicated by the influence of the producing cell type, the responding cell type and the phase of the immune response. ILs can also have pro- and anti-inflammatory effects, further complicating their characterisation. These molecules are under constant pressure to evolve due to continual competition between the host's immune system and infecting organisms; as such, ILs have undergone significant evolution. This has resulted in little amino acid conservation between orthologous proteins, which further complicates the gene family organisation. Within the literature there are a number of overlapping nomenclature and classification systems derived from biological function, receptor-binding properties and originating cell type. Determining evolutionary relationships between ILs therefore can be confusing. More recently, crystallographic data and the identification of common structural motifs have led to a more accurate classification system. To date, the known ILs can be divided into four major groups based on distinguishing structural features. These groups include the genes encoding the IL1-like cytokines, the class I helical cytokines (IL4-like, γ-chain and IL6/12-like), the class II helical cytokines (IL10-like and IL28-like) and the IL17-like cytokines. In addition, there are a number of ILs that do not fit into any of the above groups, due either to their unique structural features or lack of structural information. This suggests that the gene family organisation may be subject to further change in the near future.
Similar content being viewed by others
Introduction
ILs are a large group of immunomodulatory proteins that elicit a wide variety of responses in cells and tissues. These cytokines comprise a large number of the known immunological 'second-messenger' molecules within mammals [1]. Interleukins initiate a response by binding to high-affinity receptors located on the surface of cells; ILs act in a paracrine or autocrine fashion, rather than as an endocrine signal, which is more common with steroidal and amino acid-derived hormones. The response of a particular cell to these cytokines depends on the ligands involved, specific receptors expressed on the cell surface and the particular signalling cascades that are activated. ILs modulate growth, differentiation and activation during an immune response [1]. This distinguishes them from chemokines -- the main function of which is to direct immune cells to the site of inflammation via chemotaxis -- and interferons (IFNs), which predominantly mediate cellular response to viral infection [1]. Despite attempts to separate these three groups based on function, there is a degree of overlap. ILs can exert both inflammatory and anti-inflammatory actions. A few members act as che-moattractants for helper T cells, paralleling the actions of chemokines. Others are intimately involved in the cellular response to viral pathogens, making them akin to IFNs. ILs are very important mediators of the physiological response to infection and also contribute significantly to the pathophy-siology of a wide range of disorders. As such, they represent a group of proteins with potential importance as therapeutic targets.
In this review, IL-containing cytokine groups and their closest homologues are described (Table 1). The grouping of genes is based primarily on genomic architecture and characteristic protein structural features (Table 2). Groups that are discussed include IL1-like cytokines, class I helical cytokines (IL4-like, γ-chain and IL6/12-like), class II helical cytokines (IL10-like and IL28-like) and IL17-like cytokines. A number of 'non-classified' ILs that also act as immunomodulating agents are also reviewed. All official gene and protein names discussed in this review can be found at http://www.genenames.org.
Nomenclature
The term 'interleukin' was first coined in 1979 in a letter to the editor of the Journal of Immunology to describe a number of secreted molecules produced by leukocytes [2]. ILs were also described as originating from lymphocytes; as a result, they are sometimes referred to as lymphokines [3]. A number of groups have tried to classify members of this gene family by a variety of characteristics, including biological function, receptor-binding properties, sequence homology, common structural motifs and the cell types responsible for their production. This has resulted in a number of nomenclature systems and has led to a great deal of confusion when describing individual genes and gene families.
A common interleukin nomenclature system was approved by the International Union of Immunological Societies (IUIS) and the World Health Organization (WHO) Nomenclature Subcommittee for IL1-IL10, including IL1A and IL1B [4]. Designations for IL11-IL13 were approved and updated by IUIS/WHO in 1994, IL16 in 1996 and IL14-IL15 in 1997 [5–7]. Sims and colleagues recommended a new nomenclature system in 2001 for IL1 family genes [8]. This nomenclature scheme was based on peptide sequence conservation, common structural elements, gene architecture and publication date.
The designation of cytokine and IL subfamilies further complicates IL nomenclature. Boulay and colleagues grouped IL3, IL4, IL5 and colony-stimulating factor 2 (CSF 2) into the 'IL4 family' of T cell-derived lymphokines [9]. This classification was based on shared genetic and structural elements, biological functions and receptor-binding properties. Liles and Van Voorhis separated the 42 cytokine genes known at the time into six groups (namely, haematopoietic growth factors, IFNs, lymphokines, monokines, chemokines and other cytokines) -- based primarily on biological function and originating cell type [3]. This classification system placed the majority of interleukins (IL2-IL6, IL9-IL10 and IL13-IL14) within the 'lymphokine' group. IL1A, IL1B, IL1RA, IL12 and IL15 were classified as 'monokines'. IL8 was considered a 'chemokine' and IL7, IL11 and the leukaemia inhibitory factor (LIF) gene were considered 'other cytokines'.
More recently, sequence and structural comparisons have come to the forefront of evolutionary analysis. Using these methods, evolutionary relationships can be accurately determined between genes and gene families. Such a system has been applied successfully to many gene families, using conserved nucleotide and amino acid sequences to decipher evolutionary ties between genes, which continues to be the underlying theme of these 'Updates on Gene Completions and Annotations' papers in Human Genomics.
Unfortunately, sequence analysis proves difficult when analysing highly divergent gene families, and the ILs are a perfect example of such a family. In these cases, structural comparisons often provide better insight into homologous protein groups. As with many of the IL groups, it has been hypothesised that a few invariable amino acids are required to maintain the overall conserved fold structures, leading to little sequence homology between evolutionarily related group members. This review focuses on IL classification using known structural motifs. The ILs can be separated into groups and subgroups using common structural elements identified from three-dimensional protein structures. This type of classification is useful, especially when applied to cytokines that share very little sequence homology. The accuracy is, obviously, solely dependent on the amount of structural information available within existing databases and publications, however.
Interleukin structure
Structural comparisons are a very useful way to classify evolutionarily related proteins. The IL1-like cytokines can be distinguished from other cytokines by the presence of a fold rich in β-strands [10]. Crystallographic comparison of IL1A, IL1B and IL1RA identified a shared fold structure comprising 12 packed β-sheets, known as a β-trefoil [11–13].
A number of ILs are classified as either class I or class II helical cytokines. Class I (or type I) cytokines share a common structural fold characterised by four tightly packed α-helices arranged in an 'up-up-down-down' orientation, often referred to as a 'four-helix bundle'. Class II (or type II) proteins are distinguished by a similar 'bundled helix' structural motif containing six or seven stacked helices.
The class I cytokines can then be further subdivided into either 'long-chain' or 'short-chain', based on the length of the core helices making up the bundle [14]. The length of the internal bundles affects overall peptide length; consequently, longchain cytokines are generally > 165 amino acids in length and short-chain usually contain < 165 residues. IL6, IL11, IL12A, IL23A, IL27A, IL31, cardiotrophin-like cytokine factor 1 (CLCF1), ciliary neurotrophic factor (CNTF), cardiotrophin 1 (CTF1), LIF, oncostatin M (OSM) and CSF3 are long-chain class I helical cytokines. The short-chain helical cytokines include IL2, IL3, IL4, IL5, IL7, IL9, IL13, IL15 and IL21. Thymic stromal lymphopoietin (TSLP) and CSF2 also fall into the short-chain class I cytokines.
The IL10-like and IL28-like (as well as some IFNs) are class II cytokines and are structurally related through a shared characteristic fold that contains six or seven stacked α-helices arranged in an anti-parallel conformation [15–17]. The IL28-like genes also share similar intron-exon structure with the IL10-like cytokines [18]. The IL10-like and IL28-like ILs, together with the type I and type II IFNs, have also been called the IL10-IFN gene family [19]. The IL10-like proteins including IL10, IL19, IL20, IL22, IL24 and IL26. IL28A, IL28B and IL29 comprise the IL28-like group.
IL17A and its homologues are structurally unrelated to other IL groups. IL17F shares a high degree of sequence homology with IL17A. IL17F was recently crystallised and structural analysis revealed the presence of a cysteine-knot fold [20]. This structural motif is characteristic of neurotrophins, which are a distinct class of secreted growth factors involved in the growth, differentiation and survival of particular cell types, including neurones [21].
Evolution
Genes associated with the immune system are under constant evolutionary pressure to change as a result of host-parasite co-evolution, where advantageous mutations are heavily favoured [22]. Consequently, cytokines are some of the most rapidly evolving genes within mammals. A study comparing the degree of evolutionary divergence between mouse and human orthologues showed that seven of the top 25 most highly divergent genes are cytokines [23]. In spite of a high degree of divergence within mammalian cytokines, a pheromone isolated from the marine protozoan, Euplotes raikovi, was found to share some structural similarities with IL2 [24]. Further studies revealed that the protozoan pheromone was able to bind to the mammalian IL2 receptor [25]. The same authors showed that mammalian IL2 was able to bind to the corresponding pheromone receptor located on the ciliated cell surface of the protozoan, suggesting that ILs may be evolutionarily grounded in ancient cell-signalling systems. Despite lacking a 'true' immune system, invertebrates are capable of mounting a highly effective inflammatory-like response similar to that observed in higher eukaryotes [26]. It appears that the invertebrate system arose independently, however, utilising analogous mechanisms and convergent evolution [23].
IL1 family members are clustered within 430 kilobases (kb) on human chromosome (Chr) 2q13, except for IL18 and IL33, which reside on Chr 11 and Chr 9, respectively [27]. The close proximity of IL1 family members on Chr 2 indicates that they most likely originated from a common ancestral gene that underwent multiple gene duplications [28].
Gene duplications are also common within the class I helical cytokines. IL2 and IL21 are separated by less than 244 kb on 4q26, suggesting another gene duplication event that led to multiple, functionally distinct proteins. The duplication is also found within the syntenic region on mouse Chr 3A3. Human IL3, IL4, IL5 and CSF2 localise to Chr 5q23-32 [29–31]. IL3 and CSF2 are closely linked and separated by only 9 kb [30]. Although tightly linked, the genes share relatively low amino acid homology [32]. IL5 and CSF2 also share a conserved regulatory element within their respective promoter regions [32]. The four genes in the cluster (IL3, IL4, IL5 and CSF2) have similar gene structure, biological activities and regulatory mechanisms. This same cluster is conserved within the mouse genome on Chr 11A1 [31, 33]. The more recent discovery of the IL9 and IL13 genes places these members relatively close to one another on Chr 5q31, indicating that they may also be part of the same gene cluster [34]. The genomic structure of the mouse orthologues is conserved on mouse Chr 11, further supporting origins grounded in more ancient duplication events [33, 35]. LIF and OSM are structurally similar growth factors that share many functions with IL6 and CNTF. The LIF and OSM genes are separated by < 16 kb on Chr 22q12 [36, 37]. The close physical linkage of these two related genes suggests that they are evolutionarily related and, like many other cytokines, are likely to be the product of gene duplication. The relatively low degree of sequence homology suggests rapid divergent evolution subsequent to the ancient gene duplications.
The class II helical cytokine genes, IL10, IL19, IL20 and IL24, form a tightly linked cluster on Chr 1q32 [17]. IL22, IL26 and a related gene, IFN gamma (IFNG), form an additional gene cluster on Chr 12q15 [38]. IL28A, IL28B and IL29 form a tight gene cluster on Chr 19q3 similar to the mouse genes, Il28a, Il28b and Il29, which cluster on Chr 7A3 [39].
IL17A and IL17F are linked on Chr 6 in humans and in the syntenic region on Chr 1 in mice in a tail-to-tail configuration [40]. The remaining IL17-like cytokines are spread throughout the genome. IL17B, IL17C, IL17D and IL25 are located on human Chr 5q33, 16q24, 13q11 and 14q11, respectively [41].
Interestingly, a number of groups have suggested a high degree of co-evolution between ILs and their receptors. One example is IL8, which is clustered with a number of structurally related chemokines on human Chr 4 [42]. The secretory proteins in this cluster bind to a specific family of G-protein-coupled receptors found within another gene cluster located on Chr 2 [43]. A similar situation has also been observed with IL6 ligand/receptor pairs, which appear also to have undergone co-evolution [44]. Evolutionary expansion through the tandem duplication of ligand and receptor genes located on these chromosomes, followed by divergence and co-evolution, led to the diversification of ligands and their cognate receptor gene families [42].
Gene duplication events and divergent evolution have played an important role in the expansion and diversity of this gene family. This expansion is fairly apparent, especially in vertebrates, where some genes underwent multiple rounds of duplication [14, 45, 46]. The gene clusters for known human ILs are summarised in Table 3.
IL1-like cytokines
There are currently 11 IL1-like proteins and each is encoded by a distinct gene - that is, IL1A, IL1B, IL1RN, IL1F5, IL1F6, IL1F7, IL1F8, IL1F9, IL1F10, IL18 and IL33 (Table 2) [47, 48]. IL1-like gene products are primarily secreted by stimulated monocytes or macrophages;[49] however, other immune- and non-immune-related cell types (including neutrophils, lymphocytes, keratinocytes, chondrocytes, fibroblasts and endothelial, epithelial and smooth muscle cells) appear also to produce these proteins [50, 51]. Receptor activation by IL1-like proteins plays a role in immune regulation and inflammatory processes by triggering transcriptional activation of downstream effector proteins. IL1A and IL1B rapidly induce transcription of hundreds of downstream targets, including other cytokines/chemokines, nitric oxide synthase (NOS), matrix metalloproteinases (MMPs), as well as activating their own genes [52–54]. Dysregulation of IL1-like cytokine signalling is associated with many inflammatory and autoimmune disorders, including inflammatory bowel disease (IL22), rheumatoid arthritis (IL20 and IL22) and psoriasis (IL19, IL20 and IL22) [55–58]. As a result, repression of the actions of IL1-like proteins is regarded as a promising therapeutic intervention to treat these and other common inflammatory disorders involving IL1-like cytokines [59, 60].
IL1A, IL1B, IL18 and IL33 are expressed as precursor (pro-) proteins that are proteolytically cleaved to form mature secretory peptides. The cleavage of pro-IL1A and pro-IL33 is mediated by calpain, whereas pro-IL1B and pro-IL18 cleavage is dependent on caspase-1 [61–63]. IL1A and IL1B initiate a strong pro-inflammatory signalling cascade. Both ligands bind to IL1 receptor type I (IL1R1) and signal transduction is initiated after recruitment of the IL1 receptor accessory protein (IL1RAP) [48]. They can also bind to the IL1 receptor type II (IL1R2), which is a non-signalling complex that serves as a 'decoy receptor'. IL1 receptor antagonist (IL1RN; also known as IL1RA) directly competes with IL1A and IL1B for binding to IL1R1 but does not activate the receptor, therefore producing anti-inflammatory effects [64]. IL18 and IL33 are both pro-inflammatory and each uses a related but distinct receptor and accessory protein pair for signal transduction. IL18 binds to a hetero-dimeric receptor complex containing IL18 receptor 1 (IL18R1) and the IL18R accessory protein (IL18RAP). IL33 serves as a ligand for IL1 receptor-like 1 (IL1RL1; also known as ST2) and the same co-receptor used by IL1A/B -- namely, IL1RAP [65]. It is interesting to note that five of the six IL1-like receptors form a closely linked gene cluster on Chr 2 [66].
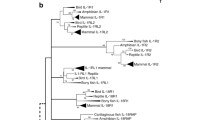
The IL1-like genes have been grouped on the basis of sequence homology, genomic architecture and chromosomal location (Table 1) [47, 48, 51, 67]. They also share common signal transduction mechanisms [68]. The 'classical' IL1-like genes include IL1A, IL1B and IL1RN. All three genes have been mapped to the same chromosomal region in both humans and mice [69]. Further investigation led to the discovery of an additional six IL1-like genes (IL1F5, IL1F6, IL1F7, IL1F8, IL1F9 and IL1F10) located within the same gene cluster (Table 3) [47]. IL18 and IL33 are found on Chr 11 and 9, respectively. Amino acid sequence identity within the group is quite varied, with identities ranging from 10-54 per cent (Table 4). IL18 and IL33 exhibit considerably less sequence homology with the other family members and are considered more distantly related [48, 70]. The close evolutionary relationship that exists between these proteins is exemplified in the tightly grouped branched structure formed within the IL/cytokine phylogram (Figure 1).
Phylogram of human ILs and closely related proteins. The phylogenetic tree was created using ClustalW2 and edited using TreeIllustrator. IL1-like (red), γ-chain utilising (light blue), IL4-like (dark blue), IL6/12-like (purple), IL10-like (light green), IL28-like (dark green) and IL17-like (brown), as well as non-classified cytokines (black) are included.
γ-chain cytokines
The common γ-chain (γc)-utilising cytokines include IL2, IL4, IL7, IL9, IL15, IL21 and TSLP (Table 2). Each gene encodes a protein of < 180 amino acids and they are considered to be short-chain class I cytokines [14]. All members of this group bind to a receptor complex that shares the γc chain (also known as IL2Rγ), with the exception of TSLP, which instead uses a closely related chain known as cytokine receptor-like factor 2 (CRLF2). The IL4, IL7, IL9, IL21 and TSLP proteins bind as heterodimeric receptor complexes, whereas IL2 and IL15 utilise a receptor with three subunits [14]. In addition to γc, the heterodimeric receptors contain a cytokine-specific receptor chain (IL4Rα, IL7Rα, IL9Rα or IL21Rα) [14, 45]. TSLP, similarly to IL7, utilises IL7Rα [71]. The IL2 and IL15 receptors contain either IL2Rβ or IL15Rα, respectively, as well as both the γc and IL2Rβ chains. Ligand-binding and receptor multimerisation results in the recruitment of Janus kinases 1 (JAK1) and 3 (JAK3), which phosphorylate a ligand-specific subset of signal transducers and activators of transcription (STAT) proteins [72]. TSLP signalling also induces STAT-dependent transcriptional activation but does not appear to involve a JAK [73].
Common γc cytokines are immunoregulatory molecules with both specific and overlapping functions. Their production influences the proliferation, differentiation and survival of innate and adaptive immune cells, including T cells, B cells, natural killer (NK) cells, natural killer T (NKT) cells and dendritic cells (DCs). IL7 is the predominant γc cytokine associated with regulating immature and mature T lymphocyte homeostasis [72]. IL9 is known to activate B cells, eosinophils, mast cells and epithelial cells [74]. The IL2, IL15 and IL21 proteins have specific functions during T cell differentiation and homeostasis [75].
Many of the γc cytokine genes are located in close genomic proximity with other class I helical cytokine genes. IL2 and IL21 are linked on Chr 4q26 and believed to be the result of gene duplication. IL15 is located relatively close at Chr 4q31, indicating that it is likely also to be the product of a distant gene duplication event. IL4 and IL9 are found within a gene cluster at Chr 5q31, which also contains many related IL4-like cytokine genes (IL3, IL5, IL13 and CSF2). TSLP also is located in close proximity to the 5q31 gene cluster at 5q22. The degree of amino acid homology observed between these proteins is not extensive, ranging from 6-22 per cent identity (Table 4). The clustering dendrogram reveals the γc cytokines to be spread across the entire interleukin/cytokine tree (Figure 1).
IL4-like cytokines
The IL4-like cytokines include IL3, IL4, IL5, IL13 and CSF2 (also known as granulocyte-macrophage CSF [GM-CSF]) (Table 2). These immunoregulatory molecules are generally < 160 amino acids in length and, like the γc ILs, are classified as short-chain class I cytokines. They are structurally very similar to the γc ILs; however, only IL4 utilises a receptor complex containing the γc chain co-receptor subunit. Although IL4 signals through γc, the high degree of homology with other IL4-like cytokines distinguishes it from the γc cytokines described above. All members in this group bind to heterodimeric receptor complexes [76]. IL4 and IL13 share signalling mechanisms through a receptor complex containing the IL4Rα chain [77]. Ligand specificity is achieved through the use of either γc or IL13Rα1 for IL4 and IL13, respectively. Ligand binding at the receptor activates the tyrosine kinase JAK1, which leads to subsequent phosphorylation of STAT1 or STAT6 [78]. The activated STATs then translocate to the nucleus, where they regulate transcription. IL3, IL5 and CSF2 modulate transcription in a similar fashion; however, they utilise the common β-chain (βc) and either IL3Rα, IL5Rα or CSF2Rα, respectively [79–81]. Subsequently, JAK2 and STAT5 mediate downstream transcriptional activation [14].
IL4 is associated with T helper type II (Th2) cell differentiation and plays an important role in allergic inflammation [82]. Along with IL13, IL4 regulates immunoglobulin class switching in B cells and mast cell recruitment [83]. IL3, IL4, IL5 and CSF2 facilitate granulocyte maturation and therefore also play an important role in regulating allergic response [84].
The five genes in this group are closely linked at Chr 5q31, most likely originating from a series of gene duplications [29]. The genomic organisation of these genes is also conserved within mice on Chr 11. Sequence homology between the proteins ranges from 20-25 per cent identity and 30-40 per cent similarity (Table 4). In spite of a relatively high degree of homology, the IL4-like cytokines are dispersed throughout the IL/cytokine dendrogram (Figure 1).
IL6/12-like cytokines
The biological functions of cytokines derived from IL6/12-like genes are diverse and comprise haematopoietic cytokines with broad physiological and pathophysiological actions. These proteins play a role in inflammation, immune response and cell differentiation and survival [85]. ILs within this group include IL6, IL11, IL12A, IL23A, IL27A and IL31 (Table 2). There are a number of other closely related cytokines, including CLCF1, CTF1, CNTF, CTF3, LIF and OSM. IL6/12-like genes are categorised as long-chain class I helical cytokines and encode proteins > 160 amino acids in length.
The group can be further subdivided into cytokines that are biologically active as monomers (IL6, IL11, IL31, CLCF1, CNTF, CTF1, LIF, OSM and CSF3) or as heterodimers (IL12, IL23, IL27A and IL35). Most of the monomeric cytokines, except IL31, signal through the common co-receptor subunit IL6 signal transducer (IL6ST), previously known as glycoprotein 130 (gp130) [14, 86]. IL31 uses the IL31RA co-receptor subunit, which is a close relative of IL6ST [87]. IL6 initially binds to a specific high-affinity receptor, IL6RA. After activation, the IL6ST receptor chain is recruited to form the active signalling complex. IL11 and OSM also use unique receptor complexes composed of IL6ST and either IL11RA or the OSM receptor (OSMR), respectively [85]. IL6 and IL11 are the only members of this group known to interact with IL6ST homodimers, where two IL-bound receptors (IL6RA or IL11RA) complex with an IL6ST dimer to create a hexameric complex that initiates subsequent signalling cascades [88, 89]. LIF and CTF1 signal through a receptor complex containing the LIF receptor (LIFR) and IL6ST [90]. CNTF and CLCF1 bind a heterotrimeric receptor complex comprising LIFR and IL6ST, as well as the CNTF receptor (CNTFR) chain [91]. IL31 signal transduction proceeds through OSMR and IL31RA [92].
The heterodimeric cytokines, composed of disulphide-linked α- and β-chains, each signal through a unique combination of dimerised receptors formed from five possible receptor subunits. The α-chain is structurally homologous to the long-chain class I cytokines (mentioned above) and contains a 'four-helix bundle' structural motif [14]. The β-chains include IL12B and Epstein-Barr virus-induced-3 (EBI3), which is also known as IL27B. These proteins are structurally homologous to soluble class I helical cytokine receptors and are distinct from other class I cytokines [93]. Biologically active IL12, a heterodimer formed between IL12A and IL12B, signals through the IL12RB1 and IL12RB2 receptor dimer [94]. Active IL23 dimer is composed of IL12B and IL23A subunits and binds to the IL23 receptor complex formed from the association of IL12RB1 and IL23R [95]. IL27A and EBI3 (IL27B) dimerisation forms the active IL27-signalling molecule, which is predominantly derived from macrophages as well as DCs [96]. IL27 signals through a receptor comprising IL27RA and IL6ST, which is also used by many of the monomeric cytokines mentioned above [97]. The IL12, IL23 and IL27A proteins are primarily expressed from antigen-presenting cells (APCs), which include DCs, macrophages and monocytes [96, 98, 99]. Another novel heterodimeric cytokine formed from the dimerisation of IL12A and EBI3 (IL27B) was recently identified and designated IL35 [100]. IL35 expression from T regulatory (Treg) cells has been found to have inhibitory effects on T cell proliferation [101]. The IL35 receptor complex has yet to be determined; however, it is hypothesised that it may use a combination of receptor subunits employed by IL12, IL23 and IL27 [100].
Monomeric and heterodimeric long-chain class I cytokine receptor activation initiate signal transduc-tion through similar JAK/STAT-signalling pathways [85, 102]. This signalling pathway is evolutionarily conserved and used by both class I and class II helical cytokines [103]. Ligand binding activates the receptor complex and promotes JAK recruitment. Activated JAKs subsequently phosphorylate STATs, including STAT3 and STAT4, which in turn translocate to the nucleus and transactivate a number of genes associated with differentiation, growth, survival and apoptosis [104, 105].
The IL6/12-like genes translate a functionally diverse group of proteins involved in the immune response to a variety of factors, including microbial and host immune stimuli [85, 106]. IL6 plays a very important role during the early immune response to infection. It causes B lymphocytes to differentiate into mature, immunoglobulin-secreting plasma B cells [107]. IL6 signalling also initiates T cell activation, growth and differentiation [108]. IL11 increases the production of proteins needed during the acute phase response and induces the differentiation of lymphocytes [109]. IL12, IL23, IL27 and IL35 are involved in Info production and helper T cell differentiation [110]. IL35 has been shown to have anti-inflammatory effects by expanding the population of anti-inflammatory cytokine-secreting cells [111]. The dysregulation and aberrant expression of IL6/12-like cytokines can result in severe inflammation and is the underlying cause of several human immunological disorders, including rheumatoid arthritis, inflammatory bowel disease and asthma [85, 112, 113].
Amino acid sequence homology between group members is rather low, ranging from 12-19 per cent (Table 4). Most of the IL6/12-like proteins cluster into a single branch within the IL phylogram, with the exception of IL12A, IL31 and OSM (Figure 1).
IL10-like cytokines
IL10, IL19, IL20, IL22, IL24 and IL26 comprise the IL10-like cytokines (Table 2). IL10 was originally described as an inhibitor of T helper type I (Th1) cell-related cytokine expression [114]. Since its discovery, the other five proteins have been added to this group. These cytokines are related through genomic organisation, intron-exon structure and a common structural fold composed of stacked α-helices [115]. The IL10-like group and closely related IL28-like group comprise the class II helical cytokines.
Despite structural similarities, the proteins display unique expression patterns and diverse functions. T lymphocytes, monocytes and B cells are the most common source of IL10 in humans [116]. IL10 is biologically active as a homodimer and activates the IL10R1 and IL10R2 receptor complex. Subsequent downstream signalling is mediated through JAK/STAT pathways, analogous to the signalling mechanisms used by class I cytokines (ie γ-chain, IL4-like and IL6/12-like cytokines) [117]. IL10 suppresses the expression of other inflammatory molecules and is an extremely important anti-inflammatory and immunosuppressive cytokine in humans. IL19 is an immunoregulatory cytokine primarily produced by monocytes. IL19 signalling proceeds through the IL20R1 and IL20R2 dimeric receptor complex, and subsequent receptor activation results in the release of IL6, tumour necrosis factor (TNF) and a number of reactive oxygen species (ROS) from target cells [118, 119]. IL20 is known for its role in haematopoiesis and inflammation [120, 121]. It was originally identified in a human keratinocyte library; studies suggest that it may play a role in the pathophysiology of some skin abnormalities, such as psoriasis and palmoplantar pustulosis [122, 123]. IL20, as well as IL24, can signal through both the IL20R1/IL20R2 and IL22R1/20R2 heterodimers [124]. IL22 is similar to other IL10-like cytokines in binding a heterodimeric receptor, albeit composed of IL22R1 and IL10R2 chains [125]. Activated T cells and NKT cells are the primary source of IL22 [126]. IL22 facilitates defence against microbial pathogens by initiating the release of antimicrobial peptides associated with host defence, such as defensin-β2 and S100A7 [127]. It is also associated with the induction of acute phase response proteins [128]. Originally named IL10-related T-cell-derived inducible factor (ILTIF), studies have also indicated that IL22, similarly to IL10, may be biologically active as a homo-dimer [129]. By contrast with IL10 and IL22, the IL19, IL20 and IL24 proteins are biologically active as monomers [130, 118]. IL24, previously known as melanoma differentiation-associated antigen 7 (MDA-7), was identified as a cytokine that was differentially expressed in healthy and metastatic mela-nocytes [131]. IL24 is predominantly secreted from melanocytes, Th2 cells and monocytes [132]. Downstream inflammatory effects of IL24 include the release of IL6, IFNG and TNF [133]. IL26 expression occurs primarily in memory Th1 cells, Th17 cells and monocytes. Akin to IL10, IL26 functions as a homodimer and activates a unique receptor complex composed of IL20R1 and IL10R2 chains [134]. The exact biological functions of IL26 have yet to be determined -- mainly due to the lack of a mouse orthologue; this limits experimental approaches commonly used to characterise other ILs, such as the generation of knockout mice [135]. Activation of either complex is linked to downstream JAK/STAT pathways.
IL10, IL19, IL20 and IL24 form a closely linked gene cluster on Chr 1q32 [132]. All four genes are found within a 195 kb region. By contrast with the other IL10-like genes, IL22 and IL26 are found in a gene cluster with IFNG on Chr 12q15 [17]. As mentioned above, a mouse homologue for IL26 has yet to be identified; however, the IL26 gene cluster is conserved within other mammals, as well as non-mammals, indicating that Il26 was lost from the murine genome [136]. Amino acid identities for members in this group range from 17-39 per cent (Table 4) and they form an isolated cluster within the IL/cytokine dendrogram (Figure 1).
IL28-like cytokines
IL28A, IL28B and IL29 were discovered in 2003 by two independent research groups [18, 137]. These cytokines exhibit IFN type I properties in response to viral or bacterial infection. As such, they have also been referred to as type III IFNs; however, genomic organisation, protein structure and receptor usage for these proteins has linked them most closely to the IL10-like cytokines [19, 138]. They contain the common structural motif found in IL10-related cytokines (stacked α-helices), making them members of the class II helical cytokines.
IL28A, IL28B and IL29, like many cytokines, utilise a dimeric cell-surface receptor complex. The heterodimeric receptor belongs to the class II cytokine receptor family and comprises the IL10R1 chain, which is shared with IL10, IL22 and IL26, and a unique chain referred to as IL28R1 [18]. IL28A, IL28B and IL29 are secreted as monomeric proteins; receptor activation utilises JAK/STAT pathways for downstream signalling, analogous to other class I and class II helical cytokines [137, 139, 140]. It is thought that most, if not all, nucleated cell types are capable of IL28-like cytokine expression following viral infection [19]. Expression is also observed after activation with bacterial components such as lipopolysaccharide (LPS) [141]. The proteins exhibit direct anti-viral activity by inhibiting viral replication [142, 143]. They also have direct anti-growth activity on many cell types, including tumour cells [19]. Recently, a great deal of attention has been given to these proteins due to their potential therapeutic benefits.
IL28A and IL28B form a tight gene cluster on Chr 19q13. Gene architectures are similar to the IL10-like genes and contain either six (IL28A and IL28B) or five (IL29) exons [39]. Only the mouse Il28a and Il28b genes translate functional gene products. Il29 contains an in-frame stop codon within the first exon and is considered a pseudogene [144]. The amino acid identities for the IL28-like cytokines are very high, ranging from 66-94 per cent (Table 4). The high degree of homology between the proteins is further supported by the tight cluster formed on the IL/cytokine dendrogram (Figure 1).
IL17-like cytokines
IL17A, IL17B, IL17C, IL17D, IL17F and IL25 are members of the IL17-like cytokine group. IL17A was originally identified within a murine hybridoma cDNA library in 1993 [145]. Genomic sequencing led to the identification of five IL17A homologues, including IL17B, IL17C, IL17D, IL25 and IL17F. IL17A and IL17F are the most well characterised of the six IL17-like cytokines. IL17F was recently crystallised; the results indicate that the protein adopts a cysteine-knot fold architecture, which is a structural motif associated with neurotrophins [20]. IL17A and IL17F are biologically active as homodimers but are also capable of forming IL17A-IL17F heterodimers [146]. IL17A and IL17F homodimers, as well as the IL17A-IL17F heterodimer, bind with varying affinities to a heterodimeric receptor complex composed of IL17RA and IL17RC [147]. IL25 also activates a dimeric receptor, comprising IL17RA and IL17RB. IL17B and IL17C have been found to bind to the IL17RB and IL17RE receptors, respectively [148, 149]. A receptor has not yet been identified for IL17D.
The functions of IL17-like cytokines are not completely understood. IL17A expression has been found to originate from a new subset of helper T cells, Th17 cells [150]. These cells are potent inducers of tissue inflammation through the release of IL17A, IL17F, IL21 and IL22. IL25 expression has not been detected in Th17 cells but secretion from Th2 cells has been identified [151]. It is hypothesised that the Th17 response and IL17A release is triggered when a specific pathogen does not elicit a strong response from either Th1 or Th2 cells [150]. In general, the specific functions of IL17-like cytokines are not well established. Nevertheless, IL17A appears to play an important role in the immune response to bacterial and fungal pathogens. IL17A is also associated with many autoimmune disorders, including rheumatoid arthritis, inflammatory bowel disease, multiple sclerosis, as well as many types of cancer [152, 153].
The IL17-like cytokines appear to form a novel family of signalling molecules. Not only are the proteins structurally unlike other immunomodulatory cytokines, but the corresponding IL17 receptor family contains five members, IL17RA-E, which are also structurally distinct from those used by other cytokines [149]. Perhaps not surprisingly, these receptors are coupled to signal transduction pathways that differ from those used by class I and II cytokines -- that is, TNF receptor-associated factor (TRAF) proteins, TRAF3 interacting protein 2 (TRAF3IP2) and nuclear factor-κB (NF-κB) [154].
IL17-like cytokines share 18-45 per cent amino acid identity and are dispersed throughout the human genome (Table 4). IL17A and IL17F are the most closely related homologues in this group, sharing > 45 per cent sequence identity and 59 per cent sequence similarity. The high degree of sequence homology can be explained by the two genes being closely linked at Chr 6p12, again most likely the product of an evolutionarily recent gene duplication. IL17B, IL17C, IL17D and IL25 are located on Chr 5q33, 16q24, 13q11 and 14q11, respectively. Pairwise comparisons between each of the other members indicate that they share between 18-26 per cent sequence identity and 24-40 per cent sequence similarity. The close relationship between these proteins is evident in the tight cluster they form within the IL/cytokine dendrogram (Figure 1).
Additional 'non-classified' interleukins
There are a number of ILs that cannot be definitively classified into any of the groups described above (Table 2). The inability to classify these molecules is generally associated with their unique structural characteristics or lack of structural information; however, their genes encode proteins with verified immunomodulatory actions.
IL8 contains a CXC structural motif, which is a defining characteristic of the CXC chemokine family. This family is characterised by the presence of three or four highly conserved cysteine residues found within the N-terminus. The CXC chemo-kines contain a variable residue between the first two conserved cysteines [1]. IL8 has been shown to participate in leukocyte recruitment during inflammation -- supporting its role as a chemokine [74]. IL8 is released from many cell types, including T cells, monocytes and endothelial cells [69–71]. Its expression is induced after exposure to a variety of inflammatory stimuli -- including bacteria, oxidative stress, LPS, TNF and IL1B [155–158]. As expected, IL8 shares little sequence homology with known ILs; however, it shares a high degree of sequence identity with other CXC chemokines [159]. Chemokine (CXC motif) ligands-1 (CXCL1), -2 (CXCL2) and -3 (CXCL3), and pro-platelet basic protein (PPBP), are all well-characterised chemo-kines, exhibiting 36 per cent, 36 per cent, 34 per cent and 33 per cent sequence identity, respectively.
Taxilin alpha (TXLNA), also known as IL14, was originally identified as a factor produced by human B cell lymphoma cells that caused enhanced proliferation of activated B lymphocytes [160]. The factor was initially called high-molecular-weight B cell growth factor. The cDNA was cloned and expression produced a 54-kilodalton (kDa) protein that was re-named IL14. Produced by B cells, T cells and DCs, IL14 enhances B cell proliferation, increases the subpopulation of memory B cells and prevents the secretion of immunoglobulins [161]. Interestingly, IL14 was also identified as a novel syntaxin-binding protein involved in vesicle transport and was re-named taxilin [162]. Database searches have revealed two closely related homologues, resulting in taxilin being re-named as taxilin-α (TXLNA), with its homologues named taxilin-β (TXLNB) and taxilin-γ (TXLNG) [163]. None of the taxilin isoforms appears to be structurally related to known ILs; their exact functions remain unknown. IL14 has been implicated in the pathophysiology of Sjögren's disease, an autoimmune disorder affecting exocrine glands [164]. TXLNA, TXLNB and TXLNG are located on Chr 1p34, 6q24 and Xp22, respectively [163]. Sequence analysis reveals that TXLNA shares little homology (< 15 per cent) with any of the cytokines listed in Table 1.
IL16 was originally described as a T helper cell chemoattractant. The compound is also described as a chemotactic cytokine but not a chemokine because it lacks characteristic structural motifs [165]. IL16 is expressed as a large precursor protein that is cleaved by caspase-3 at a serine residue within the C-terminus to produce a larger intracellular and a shorter 121-amino acid secretory protein [166]. The shorter, secreted peptide assembles into homotetramers, which are believed to be the biologically active unit [167]. The larger cleavage product remains within the cell, and its exact function has yet to be determined. Some evidence suggests that it is able to translocate into the nucleus and arrest the cell cycle [168]. Studies have shown that many cell types are capable of IL16 secretion, including T cells, eosinophils, DCs, mast cells and neuronal cells [168]. Secreted IL16 binds to the CD4 receptor to activate a downstream signalling cascade that involves STAT6 activation [169]. IL16 activates many cell types, including T cells, monocytes, macrophages, eosinophils and DCs [170, 171]. Downstream effects include the release of inflammatory cytokines including IL1B, IL6, IL15 and TNF [172]. IL16 is structurally distinct from other cytokines in that it contains multiple PDZ domains, including one that lies within the mature secretory peptide [173]. PDZ domains are a common structural motif associated with protein-protein interactions [174].
IL32 was originally identified in 1992 as a cytokine secreted from IL2-activated NK and T cells [175]. The protein does not exhibit significant sequence similarity with other cytokines and, to date, there is little structural information in the literature --making its classification very difficult. Further studies have shown that IL32 is also released from monocytes and epithelial cells [176, 177]. It stimulates the release of pro-inflammatory cytokines, including IL1B, IL6, IL8, TNF and macrophage inflammatory protein 2 (MIP2) [176]. The signalling pathways for IL32 have yet to be fully characterised; however, studies have shown that they involve the NF-κB and p38 mitogen-activated protein kinase pathways, which are commonly associated with inflammation [178]. A number of diseases associated with IL32 expression, including inflammatory bowel disease, myelodysplastic syndrome and HIV infection [179–181]. IL32 lacks significant amino acid identity (< 15 per cent) with the other cytokines listed in Table 1. Furthermore, BLAST analyses failed to identify any significantly homologous proteins.
IL34 was identified in 2008 by Lin et al [182]. It binds to the homodimeric CSF1 receptor (CSFR1), which also binds CSF1 [182]. CSF1 is a cytokine that controls the division, differentiation and function of monocytes, macrophages and osteoblasts [183]. Similar to CSF1, human IL34 has been found to function as a homodimer [182]. Furthermore, modelling of both IL34 and CSF1 identified a stacked 'four-helix bundle' structure [184]. The CSF1 monomer is structurally very similar to CSF2, a short-chain class I helical cytokine [185]. There is a high likelihood that both CSF1 and IL34 are evolutionarily related to the class I helical cytokines. BLAST analysis of the IL34 peptide sequence failed to identify any significantly homologous proteins within the human genome. Pairwise alignments revealed that IL34 shares 19 per cent sequence identity with OSM and CNTF, both of which are long-chain class I helical cyto-kines. Sequence homology between other ILs and cytokines was not significant (ie < 15 per cent).
Conclusions
To date, the human IL gene family contains 55 known IL and IL-related genes. The majority of ILs can be classified into seven distinct groups and subgroups using characteristic structural features. These groups include genes encoding the IL1-like cytokines, the class I helical cytokines (IL4-like, γ-chain and IL6/12-like), the class II helical cytokines (IL10-like and IL28-like) and the IL17-like cytokines. Because evolutionary divergence occurs so rapidly in these genes, it is impossible in many cases to assign a member to a specific 'gene family' or 'gene subfamily'; therefore, structural analysis has become the principle approach for classifying. A number of 'interleukin' genes currently lack the structural information required for classification. It is therefore likely that these groups will expand as crystallisation data and structural analyses become available.
References
Commins SP, Borish L, Steinke JW: Immunologic messenger molecules: Cytokines. interferons, and chemokines. J Allergy Clin Immunol. 2010, 125: S53-S72. 10.1016/j.jaci.2009.07.008.
Aarden LA, Brunner TK, Cerottini JC, Dayer JM, et al: Revised nomenclature for antigen-non-specific T cell proliferation and helper factors. J Immunol. 1979, 123: 2928-2929.
Liles WC, Van Voorhis WC: Review: Nomenclature and biologic significance of cytokines involved in inflammation and the host immune response. J Infect Dis. 1995, 172: 1573-1580. 10.1093/infdis/172.6.1573.
Anon: Nomenclature for secreted regulatory proteins of the immune system (interleukins). WHO-IUIS Nomenclature Subcommittee on Interleukin Designation. Bull World Health Organ. 1991, 69: 483-486.
Anon: Nomenclature for secreted regulatory proteins of the immune system (interleukins): Update. IUIS/WHO Nomenclature Committee. Bull World Health Organ. 1994, 72: 811-812.
Anon: Interleukin 16 (IL-16). IUIS/WHO Standing Committee on Interleukin Designation. Bull World Health Organ. 1996, 74: 451-452.
Anon: Nomenclature for secreted regulatory proteins of the immune system (interleukins): Update. IUIS/WHO Standing Committee on Interleukin Designation. Bull World Health Organ. 1997, 75: 175-
Sims JE, Nicklin MJ, Bazan JF, Barton JL, et al: A new nomenclature for IL-1-family genes. Trends Immunol. 2001, 22: 536-537. 10.1016/S1471-4906(01)02040-3.
Boulay JL, Paul WE: The interleukin-4 family of lym-phokines. Curr Opin Immunol. 1992, 4: 294-298. 10.1016/0952-7915(92)90079-T.
Veerapandian B, Gilliland GL, Raag R, Svensson AL, et al: Functional implications of interleukin-1 beta based on the three-dimensional structure. Proteins. 1992, 12: 10-23. 10.1002/prot.340120103.
Priestle JP, Schar HP, Grutter MG: Crystallographic refinement of interleukin 1 beta at 2.0 A resolution. Proc Natl Acad Sci USA. 1989, 86: 9667-9671. 10.1073/pnas.86.24.9667.
Vigers GP, Caffes P, Evans RJ, Thompson RC, et al: X-ray structure of interleukin-1 receptor antagonist at 2.0-A resolution. J Biol Chem. 1994, 269: 12874-12879.
Graves BJ, Hatada MH, Hendrickson WA, Miller JK, et al: Structure of interleukin 1 alpha at 2.7-A resolution. Biochemistry. 1990, 29: 2679-2684. 10.1021/bi00463a009.
Boulay JL, O'Shea JJ, Paul WE: Molecular phylogeny within type I cytokines and their cognate receptors. Immunity. 2003, 19: 159-163. 10.1016/S1074-7613(03)00211-5.
Ealick SE, Cook WJ, Vijay-Kumar S, Carson M, et al: Three-dimensional structure of recombinant human interferongamma. Science. 1991, 252: 698-702. 10.1126/science.1902591.
Zdanov A, Schalk-Hihi C, Gustchina A, Tsang M, et al: Crystal structure of interleukin-10 reveals the functional dimer with an unexpected topological similarity to interferon gamma. Structure. 1995, 3: 591-601. 10.1016/S0969-2126(01)00193-9.
Kotenko SV: The family of IL-10-related cytokines and their receptors: Related, but to what extent?. Cytokine Growth Factor Rev. 2002, 13: 223-240. 10.1016/S1359-6101(02)00012-6.
Sheppard P, Kindsvogel W, Xu W, Henderson K, et al: IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003, 4: 63-68. 10.1038/ni873.
Witte K, Witte E, Sabat R, Wolk K: IL-28A, IL-28B, and IL-29: Promising cytokines with type I interferon-like properties. Cytokine Growth Factor Rev. 2010, 21: 237-251. 10.1016/j.cytogfr.2010.04.002.
Hymowitz SG, Filvaroff EH, Yin JP, Lee J, et al: IL-17s adopt a cystine knot fold: structure and activity of a novel cytokine IL-17F, and implications for receptor binding. EMBO J. 2001, 20: 5332-5341. 10.1093/emboj/20.19.5332.
McDonald NQ, Hendrickson WA: A structural superfamily of growth factors containing a cystine knot motif. Cell. 1993, 73: 421-424. 10.1016/0092-8674(93)90127-C.
Hurst LD, Smith NG: Do essential genes evolve slowly?. Curr Biol. 1999, 9: 747-750. 10.1016/S0960-9822(99)80334-0.
Scapigliati G, Buonocore F, Mazzini M: Biological activity of cytokines: An evolutionary perspective. Curr Pharm Des. 2006, 12: 3071-3081. 10.2174/138161206777947489.
Luporini P, Vallesi A, Miceli C, Bradshaw RA: Ciliate pheromones as early growth factors and cytokines. Ann NY Acad Sci. 1994, 712: 195-205. 10.1111/j.1749-6632.1994.tb33573.x.
Vallesi A, Giuli G, Ghiara P, Scapigliati G, et al: Structure-function relationships of pheromones of the ciliate Euplotes raikovi with mammalian growth factors: Cross-reactivity between Er-1 and interleukin-2 systems. Exp Cell Res. 1998, 241: 253-259. 10.1006/excr.1998.4056.
Bingulac-Popovic J, Figueroa F, Sato A, Talbot WS, et al: Mapping of mhc class I and class II regions to different linkage groups in the zebrafish Danio rerio. Immunogenetics. 1997, 46: 129-134. 10.1007/s002510050251.
Nicklin MJ, Weith A, Duff GW: A physical map of the region encompassing the human interleukin-1 alpha. interleukin-1 beta, and interleukin-1 receptor antagonist genes. Genomics. 1994, 19: 382-384. 10.1006/geno.1994.1076.
Dunn E, Sims JE, Nicklin MJ, O'Neill LA: Annotating genes with potential roles in the immune system: Six new members of the IL-1 family. Trends Immunol. 2001, 22: 533-536. 10.1016/S1471-4906(01)02034-8.
Takahashi M, Yoshida MC, Satoh H, Hilgers J, et al: Chromosomal mapping of the mouse IL-4 and human IL-5 genes. Genomics. 1989, 4: 47-52. 10.1016/0888-7543(89)90313-3.
Yang YC, Kovacic S, Kriz R, Wolf S, et al: The human genes for GM-CSF and IL 3 are closely linked in tandem on chromosome 5. Blood. 1988, 71: 958-961.
van Leeuwen BH, Martinson ME, Webb GC, Young IG: Molecular organization of the cytokine gene cluster. involving the human IL-3, IL-4, IL-5, and GM-CSF genes, on human chromosome 5. Blood. 1989, 73: 1142-1148.
Sanderson CJ, Campbell HD, Young IG: Molecular and cellular biology of eosinophil differentiation factor (interleukin-5) and its effects on human and mouse B cells. Immunol Rev. 1988, 102: 29-50. 10.1111/j.1600-065X.1988.tb00740.x.
Lee JS, Campbell HD, Kozak CA, Young IG: The IL-4 and IL-5 genes are closely linked and are part of a cytokine gene cluster on mouse chromosome 11. Somat Cell Mol Genet. 1989, 15: 143-152. 10.1007/BF01535075.
Ryan AW, Thornton JM, Brophy K, Daly JS, et al: Chromosome 5q candidate genes in coeliac disease: Genetic variation at IL4, IL5, IL9, IL13, IL17B and NR3C1. Tissue Antigens. 2005, 65: 150-155. 10.1111/j.1399-0039.2005.00354.x.
Webb GC, Lee JS, Campbell HD, Young IG: The genes for interleukins 3 and 5 map to the same locus on mouse chromosome 11. Cytogenet Cell Genet. 1989, 50: 107-110. 10.1159/000132734.
Giovannini M, Djabali M, McElligott D, Selleri L, et al: Tandem linkage of genes coding for leukemia inhibitory factor (LIF) and oncostatin M (OSM) on human chromosome 22. Cytogenet Cell Genet. 1993, 64: 240-244. 10.1159/000133586.
Jeffery E, Price V, Gearing DP: Close proximity of the genes for leukemia inhibitory factor and oncostatin M. Cytokine. 1993, 5: 107-111. 10.1016/1043-4666(93)90048-A.
Oral HB, Kotenko SV, Yilmaz M, Mani O, et al: Regulation of T cells and cytokines by the interleukin-10 (IL-10)-family cytokines IL-19, IL-20, IL-22, IL-24 and IL-26. Eur J Immunol. 2006, 36: 380-388. 10.1002/eji.200425523.
Uze G, Monneron D: IL-28 and IL-29: Newcomers to the interferon family. Biochimie. 2007, 89: 729-734. 10.1016/j.biochi.2007.01.008.
Starnes T, Robertson MJ, Sledge G, Kelich S, et al: Cutting edge: IL-17F. a novel cytokine selectively expressed in activated T cells and monocytes, regulates angiogenesis and endothelial cell cytokine production. J Immunol. 2001, 167: 4137-4140.
Moseley TA, Haudenschild DP, Rose L, Reddi AH: Interleukin-17 family and IL-17 receptors. Cytokine Growth Factor Rev. 2003, 14: 155-174. 10.1016/S1359-6101(03)00002-9.
Fryxell KJ: The coevolution of gene family trees. Trends Genet. 1996, 12: 364-369.
Ahuja SK, Ozcelik T, Milatovitch A, Francke U, et al: Molecular evolution of the human interleukin-8 receptor gene cluster. Nat Genet. 1992, 2: 31-36. 10.1038/ng0992-31.
Kohu K, Yamabe E, Matsuzawa A, Onda D, et al: Comparison of 30 immunity-related genes from the common marmoset with orthologues from human and mouse. Tohoku J Exp Med. 2008, 215: 167-180. 10.1620/tjem.215.167.
Kaiser P, Rothwell L, Avery S, Balu S: Evolution of the interleukins. Dev Comp Immunol. 2004, 28: 375-394. 10.1016/j.dci.2003.09.004.
Liongue C, Ward AC: Evolution of class I cytokine receptors. BMC Evol Biol. 2007, 7: 120-10.1186/1471-2148-7-120.
Nicklin MJ, Barton JL, Nguyen M, FitzGerald MG, et al: A sequence-based map of the nine genes of the human interleukin-1 cluster. Genomics. 2002, 79: 718-725. 10.1006/geno.2002.6751.
Arend WP, Palmer G, Gabay C: IL-1. IL-18, and IL-33 families of cytokines. Immunol Rev. 2008, 223: 20-38. 10.1111/j.1600-065X.2008.00624.x.
Barksby HE, Lea SR, Preshaw PM, Taylor JJ: The expanding family of interleukin-1 cytokines and their role in destructive inflammatory disorders. Clin Exp Immunol. 2007, 149: 217-225. 10.1111/j.1365-2249.2007.03441.x.
Gabay C, Lamacchia C, Palmer G: IL-1 pathways in inflammation and human diseases. Nat Rev Rheumatol. 2010, 6: 232-241. 10.1038/nrrheum.2010.4.
Weber A, Wasiliew P, Kracht M: Interleukin-1 (IL-1) pathway. Sci Signal. 2010, 3: cm1-10.1126/scisignal.3105cm1.
Allantaz F, Chaussabel D, Stichweh D, Bennett L, et al: Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J Exp Med. 2007, 204: 2131-2144. 10.1084/jem.20070070.
Vincenti MP, Brinckerhoff CE: Early response genes induced in chondrocytes stimulated with the inflammatory cytokine interleukin-1beta. Arthritis Res. 2001, 3: 381-388. 10.1186/ar331.
Dinarello CA: The IL-1 family and inflammatory diseases. Clin Exp Rheumatol. 2002, 20: S1-13.
Wolk K, Witte E, Hoffmann U, Doecke WD, et al: IL-22 induces lipopolysaccharide-binding protein in hepatocytes: A potential systemic role of IL-22 in Crohn's disease. J Immunol. 2007, 178: 5973-5981.
Hsu YH, Li HH, Hsieh MY, Liu MF, et al: Function of interleukin-20 as a proinflammatory molecule in rheumatoid and experimental arthritis. Arthritis Rheum. 2006, 54: 2722-2733. 10.1002/art.22039.
Ikeuchi H, Kuroiwa T, Hiramatsu N, Kaneko Y, et al: Expression of interleukin-22 in rheumatoid arthritis: Potential role as a proinflammatory cytokine. Arthritis Rheum. 2005, 52: 1037-1046. 10.1002/art.20965.
Otkjaer K, Kragballe K, Funding AT, Clausen JT, et al: The dynamics of gene expression of interleukin-19 and interleukin-20 and their receptors in psoriasis. Br J Dermatol. 2005, 153: 911-918. 10.1111/j.1365-2133.2005.06800.x.
Salvi GE, Lang NP: Host response modulation in the management of periodontal diseases. J Clin Periodontol. 2005, 32 (Suppl. 6): 108-129.
Burger D, Dayer JM, Palmer G, Gabay C: Is IL-1 a good therapeutic target in the treatment of arthritis?. Best Pract Res Clin Rheumatol. 2006, 20: 879-896. 10.1016/j.berh.2006.06.004.
Thornberry NA, Bull HG, Calaycay JR, Chapman KT, et al: A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992, 356: 768-774. 10.1038/356768a0.
Hayakawa M, Hayakawa H, Matsuyama Y, Tamemoto H, et al: Mature interleukin-33 is produced by calpain-mediated cleavage in vivo. Biochem Biophys Res Commun. 2009, 387: 218-222. 10.1016/j.bbrc.2009.07.018.
Talabot-Ayer D, Lamacchia C, Gabay C, Palmer G: Interleukin-33 is biologically active independently of caspase-1 cleavage. J Biol Chem. 2009, 284: 19420-19426. 10.1074/jbc.M901744200.
Thompson RC, Dripps DJ, Eisenberg SP: IL-1ra: Properties and uses of an interleukin-1 receptor antagonist. Agents Actions Suppl. 1991, 35: 41-49.
Kakkar R, Lee RT: The IL-33/ST2 pathway: Therapeutic target and novel biomarker. Nat Rev Drug Discov. 2008, 7: 827-840. 10.1038/nrd2660.
Dale M, Nicklin MJ: Interleukin-1 receptor cluster: Gene organization of IL1R2, IL1R1, IL1RL2 (IL-1Rrp2), IL1RL1 (T1/ST2), and IL18R1 (IL-1Rrp) on human chromosome 2q. Genomics. 1999, 57: 177-179. 10.1006/geno.1999.5767.
Taylor SL, Renshaw BR, Garka KE, Smith DE, et al: Genomic organization of the interleukin-1 locus. Genomics. 2002, 79: 726-733. 10.1006/geno.2002.6752.
O'Neill LA: Interleukin-1 signal transduction. Int J Clin Lab Res. 1995, 25: 169-177. 10.1007/BF02592694.
Steinkasserer A, Spurr NK, Cox S, Jeggo P, et al: The human IL-1 receptor antagonist gene (IL1RN) maps to chromosome 2q14-q21, in the region of the IL-1 alpha and IL-1 beta loci. Genomics. 1992, 13: 654-657. 10.1016/0888-7543(92)90137-H.
Gracie JA, Robertson SE, McInnes IB: Interleukin-18. J Leukoc Biol. 2003, 73: 213-224. 10.1189/jlb.0602313.
Park LS, Martin U, Garka K, Gliniak B, et al: Cloning of the murine thymic stromal lymphopoietin (TSLP) receptor: Formation of a functional heteromeric complex requires interleukin 7 receptor. J Exp Med. 2000, 192: 659-670. 10.1084/jem.192.5.659.
Rochman Y, Spolski R, Leonard WJ: New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009, 9: 480-490. 10.1038/nri2580.
Liu YJ, Soumelis V, Watanabe N, Ito T, et al: TSLP: An epithelial cell cytokine that regulates T cell differentiation by conditioning dendritic cell maturation. Annu Rev Immunol. 2007, 25: 193-219. 10.1146/annurev.immunol.25.022106.141718.
Chang HC, Sehra S, Goswami R, Yao W, et al: The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol. 2010, 11: 527-534.
Alves NL, Arosa FA, van Lier RA: Common gamma chain cytokines: Dissidence in the details. Immunol Lett. 2007, 108: 113-120. 10.1016/j.imlet.2006.11.006.
Nelms K, Keegan AD, Zamorano J, Ryan JJ, et al: The IL-4 receptor: Signaling mechanisms and biologic functions. Annu Rev Immunol. 1999, 17: 701-738. 10.1146/annurev.immunol.17.1.701.
Obiri NI, Leland P, Murata T, Debinski W, et al: The IL-13 receptor structure differs on various cell types and may share more than one component with IL-4 receptor. J Immunol. 1997, 158: 756-764.
Chiba Y, Todoroki M, Misawa M: Activation of signal transducer and activator of transcription factor 1 by interleukins-13 and -4 in cultured human bronchial smooth muscle cells. J Smooth Muscle Res. 2009, 45: 279-288. 10.1540/jsmr.45.279.
Kitamura T, Sato N, Arai K, Miyajima A: Expression cloning of the human IL-3 receptor cDNA reveals a shared beta subunit for the human IL-3 and GM-CSF receptors. Cell. 1991, 66: 1165-1174. 10.1016/0092-8674(91)90039-2.
Takatsu K, Takaki S, Hitoshi Y: Interleukin-5 and its receptor system: Implications in the immune system and inflammation. Adv Immunol. 1994, 57: 145-190.
Gearing DP, King JA, Gough NM, Nicola NA: Expression cloning of a receptor for human granulocyte-macrophage colony-stimulating factor. EMBO J. 1989, 8: 3667-3676.
Yang LP, Byun DG, Demeure CE, Vezzio N, et al: Default development of cloned human naive CD4 T cells into interleukin-4-and interleukin-5-producing effector cells. Eur J Immunol. 1995, 25: 3517-3520. 10.1002/eji.1830251247.
Pacor S, Gagliardi R, Spessotto P, Zabucchi G, et al: Paracrine effects of IL-4 transfection on TS/A adenocarcinoma cells mediate reduced in vivo growth. Pathol Oncol Res. 1999, 5: 110-116. 10.1053/paor.1999.0172.
Holgate ST, Polosa R: Treatment strategies for allergy and asthma. Nat Rev Immunol. 2008, 8: 218-230. 10.1038/nri2262.
Jazayeri JA, Carroll GJ, Vernallis AB: Interleukin-6 subfamily cytokines and rheumatoid arthritis: Role of antagonists. Int Immunopharmacol. 2010, 10: 1-8. 10.1016/j.intimp.2009.09.019.
Heinrich PC, Behrmann I, Haan S, Hermanns HM, et al: Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003, 374: 1-20. 10.1042/BJ20030407.
Diveu C, Lak-Hal AH, Froger J, Ravon E, et al: Predominant expression of the long isoform of GP130-like (GPL) receptor is required for interleukin-31 signaling. Eur Cytokine Netw. 2004, 15: 291-302.
Ward LD, Hammacher A, Howlett GJ, Matthews JM, et al: Influence of interleukin-6 (IL-6) dimerization on formation of the high affinity hexameric IL-6. receptor complex. J Biol Chem. 1996, 271: 20138-20144. 10.1074/jbc.271.33.20138.
Hilton DJ, Hilton AA, Raicevic A, Rakar S, et al: Cloning of a murine IL-11 receptor alpha-chain: Requirement for gp130 for high affinity binding and signal transduction. EMBO J. 1994, 13: 4765-4775.
Plun-Favreau H, Perret D, Diveu C, Froger J, et al: Leukemia inhibitory factor (LIF), cardiotrophin-1, and oncostatin M share structural binding determinants in the immunoglobulin-like domain of LIF receptor. J Biol Chem. 2003, 278: 27169-27179. 10.1074/jbc.M303168200.
Silver JS, Hunter CA: gp130 at the nexus of inflammation, autoimmunity, and cancer. J Leukoc Biol. 2010,
Chattopadhyay S, Tracy E, Liang P, Robledo O, et al: Interleukin-31 and oncostatin-M mediate distinct signaling reactions and response patterns in lung epithelial cells. J Biol Chem. 2007, 282: 3014-3026.
Trinchieri G, Pflanz S, Kastelein RA: The IL-12 family of heterodimeric cytokines: New players in the regulation of T cell responses. Immunity. 2003, 19: 641-644. 10.1016/S1074-7613(03)00296-6.
Chua AO, Chizzonite R, Desai BB, Truitt TP, et al: Expression cloning of a human IL-12 receptor component. A new member of the cytokine receptor superfamily with strong homology to gp130. J Immunol. 1994, 153: 128-136.
Parham C, Chirica M, Timans J, Vaisberg E, et al: A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit IL-23R. J Immunol. 2002, 168: 5699-5708.
Larousserie F, Bardel E, Pflanz S, Arnulf B, et al: Analysis of interleukin-27 (EBI3/p28) expression in Epstein-Barr virus- and human T-cell leukemia virus type 1-associated lymphomas: Heterogeneous expression of EBI3 subunit by tumoral cells. Am J Pathol. 2005, 166: 1217-1228. 10.1016/S0002-9440(10)62340-1.
Pflanz S, Hibbert L, Mattson J, Rosales R, et al: WSX-1 and glycoprotein 130 constitute a signal-transducing receptor for IL-27. J Immunol. 2004, 172: 2225-2231.
Brunda MJ: Interleukin-12. J Leukoc Biol. 1994, 55: 280-288.
Oppmann B, Lesley R, Blom B, Timans JC, et al: Novel pp19 protein engages IL-12p40 to form a cytokine IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000, 13: 715-725. 10.1016/S1074-7613(00)00070-4.
Niedbala W, Wei XQ, Cai B, Hueber AJ, et al: IL-35 is a novel cytokine with therapeutic effects against collagen-induced arthritis through the expansion of regulatory T cells and suppression of Th17 cells. Eur J Immunol. 2007, 37: 3021-3029. 10.1002/eji.200737810.
Collison LW, Workman CJ, Kuo TT, Boyd K, et al: The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007, 450: 566-569. 10.1038/nature06306.
Hunter CA: New IL-12-family members: IL-23 and IL-27, cytokines with divergent functions. Nat Rev Immunol. 2005, 5: 521-531. 10.1038/nri1648.
Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW: Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene. 2002, 285: 1-24. 10.1016/S0378-1119(02)00398-0.
Heinrich PC, Behrmann I, Muller-Newen G, Schaper F, et al: Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998, 334 (Pt. 2): 297-314.
Gadina M, Ferguson PR, Johnston JA: New interleukins: Are there any more?. Curr Opin Infect Dis. 2003, 16: 211-217. 10.1097/00001432-200306000-00005.
Goriely S, Cavoy R, Goldman M: Interleukin-12 family members and type I interferons in Th17-mediated inflammatory disorders. Allergy. 2009, 64: 702-709. 10.1111/j.1398-9995.2009.02039.x.
Fujihashi K, Kono Y, Kiyono H: Effects of IL6 on B cells in mucosal immune response and inflammation. Res Immunol. 1992, 143: 744-749. 10.1016/0923-2494(92)80015-D.
Van SJ, Renauld JC, Simpson RJ, Uyttenhove C, et al: Mouse Il-6. A hybridoma growth factor with multiple effects on normal B, T cells. Ann NY Acad Sci. 1989, 557: 206-13.
Schwertschlag US, Trepicchio WL, Dykstra KH, Keith JC, et al: Hematopoietic, immunomodulatory and epithelial effects of interleukin-11. Leukemia. 1999, 13: 1307-1315. 10.1038/sj.leu.2401514.
Collison LW, Vignali DA: Interleukin-35: Odd one out or part of the family?. Immunol Rev. 2008, 226: 248-262. 10.1111/j.1600-065X.2008.00704.x.
Castellani ML, Anogeianaki A, Felaco P, Toniato E, et al: IL-35. an anti-inflammatory cytokine which expands CD4+CD25+ Treg Cells. J Biol Regul Homeost Agents. 2010, 24: 131-135.
Monteleone G, Biancone L, Marasco R, Morrone G, et al: Interleukin 12 is expressed and actively released by Crohn's disease intestinal lamina propria mononuclear cells. Gastroenterology. 1997, 112: 1169-1178. 10.1016/S0016-5085(97)70128-8.
Zheng T, Zhu Z, Wang J, Homer RJ, et al: IL-11: Insights in asthma from overexpression transgenic modeling. J Allergy Clin Immunol. 2001, 108: 489-496. 10.1067/mai.2001.118510.
Fiorentino DF, Bond MW, Mosmann TR: Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med. 1989, 170: 2081-2095. 10.1084/jem.170.6.2081.
Zdanov A: Structural features of the interleukin-10 family of cytokines. Curr Pharm Des. 2004, 10: 3873-3884. 10.2174/1381612043382602.
Del PG, De CM, Almerigogna F, Giudizi MG, et al: Human IL-10 is produced by both type 1 helper (Th1) and type 2 helper (Th2) T cell clones and inhibits their antigen-specific proliferation and cytokine production. J Immunol. 1993, 150: 353-360.
Murray PJ: The JAK-STAT signaling pathway: Input and output integration. J Immunol. 2007, 178: 2623-2629.
Pletnev S, Magracheva E, Kozlov S, Tobin G, et al: Characterization of the recombinant extracellular domains of human interleukin-20 receptors and their complexes with interleukin-19 and interleukin-20. Biochemistry. 2003, 42: 12617-12624. 10.1021/bi0354583.
Liao YC, Liang WG, Chen FW, Hsu JH, et al: IL-19 induces production of IL-6 and TNF-alpha and results in cell apoptosis through TNF-alpha. J Immunol. 2002, 169: 4288-4297.
Liu L, Ding C, Zeng W, Heuer JG, et al: Selective enhancement of multipotential hematopoietic progenitors in vitro and in vivo by IL-20. Blood. 2003, 102: 3206-3209. 10.1182/blood-2003-05-1419.
Sabat R, Wallace E, Endesfelder S, Wolk K: IL-19 and IL-20: Two novel cytokines with importance in inflammatory diseases. Expert Opin Ther Targets. 2007, 11: 601-612. 10.1517/14728222.11.5.601.
Kingo K, Mossner R, Koks S, Ratsep R, et al: Association analysis of IL19, IL20 and IL24 genes in palmoplantar pustulosis. Br J Dermatol. 2007, 156: 646-652. 10.1111/j.1365-2133.2006.07731.x.
Kingo K, Mossner R, Ratsep R, Raud K, et al: Association analysis of IL20RA and IL20RB genes in psoriasis. Genes Immun. 2008, 9: 445-451. 10.1038/gene.2008.36.
Parrish-Novak J, Xu W, Brender T, Yao L, et al: Interleukins 19, 20, and 24 signal through two distinct receptor complexes. Differences in receptor-ligand interactions mediate unique biological functions. J Biol Chem. 2002, 277: 47517-47523. 10.1074/jbc.M205114200.
Xie MH, Aggarwal S, Ho WH, Foster J, et al: Interleukin (IL)-22, a novel human cytokine that signals through the interferon receptor-related proteins CRF2-4 and IL-22R. J Biol Chem. 2000, 275: 31335-31339.
Wolk K, Kunz S, Asadullah K, Sabat R: Cutting edge: Immune cells as sources and targets of the IL-10 family members?. J Immunol. 2002, 168: 5397-5402.
Liang SC, Tan XY, Luxenberg DP, Karim R, et al: Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006, 203: 2271-2279. 10.1084/jem.20061308.
Misse D, Yssel H, Trabattoni D, Oblet C, et al: IL-22 participates in an innate anti-HIV-1 host-resistance network through acute-phase protein induction. J Immunol. 2007, 178: 407-415.
de Oliveira NM, Ferreira JR, Colau D, Fischer H, et al: Interleukin-22 forms dimers that are recognized by two interleukin-22R1 receptor chains. Biophys J. 2008, 94: 1754-1765. 10.1529/biophysj.107.112664.
Chang C, Magracheva E, Kozlov S, Fong S, et al: Crystal structure of interleukin-19 defines a new subfamily of helical cytokines. J Biol Chem. 2003, 278: 3308-3313. 10.1074/jbc.M208602200.
Jiang H, Lin JJ, Su ZZ, Goldstein NI, et al: Subtraction hybridization identifies a novel melanoma differentiation associated gene, mda-7, modulated during human melanoma differentiation, growth and progression. Oncogene. 1995, 11: 2477-2486.
Commins S, Steinke JW, Borish L: The extended IL-10 superfamily: IL-10, IL-19, IL-20, IL-22, IL-24, IL-26, IL-28, and IL-29. J Allergy Clin Immunol. 2008, 121: 1108-1111. 10.1016/j.jaci.2008.02.026.
Caudell EG, Mumm JB, Poindexter N, Ekmekcioglu S, et al: The protein product of the tumor suppressor gene, melanoma differentiation-associated gene 7, exhibits immunostimulatory activity and is designated IL-24. J Immunol. 2002, 168: 6041-6046.
Hor S, Pirzer H, Dumoutier L, Bauer F, et al: The T-cell lymphokine interleukin-26 targets epithelial cells through the interleukin-20 receptor 1 and interleukin-10 receptor 2 chains. J Biol Chem. 2004, 279: 33343-33351. 10.1074/jbc.M405000200.
Dambacher J, Beigel F, Zitzmann K, De Toni EN, et al: The role of the novel Th17 cytokine IL-26 in intestinal inflammation. Gut. 2009, 58: 1207-1217. 10.1136/gut.2007.130112.
Igawa D, Sakai M, Savan R: An unexpected discovery of two interferon gamma-like genes along with interleukin (IL)-22 and -26 from teleost: IL-22 and -26 genes have been described for the first time outside mammals. Mol Immunol. 2006, 43: 999-1009. 10.1016/j.molimm.2005.05.009.
Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, et al: IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003, 4: 69-77.
Dellgren C, Gad HH, Hamming OJ, Melchjorsen J, et al: Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun. 2009, 10: 125-131. 10.1038/gene.2008.87.
Witte K, Gruetz G, Volk HD, Looman AC, et al: Despite IFN-lambda receptor expression, blood immune cells, but not keratino-cytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes Immun. 2009, 10: 702-714. 10.1038/gene.2009.72.
Dumoutier L, Tounsi A, Michiels T, Sommereyns C, et al: Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-lambda 1: Similarities with type I interferon signaling. J Biol Chem. 2004, 279: 32269-32274. 10.1074/jbc.M404789200.
Coccia EM, Severa M, Giacomini E, Monneron D, et al: Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol. 2004, 34: 796-805. 10.1002/eji.200324610.
Robek MD, Boyd BS, Chisari FV: Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005, 79: 3851-3854. 10.1128/JVI.79.6.3851-3854.2005.
Ank N, Iversen MB, Bartholdy C, Staeheli P, et al: An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol. 2008, 180: 2474-2485.
Lasfar A, Lewis-Antes A, Smirnov SV, Anantha S, et al: Characterization of the mouse IFN-lambda ligand-receptor system: IFN-lambdas exhibit antitumor activity against B16 melanoma. Cancer Res. 2006, 66: 4468-4477. 10.1158/0008-5472.CAN-05-3653.
Rouvier E, Luciani MF, Mattei MG, Denizot F, et al: CTLA-8, cloned from an activated T cell, bearing AU-rich messenger RNA instability sequences, and homologous to a herpesvirus saimiri gene. J Immunol. 1993, 150: 5445-5456.
Wright JF, Guo Y, Quazi A, Luxenberg DP, et al: Identification of an interleukin 17F/17A heterodimer in activated human CD4+ T cells. J Biol Chem. 2007, 282: 13447-13455. 10.1074/jbc.M700499200.
Chang SH, Dong C: A novel heterodimeric cytokine consisting of IL-17 and IL-17F regulates inflammatory responses. Cell Res. 2007, 17: 435-440.
Shi Y, Ullrich SJ, Zhang J, Connolly K, et al: A novel cytokine receptor-ligand pair. Identification, molecular characterization, and in vivo immunomodulatory activity. J Biol Chem. 2000, 275: 19167-19176. 10.1074/jbc.M910228199.
Gaffen SL: Structure and signalling in the IL-17 receptor family. Nat Rev Immunol. 2009, 9: 556-567. 10.1038/nri2586.
Korn T, Bettelli E, Oukka M, Kuchroo VK: IL-17 and Th17 Cells. Annu Rev Immunol. 2009, 27: 485-517. 10.1146/annurev.immunol.021908.132710.
Fort MM, Cheung J, Yen D, Li J, et al: IL-25 induces IL-4, IL-5, and IL-13 and Th2-associated pathologies in vivo. Immunity. 2001, 15: 985-995. 10.1016/S1074-7613(01)00243-6.
Ziolkowska M, Koc A, Luszczykiewicz G, Ksiezopolska-Pietrzak K, et al: High levels of IL-17 in rheumatoid arthritis patients: IL-15 triggers in vitro IL-17 production via cyclosporin A-sensitive mechanism. J Immunol. 2000, 164: 2832-2838.
Liu ZJ, Yadav PK, Su JL, Wang JS, et al: Potential role of Th17 cells in the pathogenesis of inflammatory bowel disease. World J Gastroenterol. 2009, 15: 5784-5788. 10.3748/wjg.15.5784.
Yu JJ, Gaffen SL: Interleukin-17: A novel inflammatory cytokine that bridges innate and adaptive immunity. Front Biosci. 2008, 13: 170-177. 10.2741/2667.
Shono T, Ono M, Izumi H, Jimi SI, et al: Involvement of the transcription factor NF-kappaB in tubular morphogenesis of human microvascular endothelial cells by oxidative stress. Mol Cell Biol. 1996, 16: 4231-4239.
Strieter RM, Kunkel SL, Showell HJ, Remick DG, et al: Endothelial cell gene expression of a neutrophil chemotactic factor by TNF-alpha LPS, and IL-1 beta. Science. 1989, 243: 1467-1469. 10.1126/science.2648570.
Yoshimura T, Matsushima K, Tanaka S, Robinson EA, et al: Purification of a human monocyte-derived neutrophil chemotactic factor that has peptide sequence similarity to other host defense cytokines. Proc Natl Acad Sci USA. 1987, 84: 9233-9237. 10.1073/pnas.84.24.9233.
Lakshminarayanan V, Drab-Weiss EA, Roebuck KA: H2O2 and tumor necrosis factor-alpha induce differential binding of the redox-responsive transcription factors AP-1 and NF-kappaB to the interleukin-8 promoter in endothelial and epithelial cells. J Biol Chem. 1998, 273: 32670-32678. 10.1074/jbc.273.49.32670.
Najakshin AM, Mechetina LV, Alabyev BY, Taranin AV: Identification of an IL-8 homolog in lamprey (Lampetra fluviatilis): Early evolutionary divergence of chemokines. Eur J Immunol. 1999, 29: 375-382. 10.1002/(SICI)1521-4141(199902)29:02<375::AID-IMMU375>3.0.CO;2-6.
Ambrus JL, Fauci AS: Human B lymphoma cell line producing B cell growth factor. J Clin Invest. 1985, 75: 732-739. 10.1172/JCI111754.
Ambrus JL, Chesky L, Stephany D, McFarland P, et al: Functional studies examining the subpopulation of human B lymphocytes responding to high molecular weight B cell growth factor. J Immunol. 1990, 145: 3949-3955.
Nogami S, Satoh S, Nakano M, Shimizu H, et al: Taxilin: A novel syntaxin-binding protein that is involved in Ca2+-dependent exocytosis in neuroendocrine cells. Genes Cells. 2003, 8: 17-28. 10.1046/j.1365-2443.2003.00612.x.
Nogami S, Satoh S, Tanaka-Nakadate S, Yoshida K, et al: Identification and characterization of taxilin isoforms. Biochem Biophys Res Commun. 2004, 319: 936-943. 10.1016/j.bbrc.2004.05.073.
Shen L, Suresh L, Li H, Zhang C, et al: IL-14 alpha, the nexus for primary Sjogren's disease in mice and humans. Clin Immunol. 2009, 130: 304-312. 10.1016/j.clim.2008.10.006.
Bacon K, Baggiolini M, Broxmeyer H, Horuk R, et al: Chemokine/chemokine receptor nomenclature. J Interferon Cytokine Res. 2002, 22: 1067-1068.
Zhang Y, Center DM, Wu DM, Cruikshank WW, et al: Processing and activation of pro-interleukin-16 by caspase-3. J Biol Chem. 1998, 273: 1144-1149. 10.1074/jbc.273.2.1144.
Baier M, Bannert N, Werner A, Lang K, et al: Molecular cloning, sequence, expression, and processing of the interleukin 16 precursor. Proc Natl Acad Sci USA. 1997, 94: 5273-5277. 10.1073/pnas.94.10.5273.
Wilson KC, Center DM, Cruikshank WW: The effect of interleukin-16 and its precursor on T lymphocyte activation and growth. Growth Factors. 2004, 22: 97-104. 10.1080/08977190410001704679.
Liu C, Mills J, Dixon K, Vennarini J, et al: IL-16 signaling specifically induces STAT6 activation through CD4. Cytokine. 2007, 38: 145-150. 10.1016/j.cyto.2007.05.016.
Cruikshank WW, Center DM, Nisar N, Wu M, et al: Molecular and functional analysis of a lymphocyte chemoattractant factor: Association of biologic function with CD4 expression. Proc Natl Acad Sci USA. 1994, 91: 5109-5113. 10.1073/pnas.91.11.5109.
Center DM, Kornfeld H, Cruikshank WW: Interleukin 16 and its function as a CD4 ligand. Immunol Today. 1996, 17: 476-481. 10.1016/0167-5699(96)10052-I.
Mathy NL, Scheuer W, Lanzendorfer M, Honold K, et al: Interleukin-16 stimulates the expression and production of pro-inflammatory cytokines by human monocytes. Immunology. 2000, 100: 63-69. 10.1046/j.1365-2567.2000.00997.x.
Glass WG, Sarisky RT, Vecchio AM: Not-so-sweet sixteen: The role of IL-16 in infectious and immune-mediated inflammatory diseases. J Interferon Cytokine Res. 2006, 26: 511-520. 10.1089/jir.2006.26.511.
Jemth P, Gianni S: PDZ domains: Folding and binding. Biochemistry. 2007, 46: 8701-8708. 10.1021/bi7008618.
Dahl CA, Schall RP, He HL, Cairns JS: Identification of a novel gene expressed in activated natural killer cells and T cells. J Immunol. 1992, 148: 597-603.
Kim SH, Han SY, Azam T, Yoon DY, et al: Interleukin-32: A cytokine and inducer of TNFalpha. Immunity. 2005, 22: 131-142.
Netea MG, Azam T, Ferwerda G, Girardin SE, et al: IL-32 synergizes with nucleotide oligomerization domain (NOD) 1 and NOD2 ligands for IL-1beta and IL-6 production through a caspase 1-dependent mechanism. Proc Natl Acad Sci USA. 2005, 102: 16309-16314. 10.1073/pnas.0508237102.
Conti P, Youinou P, Theoharides TC: Modulation of autoimmunity by the latest interleukins (with special emphasis on IL-32). Autoimmun Rev. 2007, 6: 131-137. 10.1016/j.autrev.2006.08.015.
Shioya M, Nishida A, Yagi Y, Ogawa A, et al: Epithelial overexpression of interleukin-32alpha in inflammatory bowel disease. Clin Exp Immunol. 2007, 149: 480-486. 10.1111/j.1365-2249.2007.03439.x.
Marcondes AM, Mhyre AJ, Stirewalt DL, Kim SH, et al: Dysregulation of IL-32 in myelodysplastic syndrome and chronic myelomonocytic leukemia modulates apoptosis and impairs NK function. Proc Natl Acad Sci USA. 2008, 105: 2865-2870. 10.1073/pnas.0712391105.
Rasool ST, Tang H, Wu J, Li W, et al: Increased level of IL-32 during human immunodeficiency virus infection suppresses HIV replication. Immunol Lett. 2008, 117: 161-167. 10.1016/j.imlet.2008.01.007.
Lin H, Lee E, Hestir K, Leo C, et al: Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science. 2008, 320: 807-811. 10.1126/science.1154370.
Wei S, Nandi S, Chitu V, Yeung YG, et al: Functional overlap but differential expression of CSF-1 and IL-34 in their CSF-1 receptor-mediated regulation of myeloid cells. J Leukoc Biol. 2010, 88: 495-505. 10.1189/jlb.1209822.
Garceau V, Smith J, Paton IR, Davey M, et al: Pivotal Advance: Avian colony-stimulating factor 1 (CSF-1), interleukin-34 (IL-34), and CSF-1 receptor genes and gene products. J Leukoc Biol. 2010, 87: 753-764. 10.1189/jlb.0909624.
Pandit J, Bohm A, Jancarik J, Halenbeck R, et al: Three-dimensional structure of dimeric human recombinant macrophage colony-stimulating factor. Science. 1992, 258: 1358-1362. 10.1126/science.1455231.
Acknowledgements
We would like to thank our colleagues for valuable discussion and critical review of this manuscript. This work was supported, in part, by the following NIH grants: R01 EY17963 (V.V.), R21 AA017754 (V.V.), F31 AA018248 (C.B.) and P30 ES06096 (D.W.N.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Brocker, C., Thompson, D., Matsumoto, A. et al. Evolutionary divergence and functions of the human interleukin (IL) gene family. Hum Genomics 5, 30 (2010). https://doi.org/10.1186/1479-7364-5-1-30
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1479-7364-5-1-30