Abstract
The DNA degradation potential and anti-cancer activities of copper nanoparticles of 4-5 nm size are reported. A dose dependent degradation of isolated DNA molecules by copper nanoparticles through generation of singlet oxygen was observed. Singlet oxygen scavengers such as sodium azide and Tris [hydroxyl methyl] amino methane were able to prevent the DNA degradation action of copper nanoparticles confirming the involvement of activated oxygen species in the degradation process. Additionally, it was observed that the copper nanoparticles are able to exert cytotoxic effect towards U937 and Hela cells of human histiocytic lymphoma and human cervical cancer origins, respectively by inducing apoptosis. The growth characteristics of U937 and Hela cells were studied applying various concentrations of the copper nanoparticles.
Similar content being viewed by others
Findings
Nanotechnology is one of the most rapidly growing disciplines with a wide range of applications, especially in electronics, information technology, sensor development, catalysis, and biomedical sciences [1–5]. Nanoparticles have a specific capacity for drug loading, efficient photoluminescence ability and are therefore important materials in the targeted delivery of imaging agents and anti-cancer drugs [6–9]. The extremely small size of the nanoparticles makes them to be utilized for potential target oriented delivery of nanomedicines in organs such as the brain, which are normally protected by specialized barriers (such as the blood-brain barrier). If these trends continue with nanomedicines, humans will be continuously benefited using exceedingly improved nanomaterials with diverse properties to act at the interface between nanotechnology and biology [10].
Continuous demand for new anti-cancer drugs has stimulated chemotherapeutic research based on the use of metals since potential drugs developed in this way may be less toxic and more prone to exhibit anti-proliferative activity against tumors [11, 12]. Transition metal complexes have been extensively studied for their nuclease-like activity using the redox properties of the metal and dioxygen to produce reactive oxygen species to promote DNA cleavage by direct strand scission or base modification [13]. More recent trend in this area has been testing of metal nanoparticles such as gold and platinum nanoparticles for DNA degradation studies [14, 15]. Use of metal nanoparticles can be in particular advantageous in generating singlet oxygen [16, 17]. A recent report by Geddes and coworkers demonstrated that the presence of metal nanoparticles can enhance singlet oxygen generation [18]. The enhanced electromagnetic fields in proximity to metal nanoparticles are the basis for the increased absorption and various computational methods are available to predict the extent of absorption and the relative increase in singlet oxygen generation from photosensitizers [19, 20]. Although a number of well-defined copper (II) complexes exhibited their DNA degradation capabilities [21, 22], there are no reports on in vitro study of DNA degradation using copper nanoparticles (CuNPs). A very recent study by Midander and coworkers reported the effect of CuNPs inducing single stranded breaks in the cultured human lung cells [23]. Earlier studies showed potent cytotoxic, genotoxic and toxicological activities of CuNPs in vivo [23, 24] and in cultured cancer cell lines [12]. However, a systematic study using CuNPs on DNA degradation and cytotoxicity towards different cancer cells are missing till to date to the best of our knowledge.
In this communication, a dose dependent DNA degradation action of copper nanoparticles (CuNPs) on isolated DNA molecules at 37°C is reported. Singlet oxygen scavengers such as sodium azide and Tris [hydroxyl methyl] amino methane were found to prevent the DNA degradation action of CuNPs and this observation confirms the involvement of activated oxygen species in the degradation process. Fluorescence quenching studies and densitometry analysis revealed the affinity of the interaction of DNA with CuNPs and the kinetics of DNA degradation by CuNPs, respectively. This study demonstrates that CuNPs can induce singlet oxygen mediated DNA damage and thus to be considered as potent cytotoxic agent to target cancer cells for the therapeutic applications. In fact, it was observed that the CuNPs could exert cytotoxic effect towards U937 and Hela cell lines of human lymphoma and cervical cancer origins, respectively by inducing apoptosis.
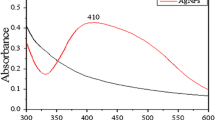
The CuNPs were prepared in aqueous solution by reducing Cu2+ ions with sodium borohydride in the presence of sodium citrate as a capping agent following a modified literature method [25]. Characterization of copper nanopaticles was carried out by UV-Vis spectroscopy and transmission electron microscopic (TEM) studies (Figure 1). The average nanoparticles size has been found to be 4-5 nm. The effect of copper nanoparticles on bacterial genomic DNA isolated from Escherichia coli was tested by treating the DNA with CuNPs of gradually increasing concentrations ranging from 50-500 μM for 100 minutes at 37°C in phosphate buffered saline (PBS) maintained at pH 7.4. After the incubation, the fate of DNA was analyzed by agarose gel electrophoresis. It was observed that the CuNPs induced DNA degradation and the degree of DNA degradation was directly proportional to the concentration of CuNPs (Figure 2a). Copper sulphate, sodium citrate, sodium borohydride solutions as well as the supernatant of CuNPs dispersion were also incubated with DNA as controls and none of these components were able to degrade DNA. This observation confirmed that CuNPs were solely responsible for DNA degradation (Figure 2a). Furthermore, the chemical scavengers which can scavenge active species in the reaction mixture were used to unravel the mechanistic pathway of degradation process. Scavengers included dimethyl sulphoxide and D-mannitol (hydroxyl free radical scavengers), sodium azide and Tris [hydroxyl methyl] amino methane (singlet oxygen scavengers) [26–28]. It was observed that sodium azide (0.1 M, 0.2 M) and tris [hydroxyl methyl] amino methane (0.1 M, 0.2 M) completely inhibited the CuNPs mediated DNA degradation (Figure 2b, Lanes 10-14). On the other hand, D-mannitol (0.1 M, 0.2 M), dimethyl sulphoxide (0.1 M, 0.2 M) were unable to prevent the DNA degradation completely (Figure 2b, Lanes 7-10). These results clearly indicate that CuNPs induced DNA degradation proceeds through a singlet oxygen mediated mechanism. In order to calculate the rate constant of the DNA degradation by copper nanoparticles, pET28b plasmid DNA was treated with 500 μM CuNPs for different time intervals and the different forms of plasmid DNA (supercoiled, circular and linear) were analyzed by agarose gel electrophoresis. The percentages of the different forms of plasmid DNA were estimated by densitometric analysis with the help of "Quantity one" software (Figure 3a and 3b). The supercoiled to circular form conversion curve was fitted in to the first order exponential decay equation [29]. The decay constant was found to be 0.0177 S-1, with an R2 value 0.99. Fluorescence quenching studies were carried out by using ethidium bromide (EB) bound herring sperm DNA with increasing concentrations of CuNPs [30]. The fluorescence quenching revealed a reasonable agreement with the classical stern-Volmer equation
Characterization of copper nanoparticles. (a) UV-Vis spectrum of the CuNPs exhibiting a Mie scattering profile and (b) TEM images: left one (I) displays a TEM image of CuNPs and right one (II) displays a higher magnification image of the same revealing the presence of well dispersed particles having size between 4-5 nm.
Singlet oxygen mediated DNA degradation by copper nanoparticles. (a) Dose dependent DNA degradation action of CuNPs. Lane 1 - Control DNA, Lane 2 - DNA + supernatant, Lanes 3 to 8 - DNA + 50, 100, 200, 300, 400, 500 μM CuNPs, respectively, Lane 9 - DNA + 500 μM CuSO4, Lane 10 - DNA + 4 mM sodium citrate, Lane 11 - DNA + 100 μM sodium borohydride and (b) Comparison of the ROS scavenging activity. Lane 1 - DNA alone, Lanes 2 to 5 - DNA + DMSO, D-mannitol, sodium azide, Tris (all 0.2 M) respectively, Lane 6 - DNA + 500 μM CuNPs, Lanes 7 to 14 - DNA + 500 μM CuNPs and DMSO 0.1 M, DMSO 0.2 M, D-mannitol 0.1 M, D-mannitol 0.2 M, azide 0.1 M, azide 0.2 M, Tris 0.1 M, Tris 0.2 M, respectively.
DNA degradation kinetics and DNA binding by Copper nanoparticles. (a) Time dependent plasmid degradation (Lane 1- DNA alone, Lane 2- DNA+ 0.2 M Tris, Lane 3-DNA+ 0.2 M Tris + 500 μM CuNPs, Lanes 4 to 14 - DNA + 500 μM CuNPs incubated for 10, 60, 120,180, 240, 300, 360, 420, 480, 540, 600 seconds followed by addition of 0.2 M Tris; (b) Time dependent change in the plasmid forms and (c) Binding of CuNPs with DNA shown by Stern-Volmer plot of DNA-EB in presence of different concentration of CuNPs.

where IFO and IF are the emission intensity in the absence and presence of the quencher, respectively, Ksv is the stern - Volmer quenching constant and [CuNPs] is the concentration of CuNPs (Figure 3c). The value of Ksv was calculated as 0.00264 M-1. This result demonstrates that there is a significant interaction of CuNPs with the DNA. The apparent binding constant (Kapp) for DNA-CuNPs interaction was also calculated as 3.137 × 104 M-1 using the following equation

Where KEB = 1.0 × 107 M-1, [EB] = 1.3 μM and [CuNPs]50 is the concentration that cause a 50% quenching of the initial EB fluorescence [30].
Additionally, the effect of CuNPs on cultured U937 and Hela cells was tested. U937 cells were grown in RPMI-1640 medium and Hela cells were grown in DMEM medium in the presence of 10% fetal bovine serum under 5% CO2 in a humidified incubator at 37°C and were treated with different concentrations of CuNPs. U937 and Hela cells were also treated with a mixture of sodium borohydride and sodium citrate, and CuSO4 solutions as controls. Initially, to check the cytotoxic effect of CuNPs on U937 and Hela cells, a number of viable cells after exposure with CuNPs were enumerated by colorimetric MTT assay [31]. Percentages of surviving cells to untreated controls were calculated by using the formula as % viability = [(At/As) × 100] %, where At and As indicate the absorbance of the sample and control, respectively. Interference of copper in MTT assay was monitored and it was found that copper has an interfering effect with a maximum value of 17% increase in color production if it is considered that all of the added CuNPs (500 μM) enter inside the treated cells and are converted to Cu+2 ions. Results of MTT assays (Figure 4a and 4b) clearly revealed the cytotoxic effect of CuNPs in a dose dependent manner for both the cell lines and CuNPs exerted slightly better cytotoxic effect towards Hela cells in comparison to U937 cells. Although, CuSO4 also showed cytotoxicity towards cancer cells, but the effect was much less compared to citrate protected CuNPs.
Cytotoxic effect and induction of apoptosis by CuNPs towards U937 and Hela cells. (a) U937 and (b) Hela cells were treated with 100, 250 and 500 μM of CuNPs, and CuSO4 for 24 hours. Cell viability based on MTT assay is shown where viability for control untreated cells was considered as 100%. Data are presented as Mean ± SE. Phase contrast microscopic pictures of (c) U937 and (e) Hela cells untreated (top) or treated with 500 μM CuNPs (bottom). Fluorescence microscopic pictures of DAPI stained (d) U937 and (f) Hela cells untreated (top) and treated with 500 μM CuNPs (bottom). Arrows indicate fragmented nuclei. (g) U937 cells were treated for 24 hours with 250 μM CuNPs (lane 3) or a mixture of sodium borohydride and sodium citrate (lane 2) and inter nucleosomal DNA fragmentation was analyzed by electrophoresis on a 1.6% Tris-Borate-EDTA agarose gel. Lane 1 represents untreated U937 cells.
Cytotoxicity of metallic copper nanoparticles, copper oxide nanoparticles and ionic copper on different cells was documented earlier [12, 32, 33]. Studer et al. specifically compared cytotoxic effect of metallic copper nanoparticles, copper oxide nanoparticles and ionic copper on Chinese Hamster Ovary (CHO) cells and Hela cells [12]. It was observed that cytotoxic effect of carbon protected copper nanoparticles (C/Cu) towards CHO cells was less compared to CuO nanoparticles, but was greater than that of CuCl2 [12]. In contrast, Studer et al. found that in case of Hela cells, C/Cu could not exert significant cytotoxicity while both CuO nanoparticles and CuCl2 exerted cytotoxic effect [12]. Interestingly, in our present study, the citrate protected copper nanoparticles were able to show significant cytotoxicity towards both U937 and Hela cells as compared to CuSO4. In addition, U937 and Hela cells, after treatment with CuNPs, exhibited ultra structure and biochemical features that are characteristic of apoptosis, as shown by chromatin condensation and inter nucleosomal DNA fragmentation. The phase-contrast microscopic pictures of altered morphology of U937 and Hela cells which is characteristic of apoptotic cell stage when treated with CuNPs are shown in Figure 4c and 4e. Fluorescent microscopic studies after 4', 6-diamidino-2-phenylindole (DAPI) staining of untreated and CuNPs treated cells clearly exhibited nuclear fragmentation in CuNPs treated U937 and Hela cells which is a hallmark of cellular apoptosis (Figure 4d and 4f). Moreover, CuNPs treated U937 cells displayed a ladder pattern of inter nucleosomal DNA fragmentation on TBE-agarose gel electrophoresis in DNA ladder assay [34] as shown in Figure 4g (lane 3) which is also another hallmark of apoptosis. All these results demonstrate that treatment with CuNPs induce apoptosis in U937 and Hela cells.
To check stability of copper nanoparticles we carried out a number of UV-Vis spectroscopy measurements and TEM studies. TEM study clearly revealed that the size of the nanoparticles remains similar after incubation of CuNPs in cell culture media indicating the stability of copper nanoparticles with respect to its agglomeration tendency in the cell culture medium (Figure 5a). However, the UV-Vis spectroscopy of CuNPs in cell culture media indicates slight agglomeration (Additional file 1 Figure S4). Confocal microscopic studies confirmed the uptake of CuNPs inside the Hela cells (Figure 5b and 5c) with the presence of agglomerated copper nanoparticles; a similar observation was also reported by Stark and coworkers with carbon coated copper nanoparticles for Hela cells [12].
Stability of Copper nanoparticles and uptake of citrate protected copper nanoparticles (CuNPs) in Hela cells. (a) TEM image of copper nanoparticles incubated in DMEM medium for 3 hours. (b) Confocal microscopic image of Hela cells treated with 500 μM of Copper nanoparticles for 14 hours. Arrows indicate agglomerated CuNPs. (c) Confocal microscopic image of Hela cells treated with 500 μM of Copper nanoparticles for 14 hours. Arrow indicates agglomerated CuNPs in a membrane bound vesicle (probably perinuclear lysosome). (d) Confocal microscopic image of control Hela cells.
In summary, it was observed for the first time that the copper nanoparticles can initiate the DNA degradation process and also can induce apoptotic cell death in cancer cells. The CuNPs degrade DNA in a singlet oxygen mediated fashion even in the absence of any external agents like hydrogen peroxide or ascorbate. This makes CuNPs as an excellent candidate for targeted therapy. The use of copper nanoparticles as therapeutic agents could be in particular advantageous because human body has an efficient system to deal with metabolism of copper since it is a micronutrient. So the residual copper expected to be produced during the nanoparticle based drug metabolism can be easily managed by the body. Furthermore, this DNA degradation potential and cytotoxic effect of CuNPs can be utilized in designing better and more active cancer drugs by chemically modifying the CuNPs with a number of macromolecules. Current efforts in our laboratory are underway to address these questions and to study the molecular mechanisms of CuNPs mediated cytotoxicity through apoptosis towards cancer cells of different origins.
References
Park H, Park J, Lim AK, Anderson EH, Alivisatos AP, McEuen PL: Nanomechanical oscillations in a single-C60 transistor. Nature. 2000, 407: 57-60. 10.1038/35024031.
De Franceschi S, Kouwenhoven L: Nanotechnology: Electronics and the single atom. Nature. 2002, 417: 701-702. 10.1038/417701a.
Toshima N: Capped bimetallic and trimetallic nanoparticles for catalysis and information technology. Macromol Symp. 2008, 270: 27-39. 10.1002/masy.200851004.
Daniel MC, Astruc D: Gold nanoparticles: assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem Rev. 2004, 104: 293-346. 10.1021/cr030698+.
Gao J, Gu H, Xu B: Multifunctional magnetic nanoparticles: design, synthesis, and biomedical applications. Acc Chem Res. 2009, 42: 1097-1107. 10.1021/ar9000026.
Horcajada P, Chalati T, Serre C, Gillet B, Sebrie C, Baati T, Eubank JF, Heurtaux D, Clayette P, Kreuz C, Chang JS, Hwang YK, Marsaud V, Bories PN, Cynober L, Gil S, Férey G, Couvreur P, Gref R: Porous metal-organic-framework nanoscale carriers as a potential platform for drug delivery and imaging. Nat Mater. 2010, 9: 172-178. 10.1038/nmat2608.
Sajja HK, East MP, Mao H, Wang YA, Nie S, Yang L: Development of multifunctional nanoparticles for targeted drug delivery and noninvasive imaging of therapeutic effect. Curr Drug Discovery Technol. 2009, 6: 43-51. 10.2174/157016309787581066.
Nam JM, Thaxton CS, Mirkin CA: Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003, 301: 1884-1886. 10.1126/science.1088755.
Na HB, Song IC, Hyeon T: Inorganic nanoparticles for MRI contrast agents. Adv Matr. 2009, 21: 2133-2148. 10.1002/adma.200802366.
Faraji AH, Wipf P: Nanoparticles in cellular drug delivery. Bioorg Med Chem. 2009, 17: 2950-2962. 10.1016/j.bmc.2009.02.043.
Sorenson JR: Use of essential metalloelement complexes or chelates in biological studies. J Free Rad Biol Med. 1992, 13: 593-594. 10.1016/0891-5849(92)90153-8.
Studer AM, Limbach LK, Van Duc L, Krumeich F, Athanassiou EK, Gerber LC, Moch H, Stark WJ: Nanoparticle cytotoxicity depends on intracellular solubility: Comparison of stabilized copper metal and degradable copper oxide nanoparticles. Toxicology Letters. 2010, 197: 169-174. 10.1016/j.toxlet.2010.05.012.
Burrows CJ, Muller JG: Oxidative nucleobase modifications leading to strand scission. Chem Rev. 1998, 98: 1109-1152. 10.1021/cr960421s.
López T, Figueras F, Manjarrez J, Bustos J, Alvarez M, Silvestre-Albero J, Rodriguez-Reinto F, Martínez-Ferre A, Martínez E: Catalytic nanomedicine: a new field in antitumor treatment using supported platinum nanoparticles. In vitro DNA degradation and in vivo tests with C6 animal model on Wistar rats. Eur J Med Chem. 2010, 45: 1982-1990.
Shen Q, Nie Z, Guo M, Zhong CJ, Lin B, Li W, Yao S: Simple and rapid colorimetric sensing of enzymatic cleavage and oxidative damage of single-stranded DNA with unmodified gold nanoparticles as indicator. Chem Commun. 2009, 28: 929-931. 10.1039/b818081d.
Lipovsky A, Tzitrinovich Z, Friedmann H, Applerot G, Gedanken A, Lubart R: EPR Study of Visible Light-Induced ROS Generation by Nanoparticles of ZnO. J Phys Chem C. 2009, 113: 15997-16001. 10.1021/jp904864g.
Portolés MJL, Gara PMD, Kotler ML, Bertolotti S, Román ES, Rodríguez HB, Gonzalez MC: Photophysical properties of blue-emitting silicon nanoparticles. Langmuir. 2010, 26: 10953-10960.
Zhang Y, Aslan K, Previte MJ, Geddes CD: Plasmonic engineering of singlet oxygen generation. PNAS. 2008, 105: 1798-1802. 10.1073/pnas.0709501105.
Barber PW, Chang RK, Massoudi H: Electrodynamic calculations of the surface- enhanced electric intensities on large Ag spheroids. Phys Rev B. 1983, 27: 7251-7261. 10.1103/PhysRevB.27.7251.
Yang WH, Schatz GC, Duyne RPV: Discrete dipole approximation for calculating extinction and Raman intensities for small particles with arbitrary shapes. J Chem Phys. 1995, 103: 869-875. 10.1063/1.469787.
Patra AK, Bhowmick T, Roy S, Ramakumar S, Chakravarty AR: Copper(II) complexes of L-arginine as netropsin mimics showing DNA cleavage activity in red light. Inorg Chem. 2009, 48: 2932-2943. 10.1021/ic8017425.
Tseng TS, Burstyn JN: Synthesis and DNA cleavage activity of a bifunctional intercalator-linked copper(II) macrocycle. Chem Commun. 2008, 6209-6211. 10.1039/b812183d.
Midander K, Cronholm P, Karlsson HL, Elihn K, Möller L, Leygraf C, Wallinder IO: Surface characteristics, copper release, and toxicity of nano- and micrometer-sized copper and copper(II) oxide particles: a cross-disciplinary study. Small. 2009, 5: 389-399. 10.1002/smll.200801220.
Chen Z, Meng H, Xing G, Chen C, Zhao Y, Jia G, Wang T, Yuan H, Ye C, Zhao F, Chai Z, Zhu C, Fang X, Ma B, Wan L: Acute toxicological effects of copper nanoparticles in vivo. Toxicol Lett. 2006, 163: 109-120. 10.1016/j.toxlet.2005.10.003.
Samim M, Kaushik NK, Maitra A: Effect of size of copper nanoparticles on its catalytic behaviour in Ullman reaction. Bull Mater Science. 2007, 30: 535-540. 10.1007/s12034-007-0083-9.
Basu-Modak S, Tyrrell RM: Singlet oxygen: a primary effector in the ultraviolet A/near-visible light induction of the human heme oxygenase gene. J Can Res. 1993, 53: 4505-4510.
Jain A, Alvi NK, Parish JH, Hadi SM: Oxygen is not required for degradation of DNA by glutathione and Cu(II). Mutat Res. 1996, 357: 83-88. 10.1016/0027-5107(96)00085-1.
Yamamoto K, Kawanishi S: Hydroxyl free radical is not the main active species in site- specific DNA damage induced by copper (II) ion and hydrogen peroxide. J Biol Chem. 1989, 264: 15435-15440.
Ren R, Yang P, Zheng W, Hua Z: A simple copper(II)-L-histidine system for efficient hydrolytic cleavage of DNA. Inorg Chem. 2000, 39: 5454-5463. 10.1021/ic0000146.
Rahban M, Divsalar A, Saboury AA, Golestani A: Nanotoxicity and spectroscopy studies of silver nanoparticle: Calf thymus DNA and K562 as targets. J Phys Chem C. 2010, 114: 5798-5803. 10.1021/jp910656g.
Mosmann T: Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Met. 1983, 65: 55-63. 10.1016/0022-1759(83)90303-4.
Limbach LK, Wick P, Manser P, Grass RN, Bruinink A, Stark WJ: Exposure of engineered nanoparticles to human lung epithelial cells: influence of chemical composition and catalytic activity on oxidative stress. Environ Sci Technol. 2007, 41: 4158-4163. 10.1021/es062629t.
Issa Y, Brunton P, Waters CM, Watts DC: Cytotoxicity of metal ions to human oligodendroglial cells and human gingival fibroblasts assessed by mitochondrial dehydrogenase activity. Dent Mater. 2008, 24: 281-287. 10.1016/j.dental.2007.09.010.
Sengupta TK, Leclerc GM, Hsieh-Kinser TT, Leclerc GJ, Singh I, Barredo JC: Cytotoxic effect of 5-aminoimidazole-4-carboxamide-1-beta-4-ribofuranoside (AICAR) on childhood acute lymphoblastic leukemia (ALL) cells: implication for targeted therapy. Mol Cancer. 2007, 10: 46-10.1186/1476-4598-6-46.
Acknowledgements
Authors wish to thank TEM facility of Indian Association for the Cultivation of Science. Authors also want to thank Govuthami Murugesan and Pritha Dasgupta for their assistance. Authors are grateful to the Reviewers for very useful suggestions. Authors also thank Dr. P.S. Ray for valuable suggestions and Mr. Ritabrata Ghosh for technical assistance for confocal microscopy. GPJ and SS thank Council of Scientific and industrial Research, Government of India for research fellowships. SKM thanks Department of Science and Technology, India for financial support.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SS synthesized and characterized copper nanoparticles. GPJ performed experiments. SKM and TKS conceived and designed the experiments. GPJ, SS, SKM and TKS interpreted the data and prepared the manuscript. All authors read and approved the manuscript.
Electronic supplementary material
12951_2010_118_MOESM1_ESM.DOC
Additional file 1: Additional Data Files. The file is organized into two sections. Section 1 describes essential methods. Section 2 provides graphical representation of change in the percentage of the super coiled DNA on incubation with copper nanoparticles fitted in to an exponential decay function (Figure S1) and Emission spectra of Ethidium Bromide bound to DNA in the absence and presence of different concentrations of copper nanoparticles (Figure S2). Figure S3 represents UV-Vis spectroscopic profile of CuNP (250 μM) incubated in PBS, pH 7.4 for different times at 37°C. Figure S4 represents UV-Vis spectroscopic profile of (A) CuNPs (250 μM) and (B) CuSO4 (250 μM) incubated in DMEM cell culture medium for different times at 37°C. (DOC 375 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Jose, G.P., Santra, S., Mandal, S.K. et al. Singlet oxygen mediated DNA degradation by copper nanoparticles: potential towards cytotoxic effect on cancer cells. J Nanobiotechnol 9, 9 (2011). https://doi.org/10.1186/1477-3155-9-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-3155-9-9








