Abstract
Background
The adverse effects on offspring of diabetic and/or obese mothers can be passed to the next generation. However, the mechanisms behind this are still unclear. Epigenetics may play a key role during this process.
Methods
To confirm the hypothesis, we investigated the DNA methylation of several imprinted genes in spermatozoa of offspring from diabetic and/or obese mothers utilizing streptozotocin (STZ)- and high-fat-diet (HFD)-induced mouse models.
Results
We found that the DNA methylation of Peg3 was significantly increased in spermatozoa of offspring of obese mothers compared to that in spermatozoa of offspring of normal mothers. The DNA methylation of H19 was significantly higher in spermatozoa of offspring of diabetic mothers than that in spermatozoa of offspring of non-diabetic mothers.
Conclusions
These results indicate that pre-gestational diabetes and/or obesity can alter DNA methylation in offspring spermatozoa.
Similar content being viewed by others
Background
There is evidence that exposure to an adverse fetal and/or early postnatal environment may enhance susceptibility to a number of chronic diseases in the future life of offspring. Pre-existing maternal diabetes and obesity are both complications which influence the development of offspring during fetal life and postnatal development. Offspring of women with pre-gestational diabetes have a high risk of developing obesity, impaired glucose tolerance, and type 2 diabetes in adulthood [1]. Epidemiological and clinical studies have shown that maternal type 1 diabetes during pregnancy is an important risk factor for the development of obesity and diabetes in offspring [2, 3]. In animal models, a fetus exposed to poorly controlled female diabetes mellitus has a higher incidence of abortion, metabolic diseases, malformation, and stillbirth [4, 5]. Furthermore, offspring from females with pregestational diabetes are susceptible to onset of obesity, glucose intolerance, and type 2 diabetes [1, 6]. Such metabolic consequences persist through the F1 and F2 generations [7, 8].
It is widely recognized that obesity is becoming a big problem for the world-wide. Offspring of women with pre-pregnant obesity and overweight are prone to onset of obesity, type 2 diabetes, hypertension, and cardiovascular diseases [9]. A retrospective study shows that children of obese mothers are twice as likely to be obese at 24 months of age compared to non-obese mothers [10]. Lawlor and colleagues showed that offspring of overweight and obese mothers had more adipose tissue than those born to normal-weight mothers at 18 years of age [10, 11]. In high fat diet-induced obese rats, the offspring of obese dams exhibit increased adiposity and insulin resistance until postnatal day 130 compared to those of lean dams [12]. Bayol et al. showed that 10-week-old rats born to mothers fed a high-fat diet exhibited an increased body weight and fat mass when compared to control offspring [13]. Obesity, insulin resistance, glucose intolerance and metabolic syndrome observed in offspring born by obese mothers are still apparent in the second generation of offspring [14].
The intergenerational transmission of adverse health effects from mothers with pre-existing diabetes and/or obesity cannot be completely explained by genetics [15]. Epigenetics may perfectly illustrate how the effect is inherited by the next generations. It has been reported that maternal and/or paternal lineages imprinted genes may play a key role in defects perpetuated to the next generations [16, 17]. In our study, we have previously shown that the methylation patterns of several imprinted genes in oocytes of offspring from mothers with pregestational diabetes [18] or obesity [19] were not altered. However, the overall methylome patterns in male pup’s sperm of pre-diabetic fathers were altered [20]. Whether the DNA methylation status in male offspring spermatozoa of pre-diabetic/-obese mothers is affected is still unknown. Therefore, we investigated the DNA methylation patterns in DMRs (differential methylation regions) of paternally imprinted genes H19, Gtl2, Rasgrf1, and maternally imprinted genes Peg3, Snrpn in spermatozoa of offspring born to obese mothers. In this study we further investigated DNA methylation patterns of H19, Gtl2 and Peg3 in spermatozoa from offspring of diabetic mothers.
Materials
CD-1 mice were provided by Beijing Vital River Experimental Animals Centre and fed in a temperature controlled room with a light cycle of 12 L:12D (light:dark). All procedures described were reviewed and approved by the ethical committee of the Institute of Zoology, Chinese Academy of Sciences.
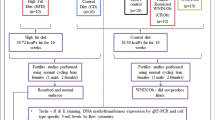
Offspring born by maternal diabetic and obese mice
CD-1 mice 6-7-weeks old whose weights were 26.5-27.5 g were divided into two groups, randomly. One group received a single intraperitoneal injection of streptozotocin (STZ), which can impair islets and reduce the synthesis of insulin, at a dose of 230 mg/kg and the other group received buffer. The blood glucose level was checked utilizing a glucometer, Blood Testing Equipment, Accu-CHEK Active (Roche Diagnostic, Germany) by cutting the tip of the tail on day 4 after injection of STZ. If the glucose concentration was higher than 17.0 mmol/l, the mice were selected. The blood glucose level of mice injected with buffer was also checked and the blood glucose level was lower than 7.0 mmol/l. The diabetic and nondiabetic mice were mated with normal male mice at 15 days of injection with STZ/buffer. Twenty one days later, offspring were born. At the age of 7–8 weeks, nine male pups of 3–4 litters of diabetic and nondiabetic group were randomly selected and analyzed.
The weaned CD-1 mice were divided randomly into two groups. One group was fed with high-fat-diet (HFD, D12492, fat: 60% kcal, carbohydrate: 20% kcal, protein: 20% kcal, Research Diets, America) and the other was fed with control diet (CD, fat: 9.3% kcal, carbohydrate; 70% kcal, protein: 20% kcal) for 12 weeks. Then the mice fed with HFD or CD was mated with male mice fed normal diets to produce offspring, respectively. During gestational and lactational periods, the females were fed with HFD and/or CD as pre-pregnancy. After weaning, all offspring were fed CD. At the age of 7–8 weeks (Figure 1A), a total of 9–10 male pups of 3–4 litters were randomly selected and analyzed for HFD and CD.
Average weight and blood glucose level in high-fat-diet (HFD)-induced mouse models. (A) At age of 21d (day), the female mice were randomly divided into two groups and fed with CD (control diet) and HFD (high-fat-diet), respectively. Twelve weeks later, mice fed with CD/HFD were mated with normal male mice, respectively. The pups of them were fed with diets as pre-pregnancy until weaning. Weaned male pups were fed with CD. At age of 7–8 weeks, spermatozoa of male pups were collected. After 12 weeks of treatment, the average weight of the HFD (n = 17) group was clearly heavier than that of the CD (n = 13) group, P < 0.001 (B), but the average blood glucose level of CD (n = 13) and HFD (n = 15) was similar between the two groups (C), P = 0.8209.
Offspring sperm collection
Spermatozoa of offspring were collected according to previously described method [21]. Briefly, the cauda epididymis was separated and punctured with a sterile needle in HTF (Human Tube Fluid) for 30 min at 37°C. The motile spermatozoa that remained in the supernatant were carefully transferred to a new Eppendorf tube (EP) and this procedure was repeated 3 times. Then the supernant was centrifuged at 13,400 rpm for 10 min to pellet the sperm.
DNA bisulfite modification and PCR amplification
Spermatozoa were modified with EZ DNA Methylation-Direct™ Kit (ZYMO RESEARCH, USA) according to the manufacturer’s direction. Modified DNA was used as template for nested-PCR amplification. Briefly, nested PCR was carried out using 0.5 μl solution with modified DNA in the first round PCR and 2 μl product of first round PCR was added into the second reaction system as template. The relative primers are shown in Table 1.
Combined bisulfite restriction analysis (COBRA) and bisulfite sequencing (BS)
When DNA was treated by bisulfite, the unmethylated CG was translated to TG. So we can use restriction endogenous enzymes and sequencing to analyze the methylation patterns at CpG (cytosine-phosphate-guanine) sites [18]. The product of nested-PCR was digested by one or two restriction endogenous enzymes of which there were CpG loci included in the recognition sites. The recognition sites of enzymes used in the assay were TaqαI (T/CGA), RsaI (GTAC/), and BstUI (CG/CG). To further investigate the methylation patterns of relative genes, all samples were pooled together and was cloned using T vector (TAKARA). Then it was sequenced. At least ten clones per gene were sequenced.
Statistical analysis
Body weight and blood glucose are represented as Mean ± SD and the significance between groups was evaluated by One-way ANOVA. The Chi-square test was used to evaluate whether significant difference exists in methylation density between different groups. A probability level of P < 0.05 was considered significant.
Results
DNA methylation in spermatozoa of offspring of obese mothers is altered
After 12 weeks fed with CD/HFD, the average weight of mice fed with HFD (49.37 ± 0.6037) was obviously heavier than those fed with CD (35.81 ± 0.6295, Figure 1B). The average blood glucose level between HFD and CD groups was similar (Figure 1C). A total of 9–10 pups of obese/non-obese mothers were analyzed in each group. The pups were randomly selected from 3–4 litters. For paternally imprinted genes, results of COBRA showed that all of them were digested by relative enzymes (Figure 2A-C). Compared to oocytes, the bands of spermatozoa of the maternally imprinted gene Snrpn was completely undigested by BstuI (Figure 2E). This also confirmed that our samples were not contaminated by somatic cells. However, for Peg3, we observed that several samples were partly digested by TaqαI and/or BstuI (Figure 2D, shown by red arrowheads). To further prove that these results were not attributed to contamination by somatic cells, we carried out first-round PCR with primers of Peg3 and sH19 (a shorter fragment in DMR of H19, 176 bp). Then the products of PCR were analyzed by COBRA and the results showed that products of sH19 were completely digested (Figure 2F).
DNA methylation status in DMRs of imprinted genes in spermatozoa of OHFD analyzed by COBRA. DNA methylation patterns in DMRs of paternally imprinted genes H19 (423 bp), Gtl2 (425 bp), Rasgrf1 (284 bp) and maternally imprinted genes Peg3 (444 bp) and Snrpn (420 bp) in spermatozoa of OHFD (n = 9-10) and OCD mice (n = 10) were evaluated by COBRA. Oocytes were utilized as a control. Enzymes used are shown in the right column; Oo, oocyte. (A) H19; (B) Gtl2; (C) Rasgrf1; (D) Peg3, red arrowheads showed the digested bands; (E) Snrpn; (F) a shorter region located in DMR of H19 was amplified with Peg3 in the first-round PCR and the methylation pattern was analyzed by COBRA.
Next, we used BS to further investigate the methylation patterns of paternally imprinted gene H19 and maternally imprinted gene Peg3. We pooled the products of all samples together for H19 and Peg3, respectively. 10–20 clones were sequenced. We evaluated the bisulfite conversion rates and it ranged from 95% to 99% (the average conversion rate was 97.74%). Sequencing results showed that DNA methylation in DMR of H19 was not affected in sperm, either (Figure 3A). For Peg3, the methylation level in the OHFD group was 28.05%, which is significantly higher (P < 0.01) than that in the OCD (male offspring of non-obese mothers, 0.5%) group (Figure 3B).
DNA methylation in spermatozoa of OHFD analyzed by BS. DNA methylation of paternally imprinted gene H19 and maternally imprinted gene Peg3 was further analyzed by BS. All the samples were pooled together for each group. (A) represents H19 methylation status; the number shows the methylation%; (B) shows the methylation patterns of Peg3. Black circle, methylated; white circle, unmethylated; blank loci, CpG lost.
DNA methylation status in DMR of paternally imprinted gene H19is altered in spermatozoa of offspring from diabetic mothers (OD)
We checked the blood glucose level of diabetic and non-diabetic mothers at pre-pregnancy (nondiabetic & diabetic, 6.14 ± 0.51 & 23.53 ± 3.27) and post-partum (nondiabetic & diabetic, 6.46 ± 0.61 & 25.86 ± 1.34). Dlood glucose level was similar between ON (offspring of nondiabetic mothers, 6.72 ± 0.46) and OD (offspring of diabetic mothers, 7.1 ± 0.74) at age of 7–8 weeks. Nine pups from 3–4 litters were analyzed in each group. As shown in Figure 4A, the samples of Peg3 were all uncut by both enzymes in the two groups, suggesting that the methylation pattern of Peg3 in spermatozoa of OD was not affected by maternal diabetes (Figure 4A). However, for H19, some of the samples were not all cut by both enzymes (Figure 4B) (shown by red arrows). In Figure 4C, some samples were still not completely digested by one or two enzymes.
Methylation patterns in DMRs of imprinted genes in sperm from offspring of diabetic (OD) and non-diabetic (ON) females. OD (n = 9)and ON (n = 9) (7–8 weeks old) mice were killed and spermatozoa were collected and treated with bisulfite and then amplified by nest-PCR. (A ~ C) The products of PCR were digested by enzymes and analyzed by 2.5% agarose gel electrophoresis. The red arrow heads point to the samples that were only partially digested by enzymes. (D, E) The methylation rates in DMRs of H19 and Gtl2 were evaluated by sequencing. Black circle, methylated; white circle, unmethylated; blank loci, CpG lost.
Although some samples were not all digested by enzymes for H19 and Gtl2, maybe the demethylation just took place on recognition sites. To further investigate the methylation status, we pooled all of the samples together and sequenced according to COBRA results. The sequencing results indicated that the methylation level of H19 in spermatozoa of OD (64.57%)was significantly lower than that of ON (97.54%, P < 0.001, Figure 4D). For Gtl2, the methylation levels were 98.47% and 99.16% in the ON and OD (Figure 4E) groups, respectively.
Discussion
Although the DNA methylation patterns in DMRs of imprinted genes in oocytes of offspring from mothers with pregestational diabetes [18] and/or obesity were not affected, DNA methylation modification was altered by maternal diabetes and/or obesity in spermatozoa of offspring. These results indicate that DNA methylation may play a role in passing the adverse effects from maternal diabetes and/or obesity on to the next generations.
Genomic imprinting involving DNA methylation is a widely existing epigenetic phenomenon in flowering plants and mammals [22–24]. Imprinted genes show parental-origin-dependent monoallelic expression controlled by differentially methylated regions (DMRs) [25–27]. The methylation at the DMRs which are erased in primordial germ cells (PGCs) and re-established during gametogenesis before being passed on to the next generation is germline-specific [28–30]. However, the time points of methylation establishment of germ cell imprints are different between males and females. In female germline, de novo methylation of imprints is initiated at the primordial germ cell (PGC) stage and mostly completed at the metaphase II stage (MII) after birth [31–33]. This process is earlier for males. De novo methylation in male PGCs of mammals takes place several days after erasure being completed at the stage of E14.5 to E16.5, and is mostly completed before the postnatal stage [34–36]. Therefore, the process of re-establishing imprints in the male germline is more easily disturbed by a deleterious uterus environment than that in the female germline. In the present study, we found the DNA methylation patterns at the DMRs of H19 and Peg3 in spermatozoa of offspring were influenced by pre-existing maternal diabetes and obesity, respectively. But they were not affected in oocytes of offspring born by mothers with diabetes and/or obesity [18].
It is well known that sexual dimorphism is very common phenomenon in mammals including humans. In humans, the expression of individual genes is different in the placenta between males and females [37, 38]. A similar phenomenon is observed in the mouse [39, 40]. McPherson and Chenoweth have discussed the evolution, benefits and costs of mammalian sexual dimorphisms [41]. We found that the effects on DNA methylation of imprinted genes were different in germ cells between female and male offspring born to mothers with diabetes and/or obesity. The sexual dimorphism may partly elucidate the different effects on DNA methylation of imprinted genes in germ cells of female and male offspring of diabetic and nondiabetic mothers. Certainly, the difference may also be induced by contamination with somatic cells. In our study, we showed that the difference was not the result of contamination with somatic cells: the methylation of Peg3 (maternally imprinted) was altered in spermatozoa of offspring from obese mothers, but H19 (paternally imprinted) was not changed. To further confirm this, we carried out the first-round PCR including primers of Peg3 and sH19. Then they were amplified for the second-round PCR. We found that the alteration of Peg3 was not induced by contamination with somatic cells.
In the present study, we found that the DNA methylation in DMR of Peg3 was altered in spermatozoa of offspring from obese mothers but it was not affected in spermatozoa of offspring from diabetic mothers. However, the DNA methylation of H19 in the offspring sperm was changed by maternal diabetes. This may be mainly because the two models have different influences on testis and sperm. It has previously been shown that maternal diabetes reduced testis weight, thickness of the testicular capsule, number of Leydig and Sertoli cells, number of spermatogonia, as well as sperm reserves and the sperm transit time through the epididymis in male offspring [42–44]. Although the offspring of obese mothers display sub-fertility, few reports are available on how maternal obesity affects testis function and sperm quality of offspring.
Conclusions
In summary, the DNA methylation in spermatozoa of offspring born by obese and/or diabetic mothers was altered, indicating that abnormal DNA methylation modification in spermatozoa may play a role in the adverse effects passed on to the next generations by the mothers.
References
Cox NJ: Maternal component in NIDDM transmission. How large an effect?. Diabetes. 1994, 43: 166-168. 10.2337/diab.43.1.166.
Evers IM, de Valk HW, Visser GH: Risk of complications of pregnancy in women with type 1 diabetes: nationwide prospective study in the Netherlands. BMJ. 2004, 328: 915-10.1136/bmj.38043.583160.EE.
Giordano C: Immunobiology of normal and diabetic pregnancy. Immunol Today. 1990, 11: 301-303.
Moley KH, Chi MM, Manchester JK, McDougal DB, Lowry OH: Alterations of intraembryonic metabolites in preimplantation mouse embryos exposed to elevated concentrations of glucose: a metabolic explanation for the developmental retardation seen in preimplantation embryos from diabetic animals. Biol Reprod. 1996, 54: 1209-1216. 10.1095/biolreprod54.6.1209.
Sadler TW, Hunter ES, Balkan W, Horton WE: Effects of maternal diabetes on embryogenesis. Am J Perinatol. 1988, 5: 319-326. 10.1055/s-2007-999717.
Yessoufou A, Moutairou K: Maternal diabetes in pregnancy: early and long-term outcomes on the offspring and the concept of “metabolic memory”. Exp Diabetes Res. 2011, 2011: 218598-
Aerts L, Holemans K, Van Assche FA: Maternal diabetes during pregnancy: consequences for the offspring. Diabetes Metab Rev. 1990, 6: 147-167. 10.1002/dmr.5610060303.
Dorner G, Plagemann A: Perinatal hyperinsulinism as possible predisposing factor for diabetes mellitus, obesity and enhanced cardiovascular risk in later life. Horm Metab Res. 1994, 26: 213-221. 10.1055/s-2007-1001668.
Howie GJ, Sloboda DM, Kamal T, Vickers MH: Maternal nutritional history predicts obesity in adult offspring independent of postnatal diet. J Physiol. 2009, 587: 905-915. 10.1113/jphysiol.2008.163477.
Whitaker RC: Predicting preschooler obesity at birth: the role of maternal obesity in early pregnancy. Pediatrics. 2004, 114: e29-36. 10.1542/peds.114.1.e29.
Lawlor DA, Lichtenstein P, Fraser A, Langstrom N: Does maternal weight gain in pregnancy have long-term effects on offspring adiposity? A sibling study in a prospective cohort of 146,894 men from 136,050 families. Am J Clin Nutr. 2011, 94: 142-148. 10.3945/ajcn.110.009324.
Shankar K, Harrell A, Liu X, Gilchrist JM, Ronis MJ, Badger TM: Maternal obesity at conception programs obesity in the offspring. Am J Physiol Regul Integr Comp Physiol. 2008, 294: R528-538.
Bayol SA, Farrington SJ, Stickland NC: A maternal ‘junk food’ diet in pregnancy and lactation promotes an exacerbated taste for ‘junk food’ and a greater propensity for obesity in rat offspring. Br J Nutr. 2007, 98: 843-851.
Pentinat T, Ramon-Krauel M, Cebria J, Diaz R, Jimenez-Chillaron JC: Transgenerational inheritance of glucose intolerance in a mouse model of neonatal overnutrition. Endocrinology. 2010, 151: 5617-5623. 10.1210/en.2010-0684.
Dabelea D, Hanson RL, Lindsay RS, Pettitt DJ, Imperatore G, Gabir MM, Roumain J, Bennett PH, Knowler WC: Intrauterine exposure to diabetes conveys risks for type 2 diabetes and obesity: a study of discordant sibships. Diabetes. 2000, 49: 2208-2211. 10.2337/diabetes.49.12.2208.
Dunn GA, Bale TL: Maternal high-fat diet promotes body length increases and insulin insensitivity in second-generation mice. Endocrinology. 2009, 150: 4999-5009. 10.1210/en.2009-0500.
Dunn GA, Bale TL: Maternal high-fat diet effects on third-generation female body size via the paternal lineage. Endocrinology. 2011, 152: 2228-2236. 10.1210/en.2010-1461.
Ge ZJ, Liang XW, Guo L, Liang QX, Luo SM, Wang YP, Wei YC, Han ZM, Schatten H, Sun QY: Maternal diabetes causes alterations of DNA methylation statuses of some imprinted genes in murine oocytes. Biol Reprod. 2013, 88: 117-10.1095/biolreprod.112.105981.
Ge ZJ, Luo SM, Lin F, Liang QX, Huang L, Wei YC, Hou Y, Han ZM, Schatten H, Sun QY: DNA methylation in oocytes and liver of female mice and their offspring: effects of high-fat-diet-induced obesity. Environ Health Perspect. 2014, 122: 159-164.
Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, Sun QY: Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci U S A. 2014, 111: 1873-1878. 10.1073/pnas.1321195111.
Stouder C, Deutsch S, Paoloni-Giacobino A: Superovulation in mice alters the methylation pattern of imprinted genes in the sperm of the offspring. Reprod Toxicol. 2009, 28: 536-541. 10.1016/j.reprotox.2009.06.009.
Li E, Beard C, Jaenisch R: Role for DNA methylation in genomic imprinting. Nature. 1993, 366: 362-365. 10.1038/366362a0.
Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H: Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004, 429: 900-903. 10.1038/nature02633.
Kohler C, Weinhofer-Molisch I: Mechanisms and evolution of genomic imprinting in plants. Heredity (Edinb). 2010, 105: 57-63. 10.1038/hdy.2009.176.
Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP: Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997, 389: 745-749. 10.1038/39631.
Thorvaldsen JL, Duran KL, Bartolomei MS: Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998, 12: 3693-3702. 10.1101/gad.12.23.3693.
Fitzpatrick GV, Soloway PD, Higgins MJ: Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet. 2002, 32: 426-431. 10.1038/ng988.
Lee J, Inoue K, Ono R, Ogonuki N, Kohda T, Kaneko-Ishino T, Ogura A, Ishino F: Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Dev. 2002, 129: 1807-1817.
Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA: Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002, 117: 15-23. 10.1016/S0925-4773(02)00181-8.
Li JY, Lees-Murdock DJ, Xu GL, Walsh CP: Timing of establishment of paternal methylation imprints in the mouse. Genomics. 2004, 84: 952-960. 10.1016/j.ygeno.2004.08.012.
Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM: Methylation dynamics of imprinted genes in mouse germ cells. Genomics. 2002, 79: 530-538. 10.1006/geno.2002.6732.
Lucifero D, Mann MR, Bartolomei MS, Trasler JM: Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet. 2004, 13: 839-849. 10.1093/hmg/ddh104.
Hiura H, Obata Y, Komiyama J, Shirai M, Kono T: Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells. 2006, 11: 353-361. 10.1111/j.1365-2443.2006.00943.x.
Davis TL, Trasler JM, Moss SB, Yang GJ, Bartolomei MS: Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics. 1999, 58: 18-28. 10.1006/geno.1999.5813.
Ueda T, Abe K, Miura A, Yuzuriha M, Zubair M, Noguchi M, Niwa K, Kawase Y, Kono T, Matsuda Y, Fujimoto H, Shibata H, Hayashizaki Y, Sasaki H: The paternal methylation imprint of the mouse H19 locus is acquired in the gonocyte stage during foetal testis development. Genes Cells. 2000, 5: 649-659. 10.1046/j.1365-2443.2000.00351.x.
Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H: Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet. 2007, 16: 2272-2280. 10.1093/hmg/ddm179.
Lehavi O, Aizenstein O, Evans MI, Yaron Y: 2nd-trimester maternal serum human chorionic gonadotropin and alpha-fetoprotein levels in male and female fetuses with Down syndrome. Fetal Diagn Ther. 2005, 20: 235-238. 10.1159/000083911.
Steier JA, Bergsjo PB, Thorsen T, Myking OL: Human chorionic gonadotropin in maternal serum in relation to fetal gender and utero-placental blood flow. Acta Obstet Gynecol Scand. 2004, 83: 170-174. 10.1080/j.0001-6349.2004.00407.x.
Gallou-Kabani C, Gabory A, Tost J, Karimi M, Mayeur S, Lesage J, Boudadi E, Gross MS, Taurelle J, Vige A, Breton C, Reusens B, Remacle C, Vieau D, Ekström TJ, Jais JP, Junien C: Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS One. 2010, 5: e14398-10.1371/journal.pone.0014398.
Mao J, Zhang X, Sieli PT, Falduto MT, Torres KE, Rosenfeld CS: Contrasting effects of different maternal diets on sexually dimorphic gene expression in the murine placenta. Proc Natl Acad Sci U S A. 2010, 107: 5557-5562. 10.1073/pnas.1000440107.
McPherson FJ, Chenoweth PJ: Mammalian sexual dimorphism. Anim Reprod Sci. 2012, 131: 109-122. 10.1016/j.anireprosci.2012.02.007.
Amorim EMP, Damasceno DC, Perobelli JE, Spadotto R, Fernandez CDB, Volpato GT, Kempinas WDG: Short- and long-term reproductive effects of prenatal and lactational growth restriction caused by maternal diabetes in male rats. Reprod Biol Endocrinol. 2011, 9: 154-10.1186/1477-7827-9-154.
Jelodar G, Khaksar Z, Pourahmadi M: Endocrine profile and testicular histomorphometry in adult rat offspring of diabetic mothers. J Physiol Sci. 2009, 59: 377-382. 10.1007/s12576-009-0045-7.
Ding GL, Wang FF, Shu J, Tian S, Jiang Y, Zhang D, Wang N, Luo Q, Zhang Y, Jin F, Leung PC, Sheng JZ, Huang HF: Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes. 2012, 61: 1133-1142. 10.2337/db11-1314.
Acknowledgements
We thank Ying-Chun Ouyang, Enkui Duan and Qi Chen for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
GZJ, ZCL, HS and SQY designed and wrote the manuscript. LQX, HY and GZJ acquired data. HZM analyzed the data. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Ge, ZJ., Liang, QX., Hou, Y. et al. Maternal obesity and diabetes may cause DNA methylation alteration in the spermatozoa of offspring in mice. Reprod Biol Endocrinol 12, 29 (2014). https://doi.org/10.1186/1477-7827-12-29
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-7827-12-29