Abstract
Background
Only a minority of patients with advanced non-small cell lung cancer (NSCLC) benefit from chemotherapy. Serum peptide profiling of NSCLC patients was performed to investigate patterns associated with treatment outcome.
Using magnetic bead-assisted serum peptide capture coupled to matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS), serum peptide mass profiles of 27 NSCLC patients treated with cisplatin-gemcitabine chemotherapy and bortezomib were obtained. Support vector machine-based algorithms to predict clinical outcome were established based on differential pre-treatment peptide profiles and dynamic changes in peptide abundance during treatment.
Results
A 6-peptide ion signature distinguished with 82% accuracy, sensitivity and specificity patients with a relatively short vs. long progression-free survival (PFS) upon treatment. Prediction of long PFS was associated with longer overall survival. Inclusion of 7 peptide ions showing differential changes in abundance during treatment led to a 13-peptide ion signature with 86% accuracy at 100% sensitivity and 73% specificity. A 5-peptide ion signature could separate patients with a partial response vs. non-responders with 89% accuracy at 100% sensitivity and 83% specificity. Differential peptide profiles were also found when comparing the NSCLC serum profiles to those from cancer-free control subjects.
Conclusion
This study shows that serum peptidome profiling using MALDI-TOF-MS coupled to pattern diagnostics may aid in prediction of treatment outcome of advanced NSCLC patients treated with chemotherapy.
Similar content being viewed by others
Background
Lung cancer is the most common cause of cancer-related death with an overall 5-year survival rate of 16% [1]. Non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung carcinomas and is frequently diagnosed in an advanced stage [2]. First-line treatment of advanced NSCLC typically consists of platinum-based chemotherapy [3, 4]. Only a minority of patients respond to this treatment, at the cost of substantial toxicity for all treated patients [5]. There is an urgent need for methods enabling outcome prediction in order to select patients likely to benefit from treatment.
The serum peptidome, that comprises peptides and proteins with a molecular weight of less than 10 kDa, represents a dynamic reflection of tissue function in health and disease [6]. Mass spectrometry (MS) is increasingly used to profile the serum peptidome [7–9]. Peaks in the serum peptide spectra correspond to peptide ions, with the amplitude of the peaks indicative of relative abundance [10]. Magnetic bead-assisted serum peptide capture coupled to matrix assisted laser desorption/ionization time-of-flight MS (MALDI-TOF-MS) is a serum peptide profiling strategy gaining in popularity compared to surface-enhanced laser desorption/ionization (SELDI)-based platforms due to superior resolution of MALDI instruments, the possibility to obtain structural (MS/MS) information of signature peptides and superior binding capacity of the magnetic beads compared to a flat SELDI-chip surface [11].
Here we report on a MALDI-TOF-MS dataset of serum samples from advanced NSCLC patients, who were treated with first-line chemotherapy, consisting of cisplatin and gemcitabine, as well as bortezomib, in a previously reported prospective clinical trial [12]. The efficacy of cisplatin-gemcitabine alone is limited, a partial tumor response being achieved in about one third of NSCLC patients and with a median progression free survival of four to five months [13]. Preclinical as well as initial clinical studies suggested combining cisplatin-gemcitabine with proteasome inhibitor bortezomib might enhance efficacy [14–17].
We hypothesized that specific serum peptidome patterns could predict clinical outcome of patients who underwent chemotherapy-based treatment. For this purpose, data analyses of serum peptide profiles were conducted. Our primary aim was to establish serum peptide signatures that could predict positive or negative clinical outcome upon treatment. Clinical endpoints used to establish these signatures were response to treatment according to the Response Evaluation Criteria in Solid Tumors (RECIST) criteria, as well as progression-free survival duration of treated patients. Furthermore, in a secondary analysis, we compared the serum peptide profiles of treated patients with those obtained from cancer-free control subjects in an initial attempt to establish cancer-specific serum peptide patterns.
Results
Pre-treatment serum peptide patterns of NSCLC patients
First, pre-treatment serum spectra of 27 NSCLC patients were determined. See Table 1 for patient characteristics. Six hundred eighty-two peaks could be distinguished. The intra-run and inter-run coefficients of variance were 16% and 18%, respectively.
Time-course analysis
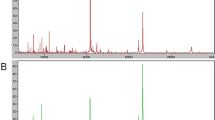
We first looked for peptides that exhibited significant changes in intensity level in three time points: (1) pre-treatment (preTx), (2) after two cycles of treatment (Post-2), and (3) at the end of treatment (EOT). Forty-four peaks were determined as significant (Table 2). The spectra overlay of the top 8 peaks is illustrated in Figure 1. Note that for example the peak at m/z 2567.3659 has a higher intensity at end of treatment (green) compared to pre-treatment (red) and the peak at m/z 1561.7288 has a lower intensity at end of treatment compared to pre-treatment.
Comparison serum profiles time course. Top 8 differential peaks that exhibit changes over the three time points: pre-treatment (PreTx) (red), after two cycles of treatment (Post-2) (blue), and at end of treatment (EOT) (green). Eight peaks randomly selected out of the remaining 638 peaks. Each overlay contains spectra with normalized intensities.
Analysis of the clinical outcome: progression-free survival
The patients were divided into two subsets according to PFS duration. Of the 27 NSCLC patients, 11 patients with the shortest PFS were nominated as "short PFS" group (PFS ≤ 127 days), 11 patients with the longest PFS were nominated as "long PFS" group (PFS ≥ 178 days). Four patients with PFS duration around the median PFS (152 days) were excluded from the analysis. A fifth patient was excluded who had a partial response but died 36 days after start of treatment due to a cause not likely due to tumor progression [12].
Six differentially expressed peptides between the two groups were detected (see Figure 2A and Table 3). Median intensity of all six peptides was higher in the "short PFS" group compared to the "long PFS" group. This 6-peptide signature was used to retrospectively divide the total study population (n = 27) into a predicted "short PFS" group and a predicted "long PFS" group. Median PFS was significantly shorter at 120 days (95% CI 54-186 days) in patients predicted to have a short PFS vs. 191 days (95% CI 154-228 days) in patients predicted to have a long PFS (p-value: 0.036). Median overall survival (OS) of patients predicted to have short PFS vs. long PFS was 144 days (95% CI 75-213 days) vs. 436 days (95% CI 292-580 days) (p-value: 0.036) (Figure 2B). For the OS analyses, two patients were censored at last known date to be alive.
Comparison serum profiles short PFS vs. long PFS. A, six peaks are differential between the two groups: short PFS (red) and long PFS (blue). Six randomly selected peaks. Each overlay contains spectra with normalized intensities. B, Kaplan-Meier (1) time to progression and (2) overall survival curve in NSCLC patients, by prediction of short or long PFS using the 6-peptide signature (intent-to-treat population, n = 27). C, median of the seven differential peaks comparing short PFS (red) and long PFS (blue) over the three time points: pre-treatment (PreTx), after two cycles of treatment (Post-2), and at end of treatment (EOT) (Y-axis: intensity).D, Kaplan-Meier (1) time to progression and (2) overall survival curve in NSCLC patients, by prediction of short or long PFS using the 13-peptide signature (intent-to-treat population, n = 27).
When considering also Post-2 and EOT samples, we found seven differential peptides distinguishing the clinical groups (see Figure 2C and Table 4). These peptides do not overlap with peptides from the pre-treatment signature. Using the combined 13-peptide signature to classify the total study population, median PFS was significantly shorter at 120 days (95% CI 70-170 days) in patients predicted to have a short PFS vs. 199 days (95% CI 181-217 days) in patients predicted to have a long PFS (p-value: < 0.001). Median overall survival of patients predicted to have short vs. long PFS was 144 days (95% CI 96-192 days) vs. 478 days (95% CI 203-753 days) (p-value: 0.002) (Figure 2D).
Finally, we carried out classification analysis using support vector machine. Using all 682 peptides, the LOOCV accuracy was very poor, at about 50%. When the six differential pre-treatment peptides were used, the LOOCV accuracy was 82% with both 82% sensitivity and 82% specificity. Selecting six different peptides randomly resulted in an average accuracy of 68% (10 runs). The LOOCV prediction accuracy improved when we used also the seven peptides that changed differently in intensity level over the three time points. Here, using the 13 peptides we could separate the two groups with a LOOCV accuracy of 86% at 100% sensitivity and 73% specificity (the average accuracy over 10 runs with random selection of 13 peptides was 71%).
Analysis of the clinical outcome: tumour response
To identify a signature associated with tumour response, we divided the patients into three groups according to tumour response following treatment: (1) partial response (PR), (2) stable disease (SD), (3) progressive disease (PD), as defined by RECIST [18]. Because there were only three patients with progressive disease, we created two groups: PR and SD/PD combined ("no PR"). The first group had 9 patients, and the second group consisted of 18 patients.
Comparing pre-treatment samples, we detected five differential peaks (see Figure 3A and Table 5). Of the five peptides, two were also present in the list of differential peaks comparing short PFS and long PFS (m/z = 2215.2849 and 2318.2202). The 5-peptide signature was used to retrospectively divide the total patient population in a PR vs. no PR group (n = 27). In the patient group classified as no PR, median PFS was significantly shorter at 125 days (95% CI 115-135 days) vs. 231 days (95% CI 85-377 days) in the group classified as PR (p-value: 0.003). Median overall survival of patients predicted to have no PR was shorter, but not significantly, at 231 days (95% CI 41-421 days) vs. 478 days (95% CI 120-836 days) for patients predicted to have a PR (p-value: 0.073) (Figure 3B).
Comparison serum profiles PR vs. no PR. A, five differential peaks in the comparison between PR (red) versus PD+SD (blue) using pre-treatment samples. Five randomly selected in the same comparison. Each overlay contains spectra with normalized intensities. B, Kaplan-Meier (1) time to progression and (2) overall survival curve in NSCLC patients, by prediction of PR or no PR, using the 5-peptide signature (intent-to-treat population, n = 27). C, median of the five differential peaks in the comparison between PR (red) versus PD+SD (blue) using three time points: pre-treatment (PreTx), after two cycles of treatment (Post-2), and at end of treatment (EOT). D, Kaplan-Meier (1) time to progression and (2) overall survival curve in NSCLC patients, by prediction of PR or no PR, using the 10-peptide signature (intent-to-treat population, n = 27).
Next we included also the Post-2 and EOT samples, resulting in five additional differential peptides (see Figure 3C and Table 6). Again, there was no overlap between these five peptides and the peptides found when using the pre-treatment samples only. However, of these five peptides, three peptides were also in the list of significant peptides when comparing short PFS and long PFS over the three time points (m/z = 1596.189, 1616.637 and 3215.194). The 10-peptide signature was used to retrospectively divide the total patient population in PR vs. no PR (n = 27). In the patient group classified as no PR, median PFS was 125 days (95% CI 91-159 days) vs. 231 days (95% CI 117-345 days) in the group classified as PR (p-value 0.038). Median overall survival of patients predicted to have no PR was shorter, but not significantly, at 231 days (95% CI 36-426 days) vs. 613 days (95% CI 0-1253 days) for patients predicted to have a PR (p-value: 0.077) (Figure 3D).
Classification analysis was performed using support vector machine with different combinations of features. The LOOCV accuracy was again poor when using all 682 peptides, at about 67%. When the five differential pre-treatment peptides were used, the LOOCV accuracy was 89% at 100% sensitivity and 83% specificity. The average accuracy over 10 runs was 74% using random feature selection for five peptides. The LOOCV prediction accuracy was reduced to 85%, at 100% sensitivity and 78% specificity, when we used also the five peptides that changed differently in intensity level over the three time points (Figure 4 provides an overview of the study and study results).
Study flowchart. Serum profiling was performed in 27 NSCLC patients. "Feature list 1" represents total number of features before intensity and statistical filters. For each comparison, "Feature list 2" represents the number of differential peptide ions comparing pre-treatment sera (white filled circle), time-course sera (black filled circle) and pre-treatment plus time-course combined (grey filled circle). Accuracy (ac), sensitivity (sn) and specificity (sp) of algorithms are indicated. SVM: support vector machine; LOOCV: leave-one-out-cross validation; pts: patients.
Peptide pattern discriminating NSCLC patients from cancer-free controls
Finally, in an exploratory additional analysis, we compared the serum peptide spectra of 13 cancer-free control subjects (median age: 38 years old; range: 27-58 years old) and the pre-treatment serum spectra of the 27 NSCLC patients included in this study. We performed a principal component analysis (PCA) analysis of the 40 profiles using all 682 peptides, see Figure 5A. While there is overlap between the two groups in the three dimensional plot, the healthy profiles (in red) are clustered at the bottom right region. Furthermore, we observed no indication of outliers in the dataset. Next we performed a supervised analysis to identify peptides that were significantly differential in intensity between the two groups. For this purpose, the Mann-Whitney U test was carried out on each of the 682 peptides using all profiles. The peptides were selected based on the criteria outlined in Methods, resulting in 47 peptides. A heat map of the intensities of the 47 peptides is shown in Figure 5B (see also Table 7). Figure 5C shows the spectra overlay of the top 8 most discriminating peaks, all of which have a p-value < 0.0001. Note that for example the peak at m/z 1777.966 has a higher intensity in NSCLC patients (blue) compared cancer-free controls (red) and the peak at m/z 1039.6249 has a lower intensity in NSCLC patients. We carried out classification analysis using support vector machine. A grid search for parameters was employed to find the best model according to LOOCV. Using all 682 peptides, an LOOCV accuracy of 93% was achieved. When the 47 peptides selected by the Mann-Whitney U test were used, the LOOCV accuracy was 98% with 100% sensitivity and 96% specificity. To substantiate the result, we compared it to a random selection of peptides. Using the same model selection mechanism for support vector machine with 47 different peptides randomly selected the average accuracy over 10 runs was 90%.
Comparison serum profiles NSCLC vs. cancer-free controls. A, Principle Component Analysis (PCA) NSCLC vs. cancer-free control comparison. B, heat map of the 47 differential peaks. The peaks are ordered by median fold change between the two groups. C, spectra overlay of the 8 most differential peaks in the healthy (red) versus NSCLC (blue) comparison. Spectra overlay of the 8 peaks randomly selected out of the remaining 635 peptides.
In this secondary analysis, control subjects were unmatched for age and gender. We therefore also considered peptides that express differently in the two gender groups. For this comparison, four peptides were differential according to our criteria described above (m/z = 1458.495, 3215.194, 2602.305, and 2789.091). Of these four peptides, two are present in the list of 47 differential peptides in the healthy versus NSCLC comparison. Ignoring these two peptides, the signature composed of the remaining 45 peptides yielded the same accuracy, sensitivity and specificity as that of the 47-peptide signature. Literature supports that serum peptidome patterns that distinguished advanced cases of cancer from cancer-free controls were unbiased by gender and age, except for the fact that healthy subjects under 35 years could be distinguished with approximately 70% accuracy [19]. All participating patients in our study were 35 years or older. However, in the cancer-free control group, 4 individuals were younger than 35 years and 9 individuals older than 35 years. Comparing these two groups, two peaks met the criteria for differential (m/z = 1020.5133 and 855.0387). These two peaks did not feature in the classifying signature between the NSCLC patients and the cancer-free controls.
Peptide identification
For structural identification of signature peptides by MS/MS, we performed an additional peptide capture on another aliquot of the sera used for profiling. Sera with highest intensity levels of signature ions were selected for MS/MS. For each eluate, a series of four spots was applied to a MALDI target plate, and candidate peaks were subjected to MS/MS in the sample spot(s) associated with the highest intensity for the pertinent peak. Seventeen peptides were positively identified by MALDI-TOF/TOF-based MS/MS analysis, see Table 8 as well as Tables 2, 4 and 6. See Figure 6 for an example of an annotated MS/MS spectrum. In agreement with results by Villanueva et al. in other tumor types, the serum peptide signatures mainly consisted of small sets of overlapping sequences, truncated in both ends in a ladder-like fashion. See Table 8 for truncation ladder examples of Fibrinopeptide alpha, Complement C3f, Complement C3 beta and Hemoglobin alpha.
Discussion
In this study, we investigated the use of serum peptide mass profiling by MALDI-TOF-MS coupled to bioinformatics pattern discovery to predict treatment outcome of advanced NSCLC patients treated with platinum-based therapy. Additionally, peptide patterns found in NSCLC patients were differential from those found in healthy volunteers.
To our knowledge we are the first to report on a serum peptide signature for response and survival prediction in NSCLC patients treated with cisplatin-based chemotherapy. For this study, serum samples were obtained not only pre-treatment, but also during treatment and after completion of treatment, whereas serum proteomics studies typically focus on pre-treatment samples only. In a study by Taguchi et al., a predictive MALDI-TOF-MS-based peptide algorithm for "good" or "poor" clinical outcome upon epidermal growth factor tyrosine kinase inhibitor (EGFR-TKI) therapy was established [20]. However, in this study none of the peptides corresponding to the classifying m/z features were identified.
In our study, for the short PFS versus long PFS classification, we achieved 82% accuracy with a 6-peptide-signature. A longer overall survival of the patients classified as "long PFS" and a shorter overall survival of the patients classified as "short PFS" was observed. Interestingly, performance of the signature was improved upon inclusion of 7 differential time-course peptides, the combined 13 peptide-ion signature achieving 86% accuracy. In this regard, it has been previously reported that histological response of locally advanced rectal cancer to radiochemotherapy could be predicted by SELDI-TOF-MS-based profiling and comparison of serum samples collected pre-treatment and during treatment [21].
For partial responders versus non-partial responders we achieved 89% accuracy with a 5-peptide signature. Inclusion of differential time-course peptides did not improve performance of this signature. Longer duration of progression-free survival was strongly associated with tumour response as seven out of eleven patients in our study with long PFS also had a partial tumour response upon treatment, compared to one out of eleven patients in the short PFS group. This suggests that the survival signature is predictive of therapy-outcome rather than prognostic. As the Kaplan-Meier analysis showed, prediction of response using the 5- or 10-peptide signatures was significantly associated with a longer median PFS of those patients, but this did not reach significance for overall survival.
In several MALDI serum peptide profiling studies involving other solid tumour types (bladder, breast, prostate, thyroid) by Villanueva et al., methodologically comparable to our study, the hypothesis was put forward that cancer-type specific changes in exopeptidase activities yield surrogate biomarkers, reflected in the differential abundance of cleavage products of common serum substrate proteins [19, 22]. The changes in exopeptidase activity were superimposed on the proteolytic events of the complement degradation and ex vivo coagulation pathways and therefore serum-specific. Regulated peptides at m/z 1690.925, 1777.966 and 1865.002 correspond to those identified in our study at 1690.90, 1777.94 and 1864.95, respectively, previously identified by MALDI-TOF-TOF-based MS/MS analysis as Complement C3f. The peptide at 2209.093 corresponds to the one we detected at 2209.08, previously identified as HMW Kininogen [19].
We show that in NSCLC, the identified differential serum peptides changing in abundance over time and those distinguishing NSCLC patients from cancer-free control subjects consisted of truncated sequence clusters. The identified peptides were derived from naturally occurring serum peptides and protein precursors, and therefore not likely to be tumor-derived, thus supporting the exoprotease hypothesis. For the predictive algorithms, it is important to realize that these might be specific for the treatment combination of bortezomib, cisplatin and gemcitabine and may not apply to other treatment regimens. For this exploratory analysis, we did not perform a power calculation beforehand as we did not know the biological variation in our patient groups, nor the number of peaks we would measure as well as other variables. We only knew the technical variation. It was therefore our approach to collect as many samples as we could accommodate. It is crucial to validate and adjust the established signatures with an independent cohort in a sufficiently powered follow-up study. Additionally, since only one report excludes an age and gender bias for a cancer-specific serum peptide signature, it is advisable to include matched cancer-free control groups for the establishment of cancer-specific peptide patterns.
Conclusion
Ideally, serum peptide mass profiling can be used to identify the therapeutic agents to which the tumour is sensitive, enabling personalized medicine. The method employed here requires readily accessible, non-invasively obtainable patient samples, is high-throughput and cost-efficient, all together important requirements for a screening platform as well as routine clinical use. The biggest challenge might very well remain lack of reproducibility related to sample collection in the clinic. In particular, maintaining a constant and precise clotting time is often difficult in clinical practice. Potentially, after initial discovery of classifying algorithms, functional proteomics tests will facilitate clinical implementation [23].
Methods
Patients and serum preparation
The training set included 27 patients with NSCLC who were treated with chemotherapy and bortezomib as well as 13 healthy volunteers [12]. All patients were treated with cisplatin 70 mg/m2 day 1 and gemcitabine 1,000 mg/m2 days 1 and 8, every 21 days for up to 6 cycles. Fifteen patients were treated with bortezomib on days 1 and 8 of every cycle (ten patients at 1.0 mg/m2 and five patients at 1.3 mg/m2). Twelve patients were treated with bortezomib on days 1,4,8 and 11 of every cycle (one patient at 0.7 mg/m2 and eleven patients at 1.0 mg/m2). There was no indication of superior clinical activity of any schedule of bortezomib in combination with cisplatin and gemcitabine [12]. Blood samples were obtained in BD Vacutainer glass "red-top" tubes (Becton Dickinson, Franklin Lakes, NJ), allowed to clot for 1 hour, and then centrifuged at 1500 g for 10 minutes. Sera were stored in polypropylene cryovials (Nunc; Roskilde, Denmark) at -80°C. Studies were performed after obtaining patient consent and under protocols approved by the institutional review board.
Serum sample processing and mass spectrometry
Samples were processed in randomized order, along with control samples to check consistency in each experiment. Magnetic Dynabeads® RPC 18 (Invitrogen, San Diego, CA) were used for serum peptide capture using the KingFischer96 platform, as described previously [11]. Briefly, in a 96-well format, magnetic beads were washed and equilibrated twice in 200 μl 200 mM NaCl/0.1% TFA, transferred to a mix of 20 μl serum sample and 2 volumes of 0.2% n-octyl glucoside/0.5% TFA (premixed for 5 min), incubated for 2 min, washed thrice with 0.1% TFA, and eluted for 2 min with 40 μl 50% acetonitrile.
Processed samples (1.5 μl eluate) were mixed with 2 volumes α-cyano-4-hydroxycinnamic acid matrix (6.2 mg/ml in 56% acetonitrile, 36% methanol; Agilent, Santa Clara, CA) and 0.7 μl of this mix was spotted on a MALDI plate. A 4800 MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Foster City, CA) was used to record with 5000 shots per spectrum (reflectron mode) serum peptide profiles in the mass range of m/z 800-4000 [11]. Internal calibration was used using a list of exact masses for fibrinogen α/fibrinopeptide A peptides as major components of serum samples. For MS/MS analysis, stepwise attempts of 5000 shots at a time generated spectra for identification by the Mascot search engine or manual identification. For Mascot searches, the SwissProt database (release 54.7) was used at a mass window of 10 ppm for MS and a 1-Da tolerance for MS/MS. Final scores were obtained by narrowing down the window. For manual identification, spectra were compared with theoretical peptide fragments of reported candidate proteins [19]. Fragments having a predicted mass differing less than 10 ppm from the mass of any of the 87 significantly regulated peaks from our profiling study were identified using FindPept http://www.expasy.ch[19]. Fragmentation patterns were predicted using MS-Product http://prospector.ucsf.edu/prospector/mshome.htm, requiring that at least 3 prominent peaks in the experimental spectrum should match b or y ions from the theoretical table.
Signal processing
Spectra were pre-processed using MarkerView, version 1.2 (Applied Biosystems) with a mass tolerance of 200.0 ppm and minimum intensity at 100.0 units. Total signal intensity of all peptide peaks was used for normalization.
Statistical analysis
Feature selection was performed using the Mann-Whitney U test on each peptide detected in the pre-processing step. We used a common threshold of 5% for the p-value. As p-values were not adjusted for multiple testing, we took additional measures to guard from false discovery. To reduce differences due to noise, each peptide was subjected to intensity filtering, requiring that the median intensity of at least one group must be greater than 80 units and the fold change of the median intensities of the two groups must be greater than 1.5. For time course analysis of the three time points, we treated the problem as three binary comparisons. For each comparison, a paired, two-sided signed rank test was carried out. Each peptide was again subjected to intensity filtering. The results of the three comparisons were merged where the significance level of each peptide was the minimum of the three p-values. Similarly, we analyzed dynamic peptide profiles (time-course), using the group information, in order to identify peptides of which the intensity level changes differently between different clinical groups. Finally, support vector machine with the Gaussian kernel was used to construct classification models. A two dimensional grid search was carried out to set model parameters using the leave-one-out cross-validation (LOOCV) measure. Analogously to Villanueva et al. [19], we used a statistical test for feature selection. This procedure is based on class label, thus bias might be introduced. Nevertheless, this strategy was shown to be effective in previous studies [19, 22]. Median duration of progression-free and overall survival was calculated using the Kaplan-Meier method; p-values according to the log-rank test (SPSS Statistics17.0, SPSS Inc., Chicago, IL).
Abbreviations
- CI:
-
confidence interval
- EOT:
-
end of treatment
- LOOCV:
-
leave-one-out cross-validation
- MALDI-TOF MS:
-
matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- MS:
-
mass spectrometry
- NSCLC:
-
non-small cell lung cancer
- OS:
-
overall survival
- PCA:
-
principal component analysis
- PD:
-
progressive disease
- PFS:
-
progression-free survival
- PR:
-
partial response
- PreTx:
-
pre-treatment
- RECIST:
-
Response Evaluation Criteria in Solid Tumors
- SD:
-
stable disease
- SELDI:
-
surface-enhanced laser desorption/ionization.
References
Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ: Cancer statistics, 2007. CA Cancer J Clin 2007, 57: 43–66. 10.3322/canjclin.57.1.43
Spira A, Ettinger DS: Multidisciplinary Management of Lung Cancer. N Engl J Med 2004, 350: 379–392. 10.1056/NEJMra035536
Pfister DG, Johnson DH, Azzoli CG, Sause W, Smith TJ, Baker SJ, Olak J, Stover D, Strawn JR, Turrisi AT, Somerfield MR, American Society of Clinical Oncology: American Society of Clinical Oncology Treatment of Unresectable Non-Small-Cell Lung Cancer Guideline: Update 2003. J Clin Oncol 2004, 22: 330–353. 10.1200/JCO.2004.09.053
Shepherd FA: Current paradigms in first-line treatment of non-small-cell lung cancer. Oncology (Williston Park) 2004, 18: 13–20.
Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH, Eastern Cooperative Oncology Group: Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 2002, 346: 92–8. 10.1056/NEJMoa011954
Lopez MF, Mikulskis A, Kuzdzal S, Bennett DA, Kelly J, Golenko E, DiCesare J, Denoyer E, Patton WF, Ediger R, Sapp L, Ziegert T, Lynch C, Kramer S, Whiteley GR, Wall MR, Mannion DP, Della Cioppa G, Rakitan JS, Wolfe GM: High-resolution serum proteomic profiling of Alzheimer disease samples reveals disease-specific, carrier-protein-bound mass signatures. Clin Chem 2005, 51: 1946–1954. 10.1373/clinchem.2005.053090
Liotta LA, Petricoin EF: Serum peptidome for cancer detection: spinning biologic trash into diagnostic gold. J Clin Invest 2006, 116: 26–30. 10.1172/JCI27467
Petricoin EF, Liotta LA: SELDI-TOF-based serum proteomic pattern diagnostics for early detection of cancer. Curr Opin Biotechnol 2004, 15: 24–30. 10.1016/j.copbio.2004.01.005
Villanueva J, Philip J, DeNoyer L, Tempst P: Data analysis of assorted serum peptidome profiles. Nat Protoc 2007, 2: 588–602. 10.1038/nprot.2007.57
Aebersold R, Mann M: Mass spectrometry-based proteomics. Nature 2003, 422: 198–207. 10.1038/nature01511
Jimenez CR, El Filali Z, Knol JC, Hoekman K, Kruyt FA, Giaccone G, Smit AB 2, Li KW: Automated serum peptide profiling using novel magnetic C18 beads off-line coupled to MALDI-TOF-MS. Proteomics Clin Appl 2007, 1: 598–604. 10.1002/prca.200600483
Voortman J, Smit EF, Honeywell R, Kuenen BC, Peters GJ, Velde H, et al.: A parallel dose-escalation study of weekly and twice-weekly bortezomib in combination with gemcitabine and cisplatin in the first-line treatment of patients with advanced solid tumors. Clin Cancer Res 2007, 13: 3642–51. 10.1158/1078-0432.CCR-07-0061
Smit EF, van Meerbeeck JP, Lianes P, Debruyne C, Legrand C, Schramel F, et al.: Three-arm randomized study of two cisplatin-based regimens and paclitaxel plus gemcitabine in advanced non-small-cell lung cancer: a phase III trial of the European Organization for Research and Treatment of Cancer Lung Cancer Group--EORTC 08975. J Clin Oncol 2003, 21: 3909–3917. 10.1200/JCO.2003.03.195
Teicher BA, Ara G, Herbst R, Palombella VJ, Adams J: The Proteasome Inhibitor PS-341 in Cancer Therapy. Clin Cancer Res 1999, 5: 2638–2645.
Mortenson MM, Schlieman MG, Virudachalam S, Bold RJ: Effects of the proteasome inhibitor bortezomib alone and in combination with chemotherapy in the A549 non-small-cell lung cancer cell line. Chemother Pharmacol 2004, 54: 343–353.
Davies AM, Lara PN Jr, Mack PC, Gandara DR: Incorporating bortezomib into the treatment of lung cancer. Clin Cancer Res 2007, 13: 4647–51. 10.1158/1078-0432.CCR-07-0334
Ludwig H, Khayat D, Giaccone G, Facon T: Proteasome inhibition and its clinical prospects in the treatment of hematologic and solid malignancies. Cancer 2005, 104: 1794–807. 10.1002/cncr.21414
Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG: New Guidelines to Evaluate the Response to Treatment in Solid Tumors. J Natl Cancer Inst 2000, 92: 205–216. 10.1093/jnci/92.3.205
Villanueva J, Martorella AJ, Lawlor K, Philip J, Fleisher M, Robbins RJ, Tempst P: Serum peptidome patterns that distinguish metastatic thyroid carcinoma from cancer-free controls are unbiased by gender and age. Mol Cell Proteomics 2006, 5: 1840–52. 10.1074/mcp.M600229-MCP200
Taguchi F, Solomon B, Gregorc V, Roder H, Gray R, Kasahara K, Nishio M, Brahmer J, Spreafico A, Ludovini V, Massion PP, Dziadziuszko R, Schiller J, Grigorieva J, Tsypin M, Hunsucker SW, Caprioli R, Duncan MW, Hirsch FR, Bunn PA Jr, Carbone DP: Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Inst 2007, 99: 838–46. 10.1093/jnci/djk195
Smith FM, Gallagher WM, Fox E, Stephens RB, Rexhepaj E, Petricoin EF III, Liotta L, Kennedy MJ, Reynolds JV: Combination of SELDI-TOF-MS and data mining provides early-stage response prediction for rectal tumors undergoing multimodal neoadjuvant therapy. Ann Surg 2007, 245: 259–266. 10.1097/01.sla.0000245577.68151.bd
Villanueva J, Shaffer DR, Philip J, Chaparro CA, Erdjument-Bromage H, Olshen AB, Fleisher M, Lilja H, Brogi E, Boyd J, Sanchez-Carbayo M, Holland EC, Cordon-Cardo C, Scher HI, Tempst P: Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest 2006, 116: 271–84. 10.1172/JCI26022
Villanueva J, Nazarian A, Lawlor K, Yi SS, Robbins RJ, Tempst P: A sequence-specific exopeptidase activity test (SSEAT) for "functional" biomarker discovery. Mol Cell Proteomics 2008, 7: 509–518.
Acknowledgements
Drs Lyle Burton and Ron Bonner (Applied Biosystems/Sciex) are acknowledged for early access to new versions of MarkerView software. The VUmc-Cancer Center Amsterdam is acknowledged for support of CRJ, TVP and JCK and proteomics infrastructure. JV was supported by a Netherlands Organization of Scientific Research ZonMW-AGIKO stipend, grant 920-03-290.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JV, TVP and JCK carried out experimental procedures and drafted manuscript. CRJ and GG conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Johannes Voortman, Thang V Pham contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Voortman, J., Pham, T.V., Knol, J.C. et al. Prediction of outcome of non-small cell lung cancer patients treated with chemotherapy and bortezomib by time-course MALDI-TOF-MS serum peptide profiling. Proteome Sci 7, 34 (2009). https://doi.org/10.1186/1477-5956-7-34
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-5956-7-34