Abstract
Background
Oocyte secreted factors (OSFs), including growth differentiation factor 9 (GDF9) and bone morphogenetic protein 15 (BMP15), play an important role in the process of follicular development and oocyte maturation. Since OSFs are expressed in oocytes and cumulus granulosa cells, the aim of the present study was to explore whether the expression levels of GDF9 and BMP15 mRNAs in cumulus granulosa cells can be used as molecular markers for predicting oocyte developmental potential.
Methods
Cumulus cells of 2426 cumulus-oocyte complexes were collected from 196 female patients who underwent intracytoplasmic sperm injection (ICSI) and were used for mRNA detection on the egg retrieval day. Pearson correlation analysis was used to analyze the correlation between OSF expression and general physiological parameters. Partial correlation analysis was used to analyze the correlation between OSF expression and oocyte developmental potential. Covariance analysis was used to compare OSF expression among different groups. Receiver operating characteristic curves were used to examine the diagnostic value of GDF9 and BMP15 mRNA for predicting pregnancy.
Results
The expression levels of GDF9 and BMP15 mRNAs were significantly associated with age, body mass index (BMI), oocyte maturation, normal fertilization, and cleavage rate (P < 0.05). The expression levels of GDF9 and BMP15 mRNAs in the group with high-quality embryos were significantly higher than those in the group without high-quality embryos (P < 0.05). The expression levels of GDF9 and BMP15 mRNAs in the pregnancy group were significantly higher than those in the nonpregnancy group (P < 0.05). The cut-off value of GDF9 mRNA for predicting pregnancy was 4.82, with a sensitivity of 82% and a specificity of 64%. The cut-off value of BMP15 mRNA for predicting pregnancy was 2.60, with a sensitivity of 78% and a specificity of 52%.
Conclusions
The expression levels of GDF9 and BMP15 mRNAs were closely associated with oocyte maturation, fertilization, embryo quality, and pregnancy outcome; therefore, GDF9 and BMP15 mRNAs in cumulus granulosa cells may be considered as new molecular markers for predicting oocyte developmental potential.
Similar content being viewed by others
Background
The oocyte developmental potential is one of the key factors for determining the successful rate of in vitro fertilization and embryo transfer (IVF-ET). The accurate evaluation of oocyte developmental potential is an important issue in assisted reproduction. The traditional method uses morphological scoring. The advantages of morphological scoring lie in its simplicity, convenience, and fast speed [1–3]. However, the main shortcoming of this method is that it depends too much on the abilities of the technician, so it is difficult to reach a uniform standard. In some cases, the morphological scoring may not accurately reflect the oocyte developmental potential and embryo quality [4].
Recently, global assessment strategies including genomic, transcriptomic, and proteomic approaches have been applied in assisted reproduction [5]. These strategies aim to present a “molecular profile” of embryo development by detecting the chemical components in the oocyte, granulosa cells, follicular fluid, and embryo culture medium. These methods pave a new way to enhance the accuracy of the oocyte developmental potential.
Granulosa cells are distributed on the follicular wall (mural granulosa cells) or closely adjacent to the oocyte (cumulus granulosa cells). The physiological function of mural granulosa cells is predominantly related to hormone secretion. Cumulus granulosa cells often exchange biological signals with oocytes through the gap junction [6–8]. There is a mutual communication between cumulus granulosa cells and the oocyte. Recent studies have shown that the expression levels of some genes in cumulus granulosa cells are helpful for predicting the oocyte developmental potential, such as hyaluronic acid synthase 2 (HAS2), gremlin 1 (GREM1), etc. [9, 10]. Since cumulus granulosa cells are by-products of intracytoplasmic sperm injection (ICSI) and the method of detecting gene expression in cumulus granulosa cells does not affect the oocyte developmental potential, it is very possible to screen suitable potential biomarkers for predicting oocyte developmental potential in IVF clinics.
Oocyte secreted factors (OSFs) include growth differentiation factor 9 (GDF9) and bone morphogenetic protein 15 (BMP15). Both factors play an important role in the process of follicular development from the recruitment of the primordial follicle to ovulation and even in corpus luteum formation [11–13]. OSFs contribute to promoting the proliferation and differentiation of granulosa cells and oocyte maturation through paracrine and autocrine signaling pathways. Previous studies have indicated that higher GDF9 and BMP15 levels in the follicular fluid are significantly associated with oocyte maturation and embryo quality [14–16]. In vitro studies have shown that GDF9 and BMP15 may stimulate M-phase-promoting factor (MPF) and mitogen-activated protein kinase (MAPK) activities in oocytes and improve oocyte quality and subsequent developmental potential [17, 18]. In addition, it has been confirmed by many studies that OSFs are expressed both in oocytes and cumulus granulosa cells [19–21]. The aims of the present study were to detect the expression levels of GDF9 and BMP15 mRNAs in cumulus granulosa cells and to analyze the correlation between their expression levels and the oocyte developmental potential.
Methods
Study design
This study was approved by the Institutional Review Board of Sun Yat-Sen University in March 2012 (NO. E2012003). All the subjects signed the informed consent. This retrospective study was conducted at the Center for Reproductive Medicine in Memorial Hospital of Sun Yat-Sen University from September 2012 to April 2013. In total, 196 women who underwent ICSI because their husbands were diagnosed as having severe oligospermia and asthenospermia (total number <1 × 106/ml, motility <5%) were recruited into this study. The general information of the patients is presented in Table 1. The inclusion criteria for all patients included a long protocol for ovarian stimulation, age ≤45 years, body mass index (BMI) of 17–35 kg/m2, and basal follicle-stimulating hormone (FSH) level ≤IU/L. The exclusion criteria included a history of previous poor response, recurrent implantation failure (failed to achieve a pregnancy after three or more cycles), submucosal fibroids, intrauterine adhesion, congenital uterine malformation, hydrosalpinx, ovarian endometriomas >3 cm in diameter, and polycystic ovarian syndrome.
Ovarian stimulation protocol
All subjects underwent a long protocol for ovarian stimulation. The blood was taken for determining the basal levels of endocrine hormones during the menstrual cycle. Endocrine hormones such as FSH, luteinizing hormone (LH), estradiol (E2), testosterone (T), and prolactin (PRL) were measured by an Axsym chemiluminescence detection system (Axsym; Abbott Laboratories, Rungis, France). A gonadotropin-releasing hormone agonist (1.25 mg, Ipsen Pharma Biotech, Paris, France) was subcutaneously injected for pituitary down regulation. Gonadotropin (Gonal-f, 150–300 IU; Merck Serono, Darmstadt, Germany) was subcutaneously injected for ovarian hyperstimulation 2 weeks after downregulation. Human chorionic gonadotropin (hCG, 10,000 IU; Northern Pharma Inc., Rostov-Na-Dony, Russia) was intramuscularly injected when the diameter of at least three dominant follicles was larger than 18 mm. The collection of cumulus-oocyte complexes (COCs) was conducted with a 17G needle under ultrasound monitoring 36 h after hCG injection.
Quantitative polymerase chain reaction (qPCR)
In total, 2426 COCs retrieved from pooled follicles of individual patients were cultured in vitro for 2 h; then, the cumulus granulosa cells were stripped off with visualization under an inverted micoscope after hyaluronidase digestion. The stripped cumulus granulosa cells (1 × 102–5 × 103) were rinsed with phosphate-buffered saline two times and mixed with Trizol (Invitrogen, Grand Island, NY, USA). Total RNA was extracted with Trizol and reversely transcribed into cDNA with the Superscript III kit (Invitrogen, Grand Island, NY, USA). The qPCR was performed with Taqman fluorescent probes and an ABI Prism7700 detection system. The primers for GDF9 (NM_005448), BMP15 (NM_005260), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were as follows: GDF9: forward 5′-GGCAAGGCCTCACAGAGGTA-3′, reverse 5′-CGGTAAACCACAGTGGCTCTAAC-3′; BMP15: forward 5′-CTGCTTTGCCTGGCTGTGT-3′, reverse 5′-CAAGGCATAGCCCCAGATTC-3′; GAPDH: forward 5′-CCTGCACCACCAACTGCTTAG-3′, reverse 5′-CAGTCTTCTGGGTGGCAGTGA-3′. GAPDH was used as the endogenous control for normalization. All the PCR conditions consisted of 93°C for 3 min, followed by 40 cycles of 93°C for 45 s and 55°C for 1 min. The 2-ΔΔCt algorithm was used to calculate the GDF9 and BMP15 mRNA levels relative to the GAPDH level.
Oocyte and embryo assessment
Oocyte maturation was examined under an inverted micoscope. If the first polar body was observed in the oocyte cytoplasm, the oocyte was regarded as being at the metaphase II (MII) stage. The oocyte maturation rate refers to the number of MII oocytes divided by the total number of all retrieved oocytes. The oocyte at the MII stage was fertilized with the help of the ICSI procedure. Oocyte fertilization was observed 18–19 h after ICSI. Normal fertilization was confirmed when two pronuclei (2PN) were found in the cytoplasm. The normal fertilization rate refers to the number of fertilized oocytes divided by the total number of all retrieved oocytes. Embryo cleavage was examined 43–45 h after ICSI. Normal embryo cleavage was defined when the fertilized egg developed into an embryo with 4–6 blastomeres. The cleavage rate refers to the number of cleaved zygotes divided by the total number of all zygotes. Embryo evaluation was conducted 67–69 h after oocyte fertilization. A high-quality embryo should consist of 7–9 blastomeres with a uniform size, and the fragment proportion should be less than 10%. If a patient had at least one embryo with the above criteria, she would be included in the group with high-quality embryos. If a patient did not have any embryos with the above criteira, she would be included in the group without high-quality embryos. No more than three embryos were transferred into the uterine cavity on day 3 of in vitro culture. Clinical pregnancy was diagnosed when the gestational sac and fetal heart beat were observed under ultrasound 5 weeks after embryo transfer.
Statistical analysis
The one-sample Kolmogorov-Smirnov test was used to examine the normal distribution of all data. The relative expression levels of GDF9 and BMP15 mRNAs were logarithmically transformed into a normal distribution. The data in the tables were expressed as means ± standard deviation (SD). Pearson correlation analysis was used to analyze the correlation between OSF expression and general physiological paratmeters. Partial correlation analysis was used to analyze the correlation between GDF9 and BMP15 expression and oocyte developmental potential after age adjustment. Covariance analysis was used to compare GDF9 and BMP15 expression in different groups after adjusting for the number of retrieved oocytes. Receiver operating characteristic (ROC) curves were used to examine the diagnostic value of GDF9 and BMP15 mRNA for predicting pregnancy. The area under the curve (AUC) represents the probability of correctly identifying the pregnant and nonpregnant cases. Data analysis was conducted with SPSS 11.5, and P < 0.05 was considered as statistically significant.
Results
Correlation between the expression of GDF9 and BMP15mRNAs and physiological parameters
The expression level of GDF9 mRNA was significantly associated with age and BMI but not FSH, and the coefficients of correlation were 0.278 (P < 0.05), 0.188 (P < 0.05), and 0.017 (P > 0.05), respectively (Figure 1a–c). Similarly, the expression level of BMP15 mRNA was also associated with age and BMI but not FSH, and the coefficients of correlation were 0.324 (P < 0.05), 0.226 (P < 0.05), and 0.024 (P > 0.05), respectively (Figure 1d–f).
Pearson analysis was used to analyze the correlation between GDF9 and BMP15 expression and general parameters in 196 patients who underwent ICSI. The numbers on the horizzontal-axes refer to the age, BMI, and FSH. The numbers on the y longitudinal axes refer to the relative mRNA expression of GDF9 or BMP15. (a) The correlation between age and the relative expression of GDF9 mRNA. (b) The correlation between BMI and the relative expression of GDF9 mRNA. (c) The correlation between FSH and the relative expression of GDF9 mRNA. (d) The correlation between age and the relative expression of BMP15 mRNA. (e) The correlation between BMI and the relative expression of BMP15 mRNA. (f) The correlation between FSH and the relative expression of BMP15 mRNA.
Correlation between the expression of GDF9 and BMP15mRNAs and oocyte developmental potential
The expression level of GDF9 mRNA was significantly associated with oocyte maturation, normal fertilization, and cleavage rate after age adjustment; and the partial correlation coefficients were 0.353 (P < 0.001), 0.489 (P < 0.001), and 0.592 (P < 0.001), respectively (Figure 2a–c). Similarly, the expression level of BMP15 mRNA was also associated with oocyte maturation, normal fertilization, and cleavage rate after age adjustment; and the partial correlation coefficients were 0.345 (P < 0.001), 0.402 (P < 0.001), and 0.593 (P < 0.001), respectively (Figure 2d–f).
Partial correlation anlysis was used to analyze the correlation between GDF9 and BMP15 expression and oocyte developmental potential in 196 patients who underwent ICSI. The numbers on the horizontal axes refer to the oocyte maturation rate (MII), normal fertilization rate (2PN), and cleavage rate. The numbers on the longitudinal axes refer to the relative mRNA expression of GDF9 or BMP15. (a) The correlation between oocyte maturation rate and the relative expression of GDF9 mRNA. (b) The correlation between normal fertilization rate and the relative expression of GDF9 mRNA. (c) The correlation between cleavage rate and the relative expression of GDF9 mRNA. (d) The correlation between oocyte maturation rate and the relative expression of BMP15 mRNA. (e) The correlation between normal fertilization rate and the relative expression of BMP15 mRNA. (f) The correlation between cleavage rate and the relative expression of BMP15 mRNA.
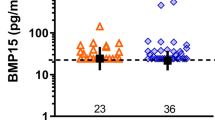
Comparison of the expression levels of GDF9 and BMP15mRNAs between the groups with and without high-quality embryos
The patient information is presented in Table 2. The relative expression levels of GDF9 and BMP15 mRNAs in the group with high-quality embryos were 4.79 ± 0.27 and 3.21 ± 0.30, respectively, while their levels in the group without high-quality embryos were 2.52 ± 0.39 and 1.69 ± 0.39, respectively. Thus, the expression levels of GDF9 (P < 0.05, F = 20.62) and BMP15 mRNAs (P < 0.05, F = 2.75) in the group with high-quality embryos were signifcantly greater than those in the group without high-quality embryos after adjusting for the number of retrieved oocytes (Figure 3a–b).
Comparison of the expression levels of GDF9 and BMP15mRNAs between the groups with or without pregnancy
The patient information is presented in Table 3. The expression level of GDF9 mRNA in the pregnancy group (4.40 ± 1.55) was significantly greater than that in the nonpregnancy group (2.65 ± 0.24) (P < 0.05, F = 18.35, Figure 4a). Also, the BMP15 mRNA level in the pregnancy group (2.79 ± 0.17) was significantly greater than that in the nonpregnancy group (1.93 ± 0.25) after adjusting for the number of retrieved oocytes (P < 0.05, F = 1.96, Figure 4b).
Diagnostic value of GDF9 and BMP15 mRNAs for predicting pregnancy
The diagnostic value of GDF9 and BMP15 mRNAs for predicting pregnancy is summarized in Table 4. The area under the ROC curve of GDF9 mRNA for predicting pregnancy was 0.816 (0.757–0.875), with a cut-off value of 4.82, a sensitivity of 82%, and a specificity of 64%. The area under the ROC curve of BMP15 for predicting pregnancy was 0.746 (0.671–0.821), with a cut-off value of 2.60, a sensitivity of 78%, and a specificity of 52%.
Discussion
In the present study, a qPCR method was used to detect the expression levels of GDF9 and BMP15 mRNAs in cumulus granulosa cells from patients who underwent ICSI. The correlation between GDF9 or BMP15 mRNA and oocyte development potential was analyzed in order to explore new biomarkers for embryo selection.
OSFs are not only autocrine but also paracrine factors. GDF9 and BMP15 were examined to predict oocyte developmental potential, while previous studies have only detected the expression levels of some downstream genes [22, 23]. Many studies have confirmed that both GDF9 and BMP15 are expressed in the oocyte cytoplasm and cumulus granulosa cells [19–21]. In animal studies, it has been shown that GDF9 and BMP15 can stimulate oocyte development [24]. Furthermore, mouse oocytes matured with exogenous GDF9 had a higher percentage of hatching blastocysts and a better blastocyst quality, and the number of viable fetuses was also increased [25]. GDF9 and BMP15 play an important role in oocyte development. Both factors contribute to promoting the proliferation and metabolism of granulosa cells, and stimulate the expression of kit ligand (KL) on granulosa cells. The KL acts on its receptor on the oocyte and modulates oocyte development [26]. To date, the expression of GDF9 and BMP15 mRNAs has not been used to predict oocyte quality. Here, we found that the expression of GDF9 and BMP15 mRNAs in cumulus granulosa cells was positively correlated with oocyte maturation, normal fertilization rate, and cleavage rate. Because cumulus granulosa cells are closely associated with the oocyte, the expression of GDF9 and BMP15 mRNA in cumulus granulosa cells may reflect oocyte developmental potential.
In addition, we observed that the mRNA expression of GDF9 and BMP15 were positively associated with age and BMI. Age and BMI are important factors that may affect the ovarian response and pregancy outcome of assisted reproductive technology. However, the age and BMI may not have a direct relationship with the expression of GDF9 and BMP15; therefore, further investigation is needed. Furthermore, the rate of oocyte maturation in the group with higher expression of GDF9 and BMP15 mRNA was significantly greater than that in the group with lower GDF9 and BMP15 mRNA expression. The rate of oocyte maturation was positively related to the mRNA expression of GDF9 and BMP15. Fertilization is closely related to oocyte quality such as oocyte maturation and spindle structure [27–30]. The fertilization rate in high-quality oocytes is often greater than that in low-quality oocytes. Low-quality oocytes tend to have a higher abnormal fertilization rate due to imperfect oocyte function. Our results indicated that the fertilization rate increased along with the increased mRNA levels of GDF9 and BMP15. Moreover, oocyte quality dramatically affects subsequent embryo development. Low-quality oocytes often have a lower chance of developing into high-quality embryos [31]. Accordingly, the patients with a higher expression of OSFs also had a higher cleavage rate. This evidence helps to explain our findings that the expression of GDF9 and BMP15 mRNAs in the group with high quality embryos was significantly greater than that in the group without a high quality embryo. Clinical pregnancy is an important indicator for evaluating embryo quality. Although the pregnancy outcome is determined by several factors such as sperm quality and the uterine endometrium, the oocyte is the most important factor for clinical outcome. A high-quality oocyte more easily develops into a high-quality embryo; accordingly, the chance of clinical pregnancy is also increased [32]. The present study confirmed that the expression of GDF9 and BMP15 mRNAs in the pregnancy group was significantly greater than that in the nonpregnancy group, suggesting that the expression of GDF9 and BMP15 mRNA may be used as an indicator to predict clinical pregnancy outcome. As the ROC curve was used to evaluate the diagnostic value of GDF9 and BMP15 mRNAs, the cut-off value can be established for predicting pregnancy with a relatively high sensitivity. These data indicate that the detection of GDF9 and BMP15 mRNAs in cumulus granulosa cells may have broad application prospects as invasive biomarkers for evaluating ooctye developmental potential.
However, the limitation of this study is that the cumulus granulosa cells were not harvested from an individual oocyte, so the expression of GDF9 and BMP15 mRNA does not match with each oocyte and embryo. A future study that detects the expression of OSFs from an individual follicle is still needed to validate the conclusion. Additionally, there may be some other OSFs such as BMP6 and fibroblast growth factor (FGF) 8B that could be used as biomarkers [33, 34]. In the present study, we only detected the mRNA expression of the two well-known factors GDF9 and BMP15 in cumulus granulosa cells. It will be interesting to explore the relationship between the mRNA expression of other OSFs and oocyte developmental potential in the future.
Conclusions
The expression of GDF9 and BMP15 mRNAs was significantly correlated with oocyte maturation, fertilization, embryo quality, and clinical pregnancy outcome. Thus, the levels of GDF9 and BMP15 mRNA in cumulus granulosa cells may be considered as new biomarkers for predicting oocyte developmental potential.
Abbreviations
- E2:
-
Estradiol
- FSH:
-
Follicle-stimulating hormone
- LH:
-
Luteinizing hormone
- PRL:
-
Prolactin
- T:
-
Testosterone.
References
Scott L, Smith S: The successful use of pronuclear embryo transfers the day following oocyte retrieval. Hum Reprod. 1998, 13: 1003-1013.
Dennis SJ, Thomas MA, Williams DB, Robins JC: Embryo morphology score on day 3 is predictive of implantation and live birth rates. J Assist Reprod Genet. 2006, 23: 171-175.
Gardner DK, Lane M, Stevens J, Schlenker T, Schoolcraft WB: Blastocyst score affects implantation and pregnancy outcome: towards a single blastocyst transfer. Fertil Steril. 2000, 73: 1155-1158.
Wang Q, Sun QY: Evaluation of oocyte quality: morphological, cellular and molecular predictors. Reprod Fertil Dev. 2007, 19: 1-12.
Seli E, Robert C, Sirard MA: OMICS in assisted reproduction: possibilities and pitfalls. Mol Hum Reprod. 2010, 16: 513-530.
Gilchrist RB, Ritter LJ, Armstrong DT: Oocyte-somatic cell interactions during follicle development in mammals. Anim Reprod Sci. 2004, 82–83: 431-446.
Kidder GM, Vanderhyden BC: Bidirectional communication between oocytes and follicle cells: ensuring oocyte developmental competence. Can J Physiol Pharmacol. 2010, 88: 399-413.
Canipari R: Oocyte--granulosa cell interactions. Hum Reprod Update. 2000, 6: 279-289. Review
Cillo F, Brevini TA, Antonini S, Paffoni A, Ragni G, Gandolfi F: Association between human oocyte developmental competence and expression levels of some cumulus genes. Reproduction. 2007, 134: 645-650.
Anderson RA, Sciorio R, Kinnell H, Bayne RA, Thong KJ, de Sousa PA, Pickering S: Cumulus gene expression as a predictor of human oocyte fertilisation, embryo development and competence to establish a pregnancy. Reproduction. 2009, 138: 629-637.
Paulini F, Melo EO: The role of oocyte-secreted factors GDF9 and BMP15 in follicular development and oogenesis. Reprod Domest Anim. 2011, 46: 354-361.
Trombly DJ, Woodruff TK, Mayo KE: Roles for transforming growth factor beta superfamily proteins in early folliculogenesis. Semin Reprod Med. 2009, 27: 14-23.
Kedem A, Fisch B, Garor R, Ben-Zaken A, Gizunterman T, Felz C, Ben-Haroush A, Kravarusic D, Abir R: Growth differentiating factor 9 (GDF9) and bone morphogenetic protein 15 both activate development of human primordial follicles in vitro, with seemingly more beneficial effects of GDF9. J Clin Endocrinol Metab. 2011, 96: 1246-1254.
McNatty KP, Hudson NL, Whiting L, Reader KL, Lun S, Western A, Heath DA, Smith P, Moore LG, Juengel JL: The effects of immunizing sheep with different BMP15 or GDF9 peptide sequences on ovarian follicular activity and ovulation rate. Biol Reprod. 2007, 76: 552-560.
Gode F, Gulekli B, Dogan E, Korhan P, Dogan S, Bige O, Cimrin D, Atabey N: Influence of follicular fluid GDF9 and BMP15 on embryo quality. Fertil Steril. 2011, 95: 2274-2278.
Wu YT, Tang L, Cai J, Lu XE, Xu J, Zhu XM, Luo Q, Huang HF: High bonemorphogenetic protein-15 level in follicular fluid is associated with high quality oocyte and subsequent embryonic development. Hum Reprod. 2007, 22: 1526-1531.
Sudiman J, Ritter LJ, Feil DK, Wang X, Chan K, Mottershead DG, Robertson DM, Thompson JG, Gilchrist RB: Effects of differing oocyte-secreted factors during mouse in vitro maturation on subsequent embryo and fetal development. J Assist Reprod Genet. 2014, 31: 295-306.
Lin ZL, Li YH, Xu YN, Wang QL, Namgoong S, Cui XS, Kim NH: Effects of growth differentiation factor 9 and bone morphogenetic protein 15 on the in vitro maturation of porcine oocytes. Reprod Domest Anim. 2014, 49: 219-227.
Zhao SY, Qiao J, Chen YJ, Liu P, Li J, Yan J: Expression of growth differentiation factor-9 and bone morphogenetic protein-15 in oocytes and cumulus granulosa cells of patients with polycystic ovary syndrome. Fertil Steril. 2010, 94: 261-267.
Hosoe M, Kaneyama K, Ushizawa K, Hayashi KG, Takahashi T: Quantitative analysis of bone morphogenetic protein 15 (BMP15) and growth differentiation factor 9 (GDF9) gene expression in calf and adult bovine ovaries. Reprod Biol Endocrinol. 2011, 9: 33-
Silva JR, van den Hurk R, van Tol HT, Roelen BA, Figueiredo JR: Expression of growth differentiation factor 9 (GDF9), bone morphogenetic protein 15 (BMP15), and BMP receptors in the ovaries of goats. Mol Reprod Dev. 2005, 70: 11-19.
Adriaenssens T, Wathlet S, Segers I, Verheyen G, De Vos A, Van der Elst J, Coucke W, Devroey P, Smitz J: Cumulus cell gene expression is associated with oocyte developmental quality and influenced by patient and treatment characteristics. Hum Reprod. 2010, 25: 1259-1270.
McKenzie LJ, Pangas SA, Carson SA, Kovanci E, Cisneros P, Buster JE, Amato P, Matzuk MM: Human cumulus granulosa cell gene expression: a predictor of fertilization and embryo selection in women undergoing IVF. Hum Reprod. 2004, 19: 2869-2874.
Sudiman J, Ritter LJ, Feil DK, Wang X, Chan K, Mottershead DG, Robertson DM, Thompson JG, Gilchrist RB: Effects of differing oocyte-secreted factors during mouse in vitro maturation on subsequent embryo and fetal development. J Assist Reprod Genet. 2014, 31: 295-306.
Lin ZL, Li YH, Xu YN, Wang QL, Namgoong S, Cui XS, Kim NH: Effects of growth differentiation factor 9 and bone morphogenetic protein 15 on the in vitro maturation of porcine oocytes. Reprod Domest Anim. 2014, 49: 219-227.
Gilchrist RB, Lane M, Thompson JG: Oocyte-secreted factors: regulators of cumulus cell function and oocyte quality. Hum Reprod Update. 2008, 14: 159-177.
Bilodeau-Goeseels S, Panich P: Effects of oocyte quality on development and transcriptional activity in early bovine embryos. Anim Reprod Sci. 2002, 71: 143-155.
Wang W, Hosoe M, Li R, Shioya Y: Development of the competence of bovine oocytes to release cortical granules and block polyspermy after meiotic maturation. Dev Growth Differ. 1997, 39: 607-615.
Santos P, Chaveiro A, Simões N, Moreira da Silva F: Bovine oocyte quality in relation to ultrastructural characteristics of zona pellucida, polyspermic penetration and developmental competence. Reprod Domest Anim. 2008, 43: 685-689.
Watson AJ: Oocyte cytoplasmic maturation: a key mediator of oocyte and embryo developmental competence. J Anim Sci. 2007, 85: 1-3.
Hill GA, Freeman M, Bastias MC, Rogers BJ, Herbert CM, Osteen KG, Wentz AC: The influence of oocyte maturity and embryo quality on pregnancy rate in a program for in vitro fertilization-embryo transfer. Fertil Steril. 1989, 52: 801-806.
Check JH, Summers-Chase D, Yuan W, Horwath D, Wilson C: Effect of embryo quality on pregnancy outcome following single embryo transfer in women with a diminished egg reserve. Fertil Steril. 2007, 87: 749-756.
Juengel JL, McNatty KP: The role of proteins of the transforming growthfactor-beta superfamily in the intraovarian regulation of follicular development. Hum Reprod Update. 2005, 11: 143-160.
Sugiura K, Su YQ, Diaz FJ, Pangas SA, Sharma S, Wigglesworth K, O'Brien MJ, Matzuk MM, Shimasaki S, Eppig JJ: Oocyte-derived BMP15 and FGFs cooperate to promote glycolysis in cumulus cells. Development. 2007, 134: 2593-2603.
Acknowledgments
This work was supported by grants from the National Science Fund of China (Nos. 81200417 and 81200476), the Doctoral Fund of the Ministry of Education of China (Nos. 20120171120093 and 20120171120122), the Science Fund of Guangdong Province (S2012040007621 and S2012040007770), the Medical Research Fund of Guangdong Province (B2012104 and B2012150), the Young Teacher Fund of Sun Yat-sen University (11ykpy24), the Project of Science and Technology New Star in Zhu Jiang of Guangzhou City (2014 J2200044), and the Yat-Sen Scholarship for Young Scientists. We thank the editors from Medjaden Bioscience Limited for proofreading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YL collected the data and drafted the manuscript. R-QL, S-BO, and N-FZ performed the qRT-PCR. LR, Q-XZ, and D-ZY revised the manuscript. L-NW designed the study and validated the data. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Li, Y., Li, RQ., Ou, SB. et al. Increased GDF9 and BMP15 mRNA levels in cumulus granulosa cells correlate with oocyte maturation, fertilization, and embryo quality in humans. Reprod Biol Endocrinol 12, 81 (2014). https://doi.org/10.1186/1477-7827-12-81
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-7827-12-81