Abstract
Background
Although a lower methylation level of whole genome has been demonstrated in Tetralogy of Fallot (TOF) patients, little is known regarding changes in specific gene DNA methylation profiles and the possible associations with TOF. In current study, the promoter methylation statuses of congenital heart defect (CHD) candidate genes were measured in order to further understand epigenetic mechanisms that may play a role in the development of TOF.
Methods
The methylation levels of CHD candidate genes were measured using the Sequenom MassARRAY platform. QRT-PCR was used to analyze the mRNA levels of CHD candidate genes in the right ventricular myocardium of TOF cases and normal controls.
Results
Methylation status analysis was performed on the promoter regions of 71 CHD candidate genes (113 amplicons). We found significant differences in methylation status, between TOF cases and controls, in 26 amplicons (26 genes) (p < 0.05). Of the 26 amplicons, 17 were up regulated and 9 were down regulated. Additionally, 14 of them were located in the CpG islands, 7 were located in the CpG island shores, and 5 were covering the regions near the transcription start site (TSS). The methylation status was subsequently confirmed and mRNA levels were measured for 7 represented candidate genes, including EGFR, EVC2, NFATC2, NR2F2, TBX5, CFC1B and GJA5. The methylation values of EGFR, EVC2, TBX5 and CFC1B were significantly correlated with their mRNA levels (p < 0.05).
Conclusions
Aberrant promoter methylation statuses of CHD candidate genes presented in TOF cases may contribute to the TOF development and have potential prognostic and therapeutic significance for TOF disease.
Similar content being viewed by others
Background
A congenital heart defect (CHD) is an abnormality in the structure and great vessels of the heart, which is present at birth. About 19–75 people per 1000 are born with CHD each year, depending on which types of defects are included[1] and the incidence is higher if stillbirths were included[2]. Tetralogy of Fallot (TOF) is the most common complex congenital heart disease[3], accounting for 10% of all congenital heart diseases[4]. The four distinct anatomical features that characterize TOF are pulmonary outflow tract obstruction, ventricular septal defect, overriding aortic root and right ventricular hypertrophy[5]. TOF is considered to be a neural crest cell and/or second heart field related conotruncal heart defects that occur during embryonic development[6]. Although advances in surgical techniques, cardiopulmonary bypass and postoperative care in the last few decades have brought us today to where survival after complete repair is greater than 98% in large congenital heart surgery programs[7] and have improved the life quality of TOF patients to an excellent condition, late sudden cardiac death remains a persistent risk, with an estimated incidence of 0.5% to 6%[8], however, the exact etiology of TOF remains unclear. Some studies have reported that some of these defects related to TOF may be associated with chromosomal aneuploidy, including trisomies 13, 18 and 21[9]. In addition to chromosomal rearrangements, mutations in a number of genes have also been found to be associated with TOF. A variety of gene insertion/deletion has been detected in patients with TOF. These genes include JAG1[10, 11], NKX2-5 and GATA4[12, 13] etc. However, a single gene mutation or deletion can be identified only in a small percentage of cases[14]. Thus, we hypothesize that aberrant epigenetic regulation may be another important factor associated with TOF. Epigenetics refers to the heritable changes in genome function that occur without a change in the primary DNA sequence. These are characterized by covalent modifications of cytosine bases and histones, and changes in the positioning of nucleosomes[15]. Currently, the most widely studied epigenetic modification in humans is DNA methylation. DNA methylation can control the transcriptional activity of genes by various mechanisms, which occurs almost exclusively in the context of CpG dinucleotides[16]. The CpG dinucleotides tend to cluster in regions called CpG islands. Studies have proven that over 60% of human gene promoters are overlapped with CpG islands and are usually unmethylated in normal cells[17]. However, recent work on colon cancer has demonstrated that DNA methylation not only occurs at CpG islands, but also takes place in regions of lower CpG density that lie in close proximity (~2 kb) to the CpG islands, which is known as the CpG island shore[18]. The CpG island shores are the most enriched with functional CpG sites and have variable methylation, which are closely associated with transcriptional regulation. Most of the tissue-specific DNA methylation seems to occur, not at CpG islands, but at CpG island shores[19, 20].
Methylation within gene promoters and CpG-dense sequences (CpG islands) have the highest functional relevance to gene expression control and the aberrant methylation changes contribute to many diseases[15]. The methylation patterns of multiple genes can provide different types of useful information about cancer cells[21]. The CpG island methylator phenotype, referring to the concurrent methylation of multiple genes, is a useful marker for tumor progression and has been reported in hepatocellular carcinoma[22]. However, although a lower methylation level of global genome has been demonstrated in TOF patients[23], little is known about gene-specific DNA methylation changes in patients with TOF.
In this study, we selected 71 CHD candidate genes, based on the evidences of methylation microarray performed previously[24], transcriptional studies, mouse models and their close association with the heart development, to explore their promoter DNA methylation changes and their association with TOF development.
Materials and methods
Patients and controls
The TOF cases were recruited from the Children’s Hospital of the Fudan University, Shanghai, China. Cardiovascular diagnosis was obtained by echocardiography. All TOF cases were tested for chromosome anomalies by karyotype and 22q11 deletion by fluorescent in situ hybridization (FISH) as previously described[25]. Patients with trisomy 21, 22q11 deletion, other chromosomal anomalies, or extracardiac major or minor associated anomalies were discarded in this study. A total of 41 TOF cases were recruited, including 26 (63.4%) males and 15 (36.6%) females, ranging in age from 2.0 to 48.0 months [11.0 (7.0 - 24.0), median (interquartile range)].
The control group was comprised of necropsy specimens from normal subjects that had died as a result of an accident. Necropsy specimens were collected at the forensic medicine department of the Fudan University, Shanghai, China. Considering a delay with necropsy, the post mortem interval (PMI) for the control samples was as short as possible. Six age-matched normal controls were collected, including 4 (66.7%) males and 2 (33.3%) females, ranging in age from 6.0 to 54.0 months [15.0 (10.5 - 30.0), median (interquartile range)]. Anagraphical characteristics of the study subjects are summarized in Table 1.
To exclude the tissue heterogeneity that may affect methylation levels, all heart tissue samples from TOF patients and controls were obtained from the right ventricular myocardial tissues and saved in storage solution (RNAlater®, AMBION, Inc., Austin, USA) immediately following surgical resection or necropsy and then stored in -80°C until use.
The Fudan University’ Ethics Committee approved this study. Written informed consents were obtained from the parents or relatives of all study subjects.
DNA extraction and sodium bisulfite conversion
Genomic DNA was extracted from myocardial tissues using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The concentration and purity of the DNA were determined by absorbance at 260 and 280 nm using a NanoDrop™ 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and agarose gel. Sodium bisulfite modification was performed on the extracted DNA, using an EZ DNA Methylation Kit™ (Zymo Research, Orange, CA, USA) strictly following the manufacturer’s instructions. The bisulfite-converted DNA was re-suspended in 10 μl elution buffer and stored at -80°C until the samples were ready for analysis.
MassARRAY quantitative methylation analysis
The Sequenom MassARRAY platform was used to perform the quantitative methylation analysis for the promoter regions of 71 CHD candidate genes. The methylation status of a detected pattern was then analyzed using Epityper software version 1.0 (Sequenom, San Diego, CA, USA). The promoter regions of the 71 CHD candidate genes were chosen using the websitehttp://genome.ucsc.edu (shown in Additional file1: Table S1). The 113 amplicons and PCR primers used in this system were designed using the websitehttp://epidesigner.com (Additional file1: Table S2).
All experiments were performed as described before[23]. Non-applicable readings and their corresponding sites were eliminated from analysis. The methylation level of the genes promoter regions is expressed as the percentage of methylated cytosines over the total of methylated and unmethylated cytosines.
RNA extraction and quantitative RT-PCR
Total RNA was extracted from tested myocardial tissue samples using Trizol Reagent (Invitrogen, CA, USA) according to the manufacturer’s instructions. The extracted RNA quality and integrity were validated before use. RNA was reverse-transcribed using a PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Japan) and the integrity of synthesized cDNA was confirmed using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the endogenous control. Quantitative RT-PCR was performed in a 7900 real-time PCR system using SYBR Premix Ex Taq GC (Perfect Real Time) (TaKaRa, Japan) following the manufacturer’s instructions. Reactions were performed in triplicate and analyzed using an ABI 7900 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Relative expression levels were calculated according to the standard 2-ΔΔCt method using beta-2 microglobulin (B2M) and the GAPDH gene as the endogenous control for normalization. Primer sequences used in QRT-PCR analysis are listed in Additional file1: Table S3.
Statistical analysis
The methylation status of the CHD candidate genes’ promoter regions was analyzed using GraphPad Prism (version 5.0; GraphPad Software Inc., San Diego, CA, USA) and SPSS (version 13.0; SPSS Inc., Chicago, IL, USA). A Mann–Whitney test was used to evaluate the significance of any differences between TOF cases and normal controls. Spearman’s rank correlation was used to examine the relationship between two continuous variables. All statistical analyses were 2-sided and P < 0.05 was considered statistically significant.
Results
Methylation status analyses of the promoter region of 71 CHD candidate genes in TOF cases and controls
To comprehensively understand the promoter regions of CHD candidate genes that were analyzed, we focused on the region covering 2000 bp + 200 bases relative to transcription start sites (TSS). Based on the information regarding the promoter region in each of 71 candidate genes, we designed a total of 113 amplicons, of which 72 covered the CpG island (64%), 24 covered the CpG island shore (21%) and 17 were not associated with CpG island (15%) (shown in Table 2). Using the Sequenom MassARRAY platform, the methylation levels in the promoter regions of these genes were detected in 10 TOF cases and 6 controls for the first round of screening. Prior to analysis, we performed strict quality control to remove any potentially unreliable measurements, such as low mass, high mass, and silent peak overlap CpG units. The CpG units that failed to produce data in more than 30% of samples (unreliable CpG units) and samples that were missing more than 30% of the data points (unreliable samples) were discarded[26]. The mean and median methylation levels for the 113 amplicons (71 candidate genes) in the TOF cases and controls are shown in Additional file1: Table S4.
The results indicated that the median methylation values in 26 of 113 amplicons (26 genes) were significantly different in TOF cases compared with controls (shown in Additional file1: Table S5, p < 0.05). The methylation levels were up regulated in 17 genes, including CFC1B, DVL2, EGFR, EDNRA, EVC2, GJA5, HAND1, HAS2, HSPG2, MED13L, NFATC1, NKX2-5, NFATC2, PAX3, TBX5, TEK and ZFPM2. Whereas, the methylation levels was down regulated in 9 genes, including EDN1, HOXA3, MYH6, NR2F2, NRG1, NRP1, PDGFRA, SLC2A10, and TBX20. There were 14 amplicons (14 genes: DVL2, EDN1, EGFR, EVC2, HAND1, HAS2, HSPG2, NKX2-5, NRG1, NFATC2, PAX3, PDGFRA, SLC2A10, TBX20), located in the CpG islands; 7 amplicons (7 genes: EDNRA, MED13L, NFATC1, NR2F2, NRP1, TBX5, ZFPM2), were located in the CpG island shore, and 5 amplicons (5 genes: CFC1B, GJA5, HOXA3, MYH6 ,TEK), covered the region near transcription start site (TSS).
Validation of the methylation statuses of seven candidate genes in TOF cases and controls
On the basis of the data analyzed above, 7 CHD candidate genes, including EGFR, EVC2, NFATC2, NR2F2, TBX5, CFC1B and GJA5, were chosen and validated for their methylation levels in the samples from 41 patients with TOF. These genes were selected because they function as key factors in the development of heart as well as showing significant differences in methylation status between TOF cases and controls. Moreover, the amplicon fragments measured in these genes were distributed in different regions of the promoter: EGFR, EVC2 and NFATC2 were associated with the CpG islands, NR2F2 and TBX5 with the CpG island shores and CFC1B and GJA5 were in the regions near the TSS.
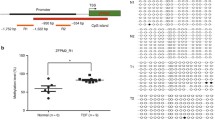
As shown in Table 3, the significant differences for methylation values of the 7 genes were confirmed in a larger number of samples. In the CpG island region of gene promoter in TOF cases, the methylation levels of the following genes were significantly up-regulated: EGFR (52.75% vs 60.50%, median, p = 0.0042, Figure 1A), EVC2 (29.63% vs 43.88%, median, p = 0.0005, Figure 1B), and NFATC2 (21.80% vs 30.29%, median, p = 0.0067, Figure 1C). The concurrent higher methylation of EGFR, EVC2 and NFATC2 might represent a CpG island methylator phenotype in TOF development.
Median methylation levels in the promoter CpG islands of candidate genes of TOF cases and controls. (A) EGFR_ M1 median methylation level; (B) EVC2_ M1 median methylation level; (C) NFATC2_ M2 median methylation level. TSS, Transcription Start Sites; red vertical line, CpG sites; red thick bars, CpG islands; M, MassARRAY amplicon; The region between two arrows, target amplicon. *P < 0.05, **P < 0.01, ***P < 0.001 (Mann–Whitney test).
In the CpG island shore region of gene promoter in TOF cases, the methylation level of NR2F2 was significantly down regulated (62.50% vs 43.40%, median, p = 0.0161, Figure 2A), though the methylation level of TBX5 was higher (42.04% vs 57.13%, median, p = 0.0207, Figure 2B) in TOF cases.
Median methylation levels in the promoter CpG island shores and the promoter region located outside of the CpG island but near the TSS between TOF cases and controls. (A) NR2F2 median methylation level; (B) TBX5 median methylation level; (C) CFC1B median methylation level; (D) GJA5 median methylation level; TSS, Transcription Start Sites; red vertical line, CpG sites; red thick bars, CpG islands; The region between two arrows, target amplicon. *P < 0.05, **P < 0.01, ***P < 0.001 (Mann–Whitney test).
Furthermore, in the TSS promoter region of gene in TOF cases, the methylation levels of CFC1B_ M2 and GJA5 were significantly up regulated (48.16% vs 63.50%, median, p = 0.0178, Figure 2C; 32.83% vs 44.00%, median, p = 0.0138, Figure 2D; respectively) compared to controls.
mRNA expression levels of seven CHD candidate genes in TOF cases and controls
QRT-PCR was performed to determine the mRNA expression levels of the 7 CHD candidate genes in TOF cases and normal controls. As shown in Table 4, the mRNA levels of the 7 candidate genes were significantly lower in TOF cases than in controls (p < 0.05).
Correlations between methylation statuses of seven CHD candidate genes and their respective mRNA levels
A correlation analysis was then done in the TOF cases to identify whether there was any relationship between the methylation statuses and mRNA levels of 7 CHD candidate genes. In the promoter CpG island region, two significant associations were found between the methylation statuses and mRNA levels for EGFR (r = - 0.531, p = 0.0015, Figure 3A) and EVC2 (r = - 0.409, p = 0.020, Figure 3B). No significant correlation was observed between the methylation status and mRNA level for NFATC2 (r = - 0.212, p = 0.183, Figure 3C).
In the CpG island shore region, no significant correlation was observed between the methylation status and mRNA level of NR2F2 (r = - 0.074, p = 0.644, Figure 4A). Interestingly, the methylation status of TBX5 was negatively correlated with its mRNA expression level (r = - 0.418, p = 0.0101, Figure 4B).
A strong correlation was found between the methylation level in the promoter region near the TSS of CFC1B and its mRNA level (r = - 0.555, p = 0.0002, Figure 4C). However, the methylation status of GJA5 was not significantly associated with its mRNA level (r = 0.068, p = 0.690, Figure 4D).
Discussion
DNA methylation constitutes an important epigenetic regulation mechanism in many eukaryotes[27]. In previous study, we have demonstrated the hypomethylation of LINE-1, which can serve as an indicator of global DNA methylation[28], in the myocardial tissue of TOF patients[23]. In the present study, quantitative methylation analysis was performed on the promoter region of 71 CHD candidate genes in the myocardial tissue of TOF cases and normal controls using the Sequenom MassARRAY platform. This system combines base-specific enzymatic cleavage with MALDI-TOF mass spectrometry and creates a highly accurate, sensitive, and high-throughput method for the quantitative analysis of DNA methylation at CpG units[29]. The robustness of this approach for quantifying methylated and unmethylated DNA has been confirmed by the Sequenom group[30]. We found that 26 of the 71 candidate genes showed significant differences in the methylation levels when comparing TOF cases to controls (p < 0.05). In which, 17 genes were up regulation and 9 genes were down regulation. DNA methylation analysis for multiple genes in TOF cases enables us to reveal the complex etiology of CHD from a novel aspect and provides potential development of new treatments for TOF disease.
The methylation statuses at different regions of the gene promoter may have different effects on the gene’s activities. The 26 genes showing significant differences in the methylation levels can be grouped to three categories according to the location of amplicons in the gene promoter. Fourteen amplicons (14 genes) were located in the CpG islands, 7 amplicons (7 genes) were located in the CpG island shore, and 5 amplicons (5 genes) were covering the region near the TSS.
Seven CHD candidate genes, including EGFR, EVC2, NFATC2, NR2F2, TBX5, CFC1B and GJA5, were chosen for further validation in a larger number of TOF cases because of their nominally significant differences in methylation levels and the important roles they play in the development of the heart. EGFR, a receptor tyrosine kinase in the ErbB family, activates several signaling cascades that convert extra-cellular cues into appropriate cellular responses and has been demonstrated to be associated with the congenital left ventricular outflow tract obstruction[31]. EVC2 plays a critical role in bone formation and skeletal development and mutations in EVC2 are associated with Ellis van Creveld syndrome in which 50-60% of congenital heart defects occur[32]. NFATC2 is a DNA binding protein with a REL-homology region (RHR) and an NFAT-homology region (NHR). It plays a central role in inducing gene transcription during the immune response. Bourajjaj M. et all, 2008, have shown that NFATC2 is a necessary mediator of calcineurin-dependent cardiac hypertrophy and heart failure[33]. NR2F2 is a ligand inducible transcription factor that is involved in the regulation of many different genes and the deletion of NR2F2 was considered to possibly contribute to congenital heart defects[34]. TBX5 is a member of the T-box transcription factor family and plays an important role in heart development and the specification of limb identity, which is very well known associated with Holt Oram syndrome, a developmental disorder affecting the heart and upper limbs[35]. The regulatory variation in the TBX5 enhancer can lead to the same phenotype as a mutation in the gene. CFC1B is an important factor in embryo development. Mutations in this gene have been reported in Chinese children with congenital heart disease[36]. The GJA5 gene encodes for the cardiac gap junction protein connexin 40. A variant in the carboxyl-terminus of connexin 40 alters GAP junctions and increases the risk for TOF[37].
Aberrant DNA methylation of CpG islands has been widely observed in human tumors and is associated with gene silencing when it occurs in promoter areas[38]. Multiple genes showing increased or decreased methylation simultaneously have been found in colorectal cancer[39] and in duodenal adenocarcinomas[40]. In the present study, we found significantly higher methylation levels in the promoter CpG islands of EGFR, EVC2 and NFATC2 in TOF cases compared with controls. The simultaneous higher methylation of EGFR, EVC2 and NFATC2 may represented a CpG island methylator phenotype in TOF development and provide useful clues as to the development of an epigenetic classification of the disease with prognostic and therapeutic potential. Moreover, EGFR and EVC2 were found to have significant negative correlations between methylation values and respective mRNA expression levels (Figure 3A,B , p < 0.05), indicating that methylation changes at the CpG island region of the two genes may have influences on gene expression, though the exact control mechanism requires further study. Although the mRNA level of NFATC 2 was significantly lower in TOF cases compared to the controls in our study (p < 0.05), no significant correlation between methylation level and mRNA expression was observed (Figure 3C, p > 0.05). We concluded that the analyzed methylation region at the promoter CpG island of NFATC2 might not be involved in regulating the gene transcription.
A recent genome-wide analysis of DNA methylation showed that 76% of differential tissue methylation regions were not located in CpG islands, but in CpG island shore[18]. In this study, we found a decreased methylation level at the CpG island shore for NR2F2 and an increased methylation level for TBX5. We cannot ascertain the factor that contributed to the lower methylation level of NR2F2. However, the methylation status of NR2F2 were found not to be associated with its mRNA level, suggesting that methylation changes at the CpG island shore of NR2F2 might not influence transcriptional activity. Further studies are needed to explore whether the methylation changes in the other region of NR2F2 promoter influences its mRNA level. Interestingly, the methylation status of the TBX5 gene was significantly negatively correlated with its mRNA level, indicating that increased methylation level at the CpG island shore of TBX5 may inhibit this gene transcriptional activity.
DNA methylation can directly inhibit transcription by precluding the recruitment of DNA binding proteins from their target sites[41]. Cao et al. found that CpG site-specific methylation can alter binding affinities of specific transcription factors, which can differentially activate or repress transcription[42]. Consistent with this, Tihomira D et al. have reported that, although methylation of all CpG sites resulted in the silencing of EphA5 promoter activity, lower levels of methylation resulted in differential activation or repression of EphA5 promoter activity, depending on the sites methylated[43]. In the current study, the methylation levels at the promoter region near the TSS of CFC1B and GJA5 were significantly higher in TOF cases compared to normal controls. In addition, other than GJA5, one significant negative correlation was observed between the methylation status and mRNA level of CFC1B, indicating that the methylation change at the promoter region near the TSS of CFC1B may play an important role in regulating the gene transcriptional activity. How the altered methylation statuses of CFC1B influence mRNA level requires further study.
However, in the present study, we have not examined the protein levels of these genes in the myocardial tissue because of the limited samples. The potential alterations of the protein levels and their associations with the transcription level would be explored in the further study.
Conclusions
Our results suggest that multiple gene-specific DNA methylation changes at the promoter regions occurred in TOF cases and may be associated with TOF development. The concurrent higher methylation of EGFR, EVC2 and NFATC2 might constitute a CpG island methylator phenotype for TOF disease and provide useful cues to understand epigenetic mechanisms in the development of TOF. The methylation values of EGFR, EVC2, TBX5 and CFC1B were significantly correlated with their respective mRNA levels (p < 0.05), indicating that aberrant methylation changes of specific genes may contributes to the development of TOF. Because heart tissue samples are difficult to collect from healthy controls and TOF patients, one limitation of this study is that we were unable to obtain enough complete matched samples. This could influence the accuracy of the methylation results. Also, we cannot be sure if the methylation patterns observed in the samples were an effect of or caused the TOF since the development of TOF long predated the measurement of methylation. Further studies with larger sample numbers are warranted to confirm our findings and to focus on whether aberrant methylation contributes to TOF development.
References
Hoffman JI, Kaplan S: The incidence of congenital heart disease. J Am Coll Cardiol. 2002, 39: 1890-1900. 10.1016/S0735-1097(02)01886-7.
Hoffman JI: Incidence of congenital heart disease: II. prenatal incidence. Pediatr Cardiol. 1995, 16: 155-165.
Child JS: Fallot's tetralogy and pregnancy: prognostication and prophesy. J Am Coll Cardiol. 2004, 44: 181-183. 10.1016/j.jacc.2004.04.009.
Bedard E, McCarthy KP, Dimopoulos K, Giannakoulas G, Gatzoulis MA, Ho SY: Structural abnormalities of the pulmonary trunk in tetralogy of Fallot and potential clinical implications: a morphological study. J Am Coll Cardiol. 2009, 54: 1883-1890. 10.1016/j.jacc.2009.06.040.
Ho S, McCarthy KP, Josen M, Rigby ML: Anatomic-echocardiographic correlates: an introduction to normal and congenitally malformed hearts. Heart. 2001, 86 (Suppl 2): II3-II11.
Ward C, Stadt H, Hutson M, Kirby ML: Ablation of the secondary heart field leads to tetralogy of Fallot and pulmonary atresia. Dev Biol. 2005, 284: 72-83. 10.1016/j.ydbio.2005.05.003.
Starr JP: Tetralogy of fallot: yesterday and today. World J Surg. 2010, 34: 658-668. 10.1007/s00268-009-0296-8.
di Felice V, Zummo G: Tetralogy of Fallot as a model to study cardiac progenitor cell migration and differentiation during heart development. Trends Cardiovasc Med. 2009, 19: 130-135. 10.1016/j.tcm.2009.07.004.
Beauchesne LM, Warnes CA, Connolly HM, Ammash NM, Grogan M, Jalal SM, Michels VV: Prevalence and clinical manifestations of 22q11.2 microdeletion in adults with selected conotruncal anomalies. J Am Coll Cardiol. 2005, 45: 595-598. 10.1016/j.jacc.2004.10.056.
Eldadah ZA, Hamosh A, Biery NJ, Montgomery RA, Duke M, Elkins R, Dietz HC: Familial Tetralogy of Fallot caused by mutation in the jagged1 gene. Hum Mol Genet. 2001, 10: 163-169. 10.1093/hmg/10.2.163.
Bauer RC, Laney AO, Smith R, Gerfen J, Morrissette JJ, Woyciechowski S, Garbarini J, Loomes KM, Krantz ID, Urban Z: Jagged1 (JAG1) mutations in patients with tetralogy of Fallot or pulmonic stenosis. Hum Mutat. 2010, 31: 594-601. 10.1002/humu.21231.
Goldmuntz E, Geiger E, Benson DW: NKX2.5 mutations in patients with tetralogy of Fallot. Circulation. 2001, 104: 2565-2568. 10.1161/hc4601.098427.
Salazar M, Consoli F, Villegas V, Caicedo V, Maddaloni V, Daniele P, Caianiello G, Pachon S, Nunez F, Limongelli G: Search of somatic GATA4 and NKX2.5 gene mutations in sporadic septal heart defects. Eur J Med Genet. 2011, 54: 306-309. 10.1016/j.ejmg.2011.01.004.
de Luca A, Sarkozy A, Ferese R, Consoli F, Lepri F, Dentici ML, Vergara P, de Zorzi A, Versacci P, Digilio MC: New mutations in ZFPM2/FOG2 gene in tetralogy of Fallot and double outlet right ventricle. Clin Genet. 2011, 80: 184-190. 10.1111/j.1399-0004.2010.01523.x.
Portela A, Esteller M: Epigenetic modifications and human disease. Nat Biotechnol. 2010, 28: 1057-1068. 10.1038/nbt.1685.
Goll MG, Bestor TH: Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005, 74: 481-514. 10.1146/annurev.biochem.74.010904.153721.
Vanderkraats ND, Hiken JF, Decker KF, Edwards JR: Discovering high-resolution patterns of differential DNA methylation that correlate with gene expression changes. Nucleic Acids Res. 2013, 41: 6816-6827. 10.1093/nar/gkt482.
Irizarry RA, Ladd-Acosta C, Wen B, Wu ZJ, Montano C, Onyango P, Cui HM, Gabo K, Rongione M, Webster M: The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009, 41: 178-186. 10.1038/ng.298.
Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S: Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009, 41: 1350-1353. 10.1038/ng.471.
Baccarelli A, Rienstra M, Benjamin EJ: Cardiovascular epigenetics: basic concepts and results from animal and human studies. Circ Cardiovasc Genet. 2010, 3: 567-573.
Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC: Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000, 24: 132-138. 10.1038/72785.
Zhang C, Li Z, Cheng Y, Jia F, Li R, Wu M, Li K, Wei L: CpG island methylator phenotype association with elevated serum alpha-fetoprotein level in hepatocellular carcinoma. Clin Cancer Res. 2007, 13: 944-952. 10.1158/1078-0432.CCR-06-2268.
Sheng W, Wang H, Ma X, Qian Y, Zhang P, Wu Y, Zheng F, Chen L, Huang G, Ma D: LINE-1 methylation status and its association with tetralogy of fallot in infants. BMC Med Genomics. 2012, 5: 20-10.1186/1755-8794-5-20.
Chowdhury S, Erickson SW, MacLeod SL, Cleves MA, Hu P, Karim MA, Hobbs CA: Maternal genome-wide DNA methylation patterns and congenital heart defects. PLoS One. 2011, 6: e16506-10.1371/journal.pone.0016506.
Lammer EJ, Chak JS, Iovannisci DM, Schultz K, Osoegawa K, Yang W, Carmichael SL, Shaw GM: Chromosomal abnormalities among children born with conotruncal cardiac defects. Birth Defects Res A Clin Mol Teratol. 2009, 85: 30-35. 10.1002/bdra.20541.
Ollikainen M, Smith KR, Joo EJ, Ng HK, Andronikos R, Novakovic B, Abdul Aziz NK, Carlin JB, Morley R, Saffery R, Craig JM: DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum Mol Genet. 2010, 19: 4176-4188. 10.1093/hmg/ddq336.
Yagi S, Hirabayashi K, Sato S, Li W, Takahashi Y, Hirakawa T, Wu G, Hattori N, Ohgane J, Tanaka S: DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res. 2008, 18: 1969-1978. 10.1101/gr.074070.107.
Zhang T, Wang L, Wang F, Guan J, Le J, Wu LH, Zou JZ, Zhao HZ, Pei LJ, Zheng XY: Relation between hypomethylation of long interspersed nucleotide elements and risk of neural tube defects. Am J Clin Nutr. 2010, 91: 1359-1367. 10.3945/ajcn.2009.28858.
Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D: Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005, 102: 15785-15790. 10.1073/pnas.0507816102.
Ehrich M, Turner J, Gibbs P, Lipton L, Giovanneti M, Cantor C, van den Boom D: Cytosine methylation profiling of cancer cell lines. Proc Natl Acad Sci U S A. 2008, 105: 4844-4849. 10.1073/pnas.0712251105.
McBride KL, Zender GA, Fitzgerald-Butt SM, Seagraves NJ, Fernbach SD, Zapata G, Lewin M, Towbin JA, Belmont JW: Association of common variants in ERBB4 with congenital left ventricular outflow tract obstruction defects. Birth Defects Res A Clin Mol Teratol. 2011, 91: 162-168. 10.1002/bdra.20764.
Shen W, Han D, Zhang J, Zhao H, Feng H: Two novel heterozygous mutations of EVC2 cause a mild phenotype of Ellis-van Creveld syndrome in a Chinese family. Am J Med Genet A. 2011, 155A: 2131-2136.
Bourajjaj M, Armand AS, da Costa Martins PA, Weijts B, van der Nagel R, Heeneman S, Wehrens XH, De Windt LJ: NFATc2 is a necessary mediator of calcineurin-dependent cardiac hypertrophy and heart failure. J Biol Chem. 2008, 283: 22295-22303. 10.1074/jbc.M801296200.
Nakamura E, Makita Y, Okamoto T, Nagaya K, Hayashi T, Sugimoto M, Manabe H, Taketazu G, Kajino H, Fujieda K: 5.78 Mb terminal deletion of chromosome 15q in a girl, evaluation of NR2F2 as candidate gene for congenital heart defects. Eur J Med Genet. 2011, 54: 354-356. 10.1016/j.ejmg.2010.12.004.
Smemo S, Campos LC, Moskowitz IP, Krieger JE, Pereira AC, Nobrega MA: Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease. Hum Mol Genet. 2012, 21: 3255-3263. 10.1093/hmg/dds165.
Wang B, Wang J, Liu S, Han X, Xie X, Tao Y, Yan J, Ma X: CFC1 mutations in Chinese children with congenital heart disease. Int J Cardiol. 2011, 146: 86-88. 10.1016/j.ijcard.2009.07.034.
Guida V, Ferese R, Rocchetti M, Bonetti M, Sarkozy A, Cecchetti S, Gelmetti V, Lepri F, Copetti M, Lamorte G: A variant in the carboxyl-terminus of connexin 40 alters GAP junctions and increases risk for tetralogy of Fallot. Eur J Hum Genet. 2013, 21: 69-75. 10.1038/ejhg.2012.109.
Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D: CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006, 38: 787-793. 10.1038/ng1834.
Karpinski P, Walter M, Szmida E, Ramsey D, Misiak B, Kozlowska J, Bebenek M, Grzebieniak Z, Blin N, Laczmanski L, Sasiadek MM: Intermediate- and low-methylation epigenotypes do not correspond to CpG island methylator phenotype (low and -zero) in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2013, 22: 201-208. 10.1158/1055-9965.EPI-12-0157.
Fu T, Pappou EP, Guzzetta AA, Jeschke J, Kwak R, Dave P, Hooker CM, Morgan R, Baylin SB, Iacobuzio-Donahue CA: CpG island methylator phenotype-positive tumors in the absence of MLH1 methylation constitute a distinct subset of duodenal adenocarcinomas and are associated with poor prognosis. Clin Cancer Res. 2012, 18: 4743-4752. 10.1158/1078-0432.CCR-12-0707.
Kuroda A, Rauch TA, Todorov I, Ku HT, Al-Abdullah IH, Kandeel F, Mullen Y, Pfeifer GP, Ferreri K: Insulin gene expression is regulated by DNA methylation. PLoS One. 2009, 4: e6953-10.1371/journal.pone.0006953.
Cao YX, Jean JC, Williams MC: Cytosine methylation of an Sp1 site contributes to organ-specific and cell-specific regulation of expression of the lung epithelial gene t1alpha. Biochem J. 2000, 350 (Pt 3): 883-890.
Petkova TD, Seigel GM, Otteson DC: A role for DNA methylation in regulation of EphA5 receptor expression in the mouse retina. Vision Res. 2011, 51: 260-268. 10.1016/j.visres.2010.09.022.
Acknowledgements
This work was supported by grants from the National Basic Research Program of China (2010CB529504, 2009CB941704), the National Natural Sciences Foundation of China (30930096), Shanghai Municipal Commission of Science and Technology Research Project (11JC1401400) and China Postdoctoral Science Foundation (2013 M540324).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SW participated in study concept and design and coordination of the study, helped with the statistical analysis and drafted the manuscript. QY, WY, WH and MX participated in TOF sample acquisition and helped to draft the manuscript. ZP participated in normal control sample acquisition and helped to draft the manuscript. CL, MD and HG conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors have read and approved the final manuscript.
Electronic supplementary material
12967_2013_1725_MOESM1_ESM.doc
Additional file 1: Table S1: Congenital heart defect (CHD) candidate gene data. Table S2. Primer sequences, position, product length, and CpG site used for MassArray quantitative methylation analysis. Table S3. Primer sequences and product length for QPT-PCR analysis. Table S4. Mean and median methylation levels for 113 amplicons (71 candidate genes) in 10 TOF cases and 6 Controls. Table S5. The median methylation levels of 26 candidate genes showing significant difference in TOF cases and controls. (DOC 510 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Sheng, W., Qian, Y., Zhang, P. et al. Association of promoter methylation statuses of congenital heart defect candidate genes with Tetralogy of Fallot. J Transl Med 12, 31 (2014). https://doi.org/10.1186/1479-5876-12-31
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1479-5876-12-31