Abstract
Background
CREBZF is a member of the mammalian ATF/CREB family of the basic region-leucine zipper (bZIP) transcription factors. Two isoforms of CREBZF have been identified from the alternative usage of initiation codons, SMILE (long isoform of CREBZF) and Zhangfei (short isoform of CREBZF). Until recently, the physiological function of CREBZF in mammalian reproductions has not been reported.
Methods
Multiple techniques were performed to investigate the spatiotemporal expression and hormonal regulation of the CREBZF gene in the mouse uterus and its role in embryo implantation.
Results
Zhangfei was not detected in the mouse uterus. SMILE immunostaining was mainly expressed in the uterine luminal and glandular epithelium, and the expression levels of both SMILE mRNA and protein gradually decreased from days 1–3 of pregnancy, peaked on day 4, and then declined again on day 6. On day 5 of pregnancy, SMILE protein expression was detected only in the luminal epithelium at implantation sites compared with the expression at inter-implantation sites. SMILE protein was not detected in decidual cells from days 6–8 of pregnancy or artificial decidualisation. Furthermore, SMILE protein was not detected in the mouse uterus on days 3–6 of pseudopregnancy, and SMILE expression was also induced in the delayed-implantation uterus, indicating that the presence of an active blastocyst was required for SMILE expression at the implantation site. Oestrogen significantly stimulated SMILE expression in the ovariectomised mouse uterus. In addition, in cycling mice, high levels of SMILE protein and mRNA expression were also observed in proestrus and oestrus uteri.
Conclusions
Taken together, these results suggested that SMILE expression was closely related to mouse implantation and up-regulated by oestrogen.
Similar content being viewed by others
Background
Successful implantation is dependent on the intricate genetic and molecular signalling dialogue between the receptive uterus and active embryo [1]. A better understanding of the molecular events underlying the regulation of embryo implantation may improve the ability to treat infertility. To date, although many molecular modulators have been identified during the implantation period, the precise molecular mechanism underlying embryo implantation is still unknown.
CREBZF is as a member of the CREB (cAMP-response-element-binding protein)/ATF (activating transcription factor) family of basic region-leucine zipper (b-ZIP) transcription factors [2]. Previously studies have found that CREBZF gene expression produces two isoforms, CREBZF-L (long isoform of CREBZF, also known as SMILE) and CREBZF-S (short isoform of CREBZF, which was previously designated as Zhangfei or ZF) [3]. However, unlike other b-ZIP proteins CREBZF lacks the ability to bind any of the consensus recognition elements for b-ZIP proteins and cannot activate promoters containing these motifs [2, 4]. Instead, CREBZF requires heterodimerisation with other factors to bind target promoters and to regulate downstream genes, which may include VP16 [5], Luman (or CREB3, L-ZIP) [6], Activating transcription factor 4 (ATF4, or CREB2) [7], X-box-binding protein-1 (XBP1) [8] and tropomyosin-related kinase A (trkA) [9, 10]. A potential reason for the inability of CREBZF to recognise these promoters may be the absence of a critical asparagine residue in its basic domain, which forms the DNA-recognition motif, NxxAAxxCR, in all b-ZIP proteins [2, 4].
Zhangfei was first identified via its interaction with the Herpes Simplex Virus-1 (HSV-1)-related cellular protein Host Cell Factor 1 (HCF-1) and has been proposed to play a role in inhibiting the replication of the herpes simplex virus [2, 5]. Recent studies have shown that Zhangfei could potentially play an important role in the mammalian unfolded protein response (UPR) [6–8]. Zhangfei might activate apoptosis in ONS-76 medulloblastoma cells via the NGF/TrkA pathway [10, 11]. Currently, the precise physiological function of SMILE remains unclear. Recently, SMILE has been reported as a novel transcriptional co-regulator of a variety of nuclear receptors, including oestrogen receptors, glucocorticoid receptor, constitutive androstane receptor, hepatocyte nuclear factor 4α and oestrogen receptor-related receptor γ [3, 12, 13]. In another study, SMILE played an important role in the development of beta cell dysfunction induced by glucolipotoxicity [14]. Zhangfei exhibits a liver- and cell-type-specific expression pattern; however, SMILE is expressed ubiquitously in mouse tissues and tumour-derived cells [3]. Thus, it is certainly possible that the two isoforms exhibit different regulatory functions in specific cellular contexts. However, until recently, the physiological function of CREBZF in mammalian reproduction has not been reported. To explore the potential function of CREBZF during pregnancy, we sought to investigate in detail the expression and regulation of CREBZF mRNA and protein in the mouse uterus during the oestrous cycle and peri-implantation period.
Methods
Animals and treatments
The experimental use of mouse for this study was performed according to the Committee for the Ethics on Animal Care and Experiments in Northwest A&F University. Mature mice (Kunming White outbred strain, 8-to 10-wk-old) were purchased from the laboratory animal centre of Xi’An JiaoTong University and housed at a temperature- (24+/−2°C) in a light-controlled room (12 h light: 12 h darkness) with free access to food and water.
Oestrous cycles were tracked by performing daily vaginal smears, and only those mice that demonstrated a regular 4-day oestrous cycle were used for these experiments. Mouse models of early pregnancy, pseudopregnancy, delayed implantation and activation, artificial decidualisation and hormonal treatment were produced as described in our previous reports [15, 16]. Eight mice were used in each stage or treatment in this study. To confirm the reproducibility of the results, each sample was analysed in triplicate. The entire uterus was collected immediately following sacrifice by cervical dislocation, and half of each uterus was immediately processed for immunohistochemistry, whereas the remaining half was frozen in liquid nitrogen for further extraction of total RNA and protein.
Embryo collection
Female mice were superovulated via an intraperitoneal injection of 5 IU of pregnant mare’s serum gonadotrophin (PMSG, Ningbo Sansheng Pharmaceutical Co., Ltd. China) at 4:00 pm followed by human chorionic gonadotrophin (hCG, Ningbo Sansheng Pharmaceutical Co., Ltd. China) 48 hr later. Treated female mice were mated with fertile males of the same strain to induce natural pregnancy. Embryos at the zygote, 2-cell and 4-cell stage were collected from oviducts at 24 hr, 46 hr and 54 hr, respectively. However, the 8-cell stage embryo, morula and blastula were collected from the uterine horns at 68 hr, 72 hr and 96 hr, respectively. A total of 160 mice were used for embryo collection and 35–40 pooled embryos per developmental stage were analyzed in each group. Three independent experiments were performed.
RNA extraction, cDNA synthesis and real-time quantitative PCR
Total RNA was extracted from the uterine horns and embryo using Trizol (Invitrogen, Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions. DNase (TaKaRa Bio, Inc., Dalian, China) was used to remove genomic DNA contamination prior to RT. Extracted RNA was dissolved in diethylpyrocarbonate (DEPC)-treated water, and the RNA concentration and purity were estimated by reading the absorbance at 260 and 280 nm on a spectrophotometer (Eppendorf, Inc., Hamburg, Germany). The cDNAs were synthesised using PrimeScriptTM RT reagent Kit (TaKaRa Bio, Inc., Dalian, China) according to the manufacturer’s instructions. The final volume of the reaction was 20 μl, including 800 ng of total RNA. The reverse transcription product was stored at −20°C.
The GenBank accession number of the mRNA, primer sequences and annealing temperatures are listed in Table 1. Real-time PCR was performed using three biological replicates and technical triplicates/duplicates of each cDNA sample in the LightCycler system (iQ5, Bio-Rad Laboratories, Inc., Hercules, USA) using SYBR® Premix Ex TaqTM II Kit (TaKaRa Bio, Inc., Dalian, China), according to the manufacturer’s protocol. Each PCR reaction (total volume of 20 μl) consisted of 2 μl reverse transcription product, 0.8 μl of each 10 μM forward and reverse primer, 10 μl SYBR® Premix Ex TaqTM II, and 6.4 μl RNase-free water. The cycling conditions included a denaturation step at 95°C for 30 sec, followed by 45 PCR cycles of 95°C for 5 sec and 60°C for 20 sec. A melting curve analysis was performed at the end of each PCR program to exclude the formation of nonspecific products. Gene mRNA quantifications were performed using the 2-△△Ct method and the amount of transcripts in each sample was normalised using RPLP0 and GAPDH as the internal control gene to correct for differences in the amount of cDNA used [17, 18].
Immunohistochemistry
Uterine tissues were fixed in 4% (v/v) paraformaldehyde (Sinopharm Chemical Reagent Co., Ltd, Shanghai, China) in phosphate-buffered saline (PBS; pH 7.4) for 24 hr, dehydrated through a graded ethanol series, and embedded in paraffin. Five μm-thick sections were mounted onto glass slides that were precoated with poly-L-Lysine solution (Sigma, St. Louis, MO, USA) and incubated overnight at 37°C. After dehydration, the samples were placed in citrate buffer (pH 6.0), and antigen retrieval was performed by treating the samples (twice) in a microwave oven at 750 W for 5 min. The slides were then washed in PBS. Sections were pretreated with 0.3% (v/v) H2O2 in methanol to quench endogenous peroxidase activity. After several washes with PBS, the sections were incubated with 10% rabbit serum for 30 min at 37°C. Following blocking, the sections were incubated with goat anti-CREBZF polyclonal antibody (Santa Cruz, sc-49328; diluted 1:200 with PBS) for 12 hr at 4°C, washed with PBS, and incubated with biotinylated anti-goat IgG antibody (MaiXin-Bio Technology Co., Ltd., Fuzhou, China) for 1 hr at 37°C. Sections were washed three times with PBS, and then incubated with HRP-labelled streptavidin (SA-HRP) for 30 min at 37°C. Next, positive reactions were visualised using a diaminobenzidine (DAB)-peroxidase substrate (Sigma-Aldrich Co. LLC, Louis, MO, USA) and 30 sec counterstaining with haematoxylin. Negative control slides without the addition of primary antibody or substitution with an appropriate dilution of normal goat IgG was performed in parallel. Slides were imaged using a digital microscope (BA400, Motic, Wetzlar, Germany).
Western blotting analyses
Proteins obtained from uterine tissues were extracted using the Total Protein Extraction Kit (Nanjing Keygen Biotech Co., Ltd., Nanjing, China) according to the supplier’s instructions. The total protein per sample was separated using 12% sodium dodecyl sulphate (SDS)-polyacrylamide gel electrophoresis (PAGE) followed by electrotransfer to polyvinylidene fluoride (PVDF) membranes (Millipore; Bedford, MA). The membrane was treated with blocking buffer (5% nonfat dried milk in Tris-buffered saline [TBS] containing 0.1% Tween 20) for 1 hr at room temperature and incubated overnight at 4°C with antibodies against CREBZF (Santa Cruz, sc-49328; 1:500 dilution) or β-actin (1:1,000; Beijing CWBIO Co., Ltd., Beijing, China) as a loading control. The membranes were washed three times with TBS, and then incubated with biotinylated anti-goat IgG antibody or biotinylated anti-mouse IgG antibody (1:5,000; MaiXin-Bio Technology Co., Ltd., Fuzhou, China) for 1 hr at room temperature. After washing, the membranes were incubated with SA-HRP for 30 min at room temperature. Finally, the immunoreactive bands were visualised by DAB staining (Sigma) at room temperature for 5 min, and the immunoreactive bands were imaged using a digital microscope (Tanon-4100, Tanon Science & Technology Co., Ltd., Shanghai, China) and densitometric analyses were processed with Quantity one v4.62 (Bio-Rad).
Immunofluorescence
Embryos were fixed in 4% paraformaldehyde for 30 min at room temperature followed by permeabilising in 0.5% Triton X-100 in PBS. After washing in PBS, the embryos were treated with blocking solution of 1% bovine serum albumin in PBS for 1 hr, followed by incubation with goat anti-CREBZF polyclonal antibody (Santa Cruz, sc-49328; 1:500 dilution) at 4°C overnight. Embryos were then washed three times with PBS, and incubated for 1 hr at room temperature in a 1:500 dilution of Cy3-labelled donkey anti-goat IgG (Beyotime Institute of Biotechnology, Jiangsu, China). The nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Embryos were viewed under a laser-scanning confocal microscope (A1R, NiKon). Additionally, negative control embryos per developmental stage were treated in the same manner as described above, but omitting the primary antibody or primary and secondary antibodies, respectively.
Statistical analysis
All experiments were replicated at least three times for each group and the data were presented as the mean ± S.E.M. Data were analysed using ANOVA, followed by Fisher’s Least Significant Different Test (Fisher LSD) and Independent-Samples T test with SPSS software (Version 13.0; SPSS, Inc., Chicago, IL). Differences were considered significant when P < 0.05.
Results
CREBZF protein expression in the mouse uterus
Changes in CREBZF protein expression in whole uterine protein extracts were detected using western blotting analyses. In all samples analysed, a single band at approximately 50–60 kDa was observed (Figure 1). Xie et al. [3] reported that SMILE and Zhangfei isoforms were apparent at molecular masses of 50–60 kDa and 40 kDa, respectively. These results indicated that SMILE was the predominant form in this study, whereas Zhangfei was not detected in the mouse uterus.
Immunoblot analysis for CREBZF proteins in the mouse uterus during oestrous cycle (A), early pregnancy (B), pseudopregnancy (C), delayed implantation (D), and steroid hormonal treatments (E). β-actin levels are shown as a loading control. The values represent the mean ± S.E.M. from three duplicates, and the bar bearing different superscript indicates a significant difference between the mean values (P < 0.05).
The positive control for SMILE and Zhangfei protein was analysed by western blotting using a CREBZF-specific antibody. Results are representative of at least three independent experiments with similar results. MCF7, human breast adenocarcinoma cell line. 293 T, human embryonic kidney cell line (Additional file 1: Figure S1).
SMILE is expressed in the mouse uterus throughout the oestrus cycle
To determine whether there are differences in the expression and localisation of SMILE mRNA and protein during the oestrus cycle, SMILE mRNA and protein expression in the mouse uterine at proestrus, oestrus, metestrus and diestrus was detected using immunohistochemistry, real-time PCR and western blotting, respectively. We found that independent of the phase of the oestrus cycle, SMILE protein was specifically expressed in the endometrial epithelial cells of both the lumen and glands, while the expression was most prominent at the oestrus phase (Figure 2A). Real-time PCR and western blotting analysis also revealed that the levels SMILE mRNA and protein at oestrus were significantly higher compared to the levels from metestrus and diestrus, although these values were not significantly different compared with those obtained from the proestrus phase (P < 0.05, Figures 1A and 2B).
Expression of SMILE in the mouse uterus during different phases of the oestrus cycle. (A) Immunolocalisation of SMILE protein in the luminal and glandular epithelial cells of the uterus during the mouse oestrous cycle. Bars =25 μm. (B) Expression of SMILE mRNA in cycle uteri as detected using real-time PCR. The values represent the mean ± S.E.M. from three duplicates, and the bar bearing different superscript indicates a significant difference between the mean values (P < 0.05).
SMILE expression during early pregnancy
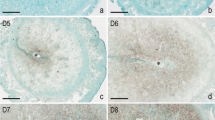
SMILE protein was mainly localised in the luminal and glandular epithelium during days 1 to 5 of pregnant uteri, whereas the staining intensity on day 4 of pregnancy was significantly increased (Figure 3 D1-D5). Compared with the inter-implantation site, the positive signal was observed in the luminal epithelium at the implantation site on day 5 of pregnancy (Figure 3 D5N and D5I). No obvious immunostaining of the SMILE protein was detected in the decidual cells, and SMILE only presented in the remaining glandular epithelium from days 6–7 of pregnancy (Figure 3 D6 and D7). Moreover, no specific signal for SMILE protein was detected in the artificial decidualisation model, and SMILE mRNA expression also did not show a significant difference in the decidualised uterus compared with the control uterus (data not shown).
Localisation of SMILE protein in the mouse uterus during early pregnancy, delayed and activated implantation and steroid hormonal treatments, examined by immunohistochemistry. D1-D7, the sections were stained with CREBZF antibody for days 1–7 pregnant uterus; D5I, implantation sites on day 5 of pregnancy; D5N, inter-implantation sites on day 5 of pregnancy. L = luminal epithelium; G = glandular epithelium; S = stromal cells; D = decidual cells; E = embryo. Bars =25 μm.
Real-time PCR was also used to detect the levels of SMILE mRNA expression in the mouse uterus on days 1–7 of pregnancy (Figure 4A). SMILE mRNA levels were gradually decreased on days 1–3 of pregnancy and were significantly lower on day 3 compared to day 1. These levels then peaked on day 4 (P < 0.05, Figure 4A). However, the level of SMILE mRNA was significantly decreased in the mouse uterus as the decidua was developing from day 6 to 7 of pregnancy. Similar with the results obtained from real-time PCR, SMILE protein was gradually decreased from day 1 to day 3, but it was up-regulated in day 4 uteri and was nearly not detectable from day 6 to day 7 uteri (Figure 1B).
Relative levels of SMILE mRNA in the mouse uterus during early pregnancy and pseudopregnancy. (A), delayed implantation (B) and steroid hormonal treatments (C) were assessed using real-time PCR. SMILE expression was normalised to RPLP0 and GAPDH. The values represent the mean ± S.E.M from three independent experiments. Bars with different letters are significantly different (P < 0.05).
SMILE expression during pseudopregnancy and activated implantation
In pseudopregnant mice, a basal level of SMILE staining was observed in luminal and glandular epithelium on days 1–2; however, no positive staining was observed in the uterus from day 3–6 (data not shown). Compared to early pregnancy, the levels of both SMILE mRNA and protein were lower on day 2 in pseudopregnant uteri (Figures 1C and 4A). As shown in Figure 3, SMILE immunostaining was abundantly expressed in activated implantation uteri, while its staining was weakly expressed in delayed implantation uteri. Moreover, significantly higher levels of SMILE mRNA and protein were also induced in the activated implantation uterus compared with the levels in the delayed uterus (P < 0.05, Figures 1D and 4B). Taken together, these observations strongly suggested that the presence of embryos affected SMILE localisation in these epithelial cells.
Steroid hormonal regulation of SMILE expression
Because E2 and P4 are essential for mouse embryo implantation, ovariectomised mice were used to examine the effects of steroid hormones on CREBZF expression. We found no visible signal in the ovariectomised mice (Figure 3). In the P4-treated group, a low level of SMILE immunostaining was present in the luminal and glandular epithelium. However, E2 significantly induced the expression of SMILE protein both in the luminal and glandular epithelium. There was only a moderate level of SMILE expression in these cells via combined E2 and P4 treatment. To further confirm SMILE expression, real-time PCR and western blotting were also performed. No significant difference in SMILE mRNA expression was observed after an injection of P4 compared to that of oil treatment (Figure 4C). In contrast, the expression levels of SMILE mRNA and protein in the uterus were significantly stimulated by E2 treatment (P < 0.05, Figures 1E and 4C). After the combined injection of E2 and P4, the SMILE mRNA and protein expression in uterus were similar to those in the E2 treatment alone, respectively.
Localisation of SMILE in mouse embryos during the peri-implantation period
To further study the role of SMILE in embryo implantation, we examined its localisation in the mouse embryos from the zygote to the blastocyst stage. These results demonstrated that SMILE was expressed at all stages of mouse pre-implantation embryos and was mainly localised in the cytoplasm and nucleus (Figure 5A). At the blastocyst stage, strong signals of SMILE protein were observed in the trophectoderm and the inner cell mass (Figure 5A). Furthermore, the levels of SMILE mRNA were gradually increased from the zygote to the blastocyst stage in the embryo (P < 0.05, Figure 5B).
Expression of SMILE mRNA and protein in the mouse embryos during pre-implantation. (A) Immunofluorescence analyses of SMILE protein in embryos from the zygote to the blastocyst stage. The red colour represents SMILE staining, and the blue colour indicates nuclear staining. Bar = 20 μm. (B) Analysis of SMILE mRNA levels using real-time PCR in the mouse embryos. The values represent the mean ± S.E.M from three independent experiments. Bars with different letters are significantly different (P < 0.05).
Discussion
This study mainly reported on the changes in the expression of two isoforms of CREBZF, as well as their cellular localisation throughout the oestrous cycle and peri-implantation period in the mouse uterus. These results indicated that SMILE was the predominant form and Zhangfei was not detected in the uterus, which was consistent with the expression pattern of SMILE in the male mouse prostate and testis [3]. Moreover, our results suggested that SMILE had a cycle-dependent expression in mouse uterus and was clearly localised in both luminal and glandular epithelial cells. A relatively higher expression of SMILE mRNA and protein at the oestrus phase accompanies the female as she reaches optimal sexual receptivity and ovulating time. Thus, we speculated that the elevated levels of SMILE might contribute to the induction of embryo earlier during development and implantation. Indeed, SMILE was localised in the cytoplasm and nucleus of mouse pre-implantation embryos, and the level of SMILE mRNA was gradually increased with embryo development. In addition, SMILE was expressed at high levels in the luminal and glandular epithelium at the proestrus and oestrus phase, and at basal levels in the metestrus and diestrus phase. These data also suggested that SMILE might be under cell-specific sex hormonal control in the mouse uterine epithelium and appeared to be important factors for implantation.
During pregnancy the uterus undergoes a series of programmed morphological changes, resulting in an extensive tissue reorganisation to accommodate the growing embryo [19]. The time from days 1 to 5 of pregnancy is critical for embryo implantation in mice, and days 4 to 5 of pregnancy is regarded as the implantation window. The embryo apposes and attaches to the uterine luminal epithelium, which is the initial step for a successful pregnancy [20]. The expression levels of SMILE mRNA and protein were gradually decreased in the mouse uterus from days 1 to 3, but were sharply up-regulated on day 4. Furthermore, the uterine expression of the SMILE protein in the luminal epithelium was predominantly localised at the implantation site on days 5 of pregnancy, demonstrating temporal and spatial specificity during the period of mouse embryo implantation. Taken together, these results suggested that SMILE might play an essential role in the initial attachment of a blastocyst to the uterine epithelia in the mouse. However, the up-regulation of SMILE was restricted to day 5 and was not expressed in decidualised cells on days 6–7 of early pregnancy or under artificial decidualisation, although SMILE was also highly expressed in glandular epithelial cells, which might not be important for decidualisation.
In the present study, SMILE was lowly expressed on days 1–2 in the pseudopregnant uteri and was not observed in the uterus from days 3–6 of pseudopregnancy, indicating that SMILE expression was dependent on the presence of the embryo. Moreover, SMILE mRNA and protein were highly expressed in the uterus when the delayed implantation was terminated by via treatment compared with the expression under conditions of delayed implantation. In addition, the localisation of SMILE protein in the trophectoderm and increased expression of SMILE mRNA and protein at the blastocyst stage might be a prerequisite for establishing contact between the blastocyst and uterine epithelium. These results suggested that SMILE expression in the mouse uterus required the presence of an active blastocyst during the peri-implantation period.
Implantation has long been known to be a steroid hormone-dependent process. The coordinated action of both E2 and P4 is necessary for the preparation and process of implantation in the mouse endometrium [21]. Because both E2 and P4 are essential for the induction of implantation in the mouse, we examined the hormonal regulation of SMILE expression. It has been reported that CREBZF isoforms regulated the inhibition of the transactivation of oestrogen receptors via a small heterodimer partner in a cell-type-specific manner, although CREBZF isoforms did not interact directly with oestrogen receptors [3]. CREBZF isoforms as a novel corepressor of nuclear receptors may interact with SIRT1 to regulate the transactivation of oestrogen receptor-related receptor γ [13]. SIRT1 plays an important role in the development of human uterine receptivity via inducing E-cadherin expression [22]. Our studies found that E2 could significantly up-regulate the expression and localisation of SMILE in the luminal epithelium and glandular epithelial cells in ovariectomised mice. In addition, P4 had no significant effects on SMILE expression in the ovariectomised mouse uterus. Similar hormonal regulation was also consistent with the spatiotemporal expression of SMILE mRNA and protein in the mouse uterus during the oestrous cycle in this study. High levels of E2 were observed in mice during proestrus and oestrus, decreasing to low levels during metestrus and diestrus [23]. During normal mouse pregnancy, higher levels of E2 were detected on day 1 and day 4, a surge in E2 on day 1 stimulated uterine epithelial cell proliferation and induced the expression of progesterone receptors. An increase in E2 levels on day 4 combined with the high P4, which further stimulated uterine stromal proliferation and induced the endometrial receptivity for the blastocyst to implant [21, 24]. The results in ovariectomised mice, oestrous cyclic mouse in pregnant mice confirmed our supposition that the expression of SMILE in the peri-implantation uterus may be mainly modulated by E2.
Conclusions
On the basis of these findings, we concluded that SMILE might play a potential role in mediating uterine receptivity during implantation and development of preimplantation embryos in mice. Moreover, SMILE expression in mouse uteri was regulated by active blastocysts and E2.
Abbreviations
- ATF:
-
Activating transcription factor
- ATF4:
-
Activating transcription factor 4
- b-ZIP:
-
Basic region-leucine zipper
- CREB:
-
cAMP-response-element-binding protein
- DAB:
-
Diaminobenzidine
- DEPC:
-
Diethylpyrocarbonate
- E2:
-
Oestrogen
- HCF-1:
-
Host cell factor 1
- hCG:
-
human chorionic gonadotrophin
- HSV-1:
-
Herpes simplex virus-1
- P4:
-
Progesterone
- PAGE:
-
Polyacrylamide gel electrophoresis
- PBS:
-
Phosphate-buffered saline
- PMSG:
-
Pregnant mare’s serum gonadotrophin
- PVDF:
-
Polyvinylidene fluoride
- SDS:
-
Sodium dodecyl sulphate
- TBS:
-
Tris-buffered saline
- trkA:
-
tropomyosin-related kinase
- UPR:
-
Unfolded protein response
- XBP1:
-
X-box-binding protein-1.
References
Cha J, Sun X, Dey SK: Mechanisms of implantation: strategies for successful pregnancy. Nat Med. 2012, 18: 1754-1767. 10.1038/nm.3012.
Lu R, Misra V: Zhangfei: a second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16. Nucleic Acids Res. 2000, 28: 2446-2454. 10.1093/nar/28.12.2446.
Xie YB, Lee OH, Nedumaran B, Seong HA, Lee KM, Ha H, Lee IK, Yun Y, Choi HS: SMILE, a new orphan nuclear receptor SHP-interacting protein, regulates SHP-repressed estrogen receptor transactivation. Biochem J. 2008, 416: 463-473. 10.1042/BJ20080782.
Cockram GP, Hogan MR, Burnett HF, Lu R: Identification and characterization of the DNA-binding properties of a Zhangfei homologue in Japanese pufferfish, Takifugu rubripes. Biochem Biophys Res Commun. 2006, 339: 1238-1245. 10.1016/j.bbrc.2005.11.139.
Akhova O, Bainbridge M, Misra V: The neuronal host cell factor-binding protein Zhangfei inhibits herpes simplex virus replication. J Virol. 2005, 79: 14708-14718. 10.1128/JVI.79.23.14708-14718.2005.
Misra V, Rapin N, Akhova O, Bainbridge M, Korchinski P: Zhangfei is a potent and specific inhibitor of the host cell factor-binding transcription factor Luman. J Biol Chem. 2005, 280: 15257-15266. 10.1074/jbc.M500728200.
Hogan MR, Cockram GP, Lu R: Cooperative interaction of Zhangfei and ATF4 in transactivation of the cyclic AMP response element. FEBS Lett. 2006, 580: 58-62. 10.1016/j.febslet.2005.11.046.
Newman JR, Keating AE: Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003, 300: 2097-2101. 10.1126/science.1084648.
Valderrama X, Rapin N, Misra V: Zhangfei, a novel regulator of the human nerve growth factor receptor, trkA. J Neurovirol. 2008, 14: 425-436. 10.1080/13550280802275904.
Valderrama X, Rapin N, Verge VM, Misra V: Zhangfei induces the expression of the nerve growth factor receptor, trkA, in medulloblastoma cells and causes their differentiation or apoptosis. J Neurooncol. 2009, 91: 7-17. 10.1007/s11060-008-9682-6.
Bodnarchuk TW, Napper S, Rapin N, Misra V: Mechanism for the induction of cell death in ONS-76 medulloblastoma cells by Zhangfei/CREB-ZF. J Neurooncol. 2012, 109: 485-501. 10.1007/s11060-012-0927-z.
Xie YB, Nedumaran B, Choi HS: Molecular characterization of SMILE as a novel corepressor of nuclear receptors. Nucleic Acids Res. 2009, 37: 4100-4115. 10.1093/nar/gkp333.
Xie YB, Park JH, Kim DK, Hwang JH, Oh S, Park SB, Shong M, Lee IK, Choi HS: Transcriptional corepressor SMILE recruits SIRT1 to inhibit nuclear receptor ERRgamma transactivation. J Biol Chem. 2009, 284: 28762-28774. 10.1074/jbc.M109.034165.
Lee KM, Seo YJ, Kim MK, Seo HA, Jeong JY, Choi HS, Lee IK, Park KG: Mediation of glucolipotoxicity in INS-1 rat insulinoma cells by small heterodimer partner interacting leucine zipper protein (SMILE). Biochem Biophys Res Commun. 2012, 419: 768-773. 10.1016/j.bbrc.2012.02.098.
Lan X, Jin Y, Yang Y, Lin P, Hu L, Cui C, Li Q, Li X, Wang A: Expression and localization of Luman RNA and protein during mouse implantation and decidualization. Theriogenology. 2013, 80: 138-144. 10.1016/j.theriogenology.2013.02.024.
Yang Y, Jin Y, Martyn AC, Lin P, Song Y, Chen F, Hu L, Cui C, Li X, Li Q, et al: Expression pattern implicates a potential role for luman recruitment factor in the process of implantation in uteri and development of preimplantation embryos in mice. J Reprod Dev. 2013, 59: 245-251. 10.1262/jrd.2012-137.
Lin P, Lan X, Chen F, Yang Y, Jin Y, Wang A: Reference gene selection for real-time quantitative PCR analysis of the mouse uterus in the peri-implantation period. PLoS One. 2013, 8: e62462-10.1371/journal.pone.0062462.
Vandesompele J, De-Preter K, Pattyn F, Poppe B, Van-Roy N, De Paepe A, Speleman F: Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3: RESEARCH0034-
Zhang S, Lin H, Kong S, Wang S, Wang H, Wang H, Armant DR: Physiological and molecular determinants of embryo implantation. Mol Aspects Med. 2013, 34: 939-980. 10.1016/j.mam.2012.12.011.
Dey SK, Lim H, Das SK, Reese J, Paria BC, Daikoku T, Wang H: Molecular cues to implantation. Endocr Rev. 2004, 25: 341-373. 10.1210/er.2003-0020.
Paulson RJ: Hormonal induction of endometrial receptivity. Fertil Steril. 2011, 96: 530-535. 10.1016/j.fertnstert.2011.07.1097.
Shirane A, Wada-Hiraike O, Tanikawa M, Seiki T, Hiraike H, Miyamoto Y, Sone K, Hirano M, Oishi H, Oda K, et al: Regulation of SIRT1 determines initial step of endometrial receptivity by controlling E-cadherin expression. Biochem Biophys Res Commun. 2012, 424: 604-610. 10.1016/j.bbrc.2012.06.160.
Campbell CS, Ryan KD, Schwartz NB: Estrous cycles in the mouse: relative influence of continuous light and the presence of a male. Biol Reprod. 1976, 14: 292-299. 10.1095/biolreprod14.3.292.
Salgado RM, Covarrubias AC, Favaro RR, Serrano-Nascimento C, Nunes MT, Zorn TM: Estradiol induces transcriptional and posttranscriptional modifications in versican expression in the mouse uterus. J Mol Histol. 2013, 44: 221-229. 10.1007/s10735-012-9476-1.
Acknowledgements
This research study was funded by the National Natural Science Foundation of China (31201966), Specialized Natural Science Basic Research Plan in Shaanxi Province of China (2012JQ3001), and Fundamental Research Funds for the Central Universities (QN2011105). We particularly thank Elsevier Language Editing Services for their help with English correction.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests with respect to the authorship and/or publication of this article.
Authors’ contributions
PL and AW designed the experiments and drafted the manuscript. NW and JZ collected the uterine samples and embryos. FC, XW, and XL performed the experiments. YJ organised and supervised the project. All authors read and approved the final manuscript.
Electronic supplementary material
12958_2013_1171_MOESM1_ESM.pdf
Additional file 1: The positive control for SMILE and Zhangfei protein was analysed by western blotting using a CREBZF-specific antibody. Results are representative of at least three independent experiments with similar results. MCF7, human breast adenocarcinoma cell line. 293 T, human embryonic kidney cell line. (PDF 35 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Lin, P., Chen, F., Wang, N. et al. CREBZF expression and hormonal regulation in the mouse uterus. Reprod Biol Endocrinol 11, 110 (2013). https://doi.org/10.1186/1477-7827-11-110
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-7827-11-110