Abstract
Background
Betaine is a methyl donor and has been considered as a lipotropic effect substance. But its mechanism remains unclear. Hepatic steatosis is associated with abnormal expression of genes involved in hepatic lipid metabolism. DNA methylation contributes to the disregulation of gene expression. Here we hypothesized that betaine supplement and subsequent DNA methylation modifications alter the expression of genes that are involved in hepatic lipid metabolism and hence alleviate hepatic triglyceride accumulation.
Methods
Male wild-type (WT) C57BL/6 mice (n = 6) were fed with the AIN-93 G diet. ApoE−/− mice (n = 12), weight-matched with the WT mice, were divided into two groups (n = 6 per group), and fed with the AIN-93 G diet and AIN-93 G supplemented with 2% betaine/100 g diet. Seven weeks after the intervention, mice were sacrificed. Liver betaine, choline, homocysteine concentration were measured by HPLC. Liver oxidants activity and triglyceride level were assessed by ultraviolet spectrophotometry. Finally, hepatic PPAR alpha gene and its target genes expression levels and the methylation status of the PPAR alpha gene were determined.
Results
ApoE−/− mice had higher hepatic triglyceride and lower GSH-Px activity when compared with the WT mice. Betaine intervention reversed triglyceride deposit, enhanced SOD and GSH-Px activity in the liver. Interestingly, mice fed on betaine-supplemented diet showed a dramatic increase of hepatic choline concentration and a decrease of betaine and homocysteine concentration relative to the WT mice and the ApoE−/− mice absent with betaine intervention. Expression of PPAR alpha and CPT1 were decreased and expression of FAS was markedly increased in ApoE−/− mice. In parallel, PPAR alpha promoter methylation level were slightly increased in ApoE−/− mice though without significance. Betaine supplement upregulated expression of PPAR alpha and its target genes (CPT1, CYP2E1) and reversed hypermethylation of PPAR alpha promoter of ApoE−/− mice. Furthermore, PPAR alpha methylation was positively correlated with hepatic betaine concentration.
Conclusions
Our findings indicate that betaine supplement could alleviate hepatic triglyceride accumulation and improve antioxidant capacity by decreasing PPAR alpha promoter methylation and upregulating PPAR alpha and its target genes mRNA expression.
Similar content being viewed by others
Background
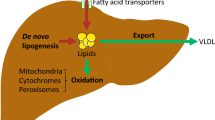
Triglyceride (TG) accumulation in hepatocytes is considered the primary manifestation of fatty liver disease[1]. In addition, the increase of liver triglyceride deposit is associated with obesity, hepatocellular carcinoma[2] and atherosclerosis[3]. Therefore, relieving hepatic lipid accumulation is viewed as potentially promising strategy for the prevention of many chronic diseases. The pathogenesis of hepatic TG deposit is incompletely understood. A previous study by others has shown that upregulation of genes expression for de novo lipogenesis (FAS, fatty acid synthase; ACC, acetyl-CoA carboxylase) and downregulation of genes expression for fatty acid oxidation (PPARα, peroxisomal proliferator-activated receptor alpha; CPT1, carnitine palmitoyl transferase I; UCP2, uncoupling proteins; ACOX, acyl-CoA oxidase; CYP2E1, cyto-chrome P450 2E1) are involved in the onsets of triglyceride accumulation in the liver[4].
Betaine, a methyl donor, is a naturally occurring compound in common foods, such as wheat bran, wheat germ, spinach, pretzels, shrimp and wheat bread etc.[5]. In vivo, betaine can also be produced by oxidation of choline, and it serves as an effective methyl donor for remethylating homocysteine (Hcy) into methionine (Met). DNA methylation in the CpG islands is one of the epigenetic mechanisms to regulate gene expression. Choline and methionine deficiency in diet are closely associated with biological functions by differentially methylated and inversely expressed genes in the tissue[6–8]. Mice with hyperhomocysteinemia, induced by heterozygous cystathionine β-synthase deficiency, show diminished methylation capacity and hypermethylation silencing of Fads2 mRNA expression which contribute to the impaired transport of TG[9]. High-fat diet can exacerbate methyl donors deficiency[10] and strikingly produce high level of serum Hcy, which may promote hypermethylation of MTTP gene and down-regulation of its expression, resulting in the hindrance to assembly lipoprotein and export lipid from liver[11]. It has become clear that PPARα can regulate the transcription of a suite of genes encoding enzymes in hepatic mitochondrial (CPT1, UCP2) and extramitochondrial (ACOX, CYP2E) fatty acid oxidation. Mice deficient in PPARα is demonstrated as a useful mouse model of fatty liver because of its important role in fatty acid oxidation and alleviation of hepatic TG[12]. Although An accumulating clinical and experimental evidences suggest that betaine is a lipotropic substance[13–15], the DNA methylation mechanism remains to be clearly defined.
In the present study, We attempt to investigate betaine supplement undergoing improvement on lipid metabolism and antioxidant capacity through changes in methylation level of PPARα promoter and expression of PPARα and its target genes(CPT1, UCP2, ACOX, CYP2E) in ApoE−/− mice .
Results
Effect of betaine supplement on body weight and liver weight
Body weight was matched before grouping. Absolute body weight of each mouse from each group was measured weekly and summarized in Figure1. As anticipated, body weight was increased after experiment initiation, gaining most rapidly in WT mice. No significant difference of body weight gain was found among groups, although the body weight gain in betaine-supplemented ApoE−/− mice was significant lower than that in WT mice after 6 weeks. Additionally, there were no significant differences in liver weight (F = 2.422, P = 0.075) and liver/body weight ratio (F = 0.533, P = 0.712) among the mice fed with or without betaine at 7 weeks (data not shown). These results suggested that betaine supplement had no significant influence on either the liver weight or the body weight.
Betaine supplement alleviates hepatic TG accumulation and improves antioxidant capacity
As anticipated, ApoE−/− mice fed with the AIN-93 G diet showed significantly higher hepatic TG content than that in the WT mice. After supplemented with betaine, hepatic TG level was significantly reduced (F = 19.040, P < 0.001) (Figure2A). Hepatic GSH-Px activity in the ApoE−/− mice was significantly lower than that in the WT mice, while betaine supplement strikingly increased GSH-Px activity (F = 9.696, P < 0.001) (Figure2B). Consistently, betaine feeding led to an significant elevation of hepatic SOD activity (F = 8.865, P < 0.001) (Figure2C). We hence suggested that betaine intervention was able to ameliorate hepatic TG deposit and improve antioxidant capacity in the ApoE−/− mice.
Effects of betaine supplement on hepatic TG levels (A) and GSH-Px (B) and SOD (C) activity. WT mice were fed with the AIN-93 G diet, while ApoE−/− mice were fed with the AIN-93 G in the absence and presence of 2% betaine. Values are means ± SE, n = 6. *P < 0.05 vs WT group, # P < 0.05 vs ApoE−/− group.
Effect of betaine supplement on liver Betaine, Choline and Hcy concentration
Hepatic betaine concentrations in the ApoE−/− mice were markedly higher and hepatic Hcy were significantly lower when compared with the WT mice. For the ApoE−/− mice, betaine supplement normalized hepatic betaine levels (F = 11.014, P < 0.001) (Figure3A), significantly increased hepatic choline levels (F = 36.492, P < 0.001) and reduced Hcy levels (F = 11.516, P < 0.001) (Figure3B and3C).
The effect of betaine supplement on the expression of lipid metabolism related genes
In the ApoE−/− mice, hepatic PPARα and CPT1 expression levels showed a trend of reduction when compared with WT mice, although the difference did not reach the statistical difference level. Significant up-regulation of PPARα (3.93 folds) and CYP2E1 (1.82 folds), however, was detected in the ApoE−/− mice by betaine supplementation (F = 12.494, P < 0.001) (Figure4). Meanwhile, hepatic FAS mRNA levels in the ApoE−/− mice were significantly higher than the WT controls, while betaine treatment significantly attenuated its expression levels. Despite these changes, betaine supplement did not exert effect on expression of other lipogenic and oxidative genes, such as ACC, UCP2, ACOX. These data suggested that betaine supplement selectively altered expression of certain genes. These results prompted us to further examine whether gene expression change in response to betaine supplement was influenced by DNA methylation.
Effect of betaine supplement on the expression of selected key genes involved in lipid metabolism. WT mice were fed with the AIN-93 G diet, while ApoE−/− mice were fed with the AIN-93 G in the absence and presence of 2% betaine. Values are means ± SE (n = 6). *P < 0.05 vs WT group, # P < 0.05 vs ApoE−/− group.
Betaine supplement decreases methylation level of hepatic PPARa promoter
We then chose the PPARα as target gene and utilized a real-time quantitative MSP method to quantitatively assess the methylation levels of PPARα promoter region in the liver. The average methylation in the PPARα gene promoter region of the ApoE−/− mice was 4.64%,compared with 2.58% in the WT mice. Betaine supplementation showed a trend of decreasing the methylation level, although the difference didn’t reach a statistical difference level (Figure5). Reduced methylation levels of PPARα promoter was accompanied by increased expression of the PPARα gene. A positive relationship was also found between the hepatic betaine levels and the methylation levels of the PPARα promoter (β = 0.572, P = 0.005) (Figure6). These results collectively suggested that the DNA methylation at the PPARα gene promoter could be regulated by betaine supplementation.
Discussion
The current study was designed to determine whether betaine intervention improves hepatic TG metabolism and antioxidant capacity in the ApoE−/− mice. There are two novel findings of this study. The first one is that betaine supplement can reduce hepatic TG content and improve antioxidant capacity,associated with the normalization of the expression of PPARα, CYP2E1,CPT1 and FAS. Secondly,we show here a tendency of increased DNA methylation in the PPARα promoter in the ApoE−/− mouse liver while betaine supplement is able to reverse the hypermethylation of the PPARα promoter.
ApoE−/− mouse is the classical modle of AS and dyslipidemia, characterized by high levels of plasma cholesterol and/or TG or low level of HDL, in addition, this mouse model indicates some incline to express fatty liver[16]. Our previous study[17] has shown that betaine has an anti-atherosclerosis effect in the ApoE−/− mice. We further evidenced that betaine intervention effectively relieved hepatic triglyceride deposit in the present study. Hepatic steatosis generated by environmental and genetic factors have been proved to influence genes expression involved in the process of lipid metabolism[4, 8, 9, 11, 18]. Fatty acids in the liver are derived from the de novo lipogenesis and the uptake from plasma, and catabolized by oxidation or exportation in the form of VLDL-TG. Fatty acid oxidation occurs mainly in the mitochondria, in which ATP is generated simultaneously. When the impairment of antioxidant capacity in mitochondria generated by excessive fatty acid occurs, the alternative peroxisomal β-oxidation and microsomal ω-oxidation will be activated which inevitably lead to the production of reactive oxygen species (ROS), including hydrogen peroxide (H2O2) and superoxide. PPARα is highly expressed in the liver and it is well-known to promote fatty catabolism by up-regulation of genes in mitochondral and extramitochondral fatty acid oxidation. The expression of hepatic PPARα expression has shown to be down-regulated in NAFLD patients[4], ApoE−/− mice also exhibited reduced hepatic PPARα expression compared with WT mice though without statistical significance in our present study. As we have expected, betaine supplement in ApoE−/− mice dramatically upregulated PPARα gene expression. The expression of CPT1, a key rate-limiting enzyme in mitochondral oxidation, just mirrored the alteration of PPARα expression in response to betaine treatment. CYP2E1 is one of the targets of PPARα, a key enzyme in the microsomal ω-oxidation. CYP2E1 overexpression in the liver is a definite factor for oxidative stress[19]. In the current study, we found that betaine treatment increased CYP2E1 expression which was paralleled by increased PPARα expression. We hence further investigated the activity of SOD and GSH-Px since these two enzymes are involved in the elimination of ROS derived from fatty acid oxidation. We observed a diminished antioxidant capacity in ApoE−/− mice, but elevated antioxidant activity in the liver of the ApoE−/− mice fed with betaine. These results indicated that CYP2E1 overexpression is unable to induce oxidative stress, but how ApoE−/− mice showed elevated hepatic TG and diminished antioxidant capacity and how betaine supplement reversed it? FAS and ACC are two key enzymes in the process of fatty acid synthesis. The current results revealed that expression of FAS was increased by five folds in the liver of the ApoE−/− mice when compared with WT controls, while betaine treatment strikingly repressed it. Accumulation of excessive hepatic TG and consequent oxidative stress have been confirmed to be the two common “hits” in the development of NAFLD[1]. Betaine treatment significantly increased PPARα, CPT1 and CYP2E1 expression and reduced the expression of FAS, the latter effect impaires fatty acid synthesis so as not to supply substrate for lipid peroxidation, and enhanced mitochondrial oxidation is sufficiently powerful to prevent TG deposit and oxidative stress.
The precise mechanisms by which betaine up regulates gene expression remain largely unclear. DNA methylation is one of the epigenetic modifications involved in the regulation of gene expression and hence versatile biological functions. Significant loss of global methylation was observed in the ApoE−/− mice by other investigators[20]. It is widely accepted that cells could undergo global DNA hypomethylation but simultaneously possess hypermethylation of a specific gene promoter, while the expression of a gene is usually silenced or attenuated when its promoter is heavily methylated. Methyl-deficient diet has been evidenced to exhibit hypermethylation promoter of genes involved in lipid metabolism which has inverse relationship with gene expression[8]. Betaine with its three chemically reactive methyl groups could serve as a methyl donor for DNA, RNA, histone and proteins, and its supplement may reverse alterations in DNA methylation induced by methyl deficiency. Here we investigated for the first time whether PPARα promoter methylation and corresponding gene expression can be affected by betaine supplement. The results of quantitative MSP assay showed that the methylation level of PPARα promoter was higher in the ApoE−/− mice when compared with that of WT mice, while betaine treatment showed a tendency to attenuate it. In attempt to explore the underlying mechanism, we did further analysis and found out that PPARα promoter methylation level was positively correlated with hepatic betaine concentration.
In present study, betaine supplement significantly decreased Hcy concentration and increased hepatic choline concentration. Unexpectedly, betaine treatment decreased hepatic betaine concentration as well. Endogenous betaine is oxidized from choline by two steps. Choline dehydrogenase (CHDH) mediates the first committed step for the formation of betaine aldehyde from choline, which is further catalyzed into betaine by betaine aldehyde dehydrogenase (BADH). Once betaine is formed, it serves as methyl donor for remethylating Hcy into Met by betaine-homocysteine methyltransferase (BHMT). Hepatic CHDH is rarely affected by dietary methionine and choline[21], but it has been shown to be competitively inhibited by both betaine aldehyde and betaine[22]. BHMT is expressed primarily in the liver of humans and mice[23]. Betaine-alone supplementation results in a 2-fold elevation of hepatic BHMT levels and activities[24]. Thus, betaine-elicited repression of CHDH as well as of activation BHMT can collectively increase the catabolism but decrease the synthesis of betaine in the liver. Our finding that betaine treatment dramaticly increased choline level and reduced Hcy level in the liver also confirmed this effect. This is among potential explainations why liver betaine levels in the betaine-supplemented ApoE−/− mice dropped to the levels that were comparable with the levels in the WT mice. We hence suggest that hepatic betaine depletion plays a role in the hypomethylation of PPARα promoter.
On the other hand, choline, another methyl donor, was observed to dramaticly increase by betaine intake. Betaine has been proved to have choline-sparing effect[25]. Choline is a basic component for the synthesis of Phosphatidylcholine (PC) which takes part in the assembly of mature VLDL. Betaine-containing diet could enhance the synthesis of hepatic SAM as well as the activity of phosphatidylethanolamine methyltransferase (PEMT), which subsequently facilitates the metabolic conversion of phosphatidylethanolamine (PE) to form PC[24]. ApoE−/− mice is characterized by lipid accumulation and the development of hepatic steatosis. As a result, more choline are available to increase PC synthesis and PC:PE ratio in VLDL, thereby promoting VLDL secretion and lipid transport[26]. These results suggest that betaine supplement exert important effects on the pool of related metabolites in the liver,whereby reduced betaine concentration in the liver connot supply enough substrate for DNA methylated modification and may selectively decrease the methylation statue of certain gene involved in lipid metabolism.
Conclusion
Taken together, the present study reports for the first time that betaine intervention reduces hepatic TG content and improves antioxidant capacity in the ApoE−/− mouse model. This is partially achieved by upregulating PPARα gene expression, as a result of hypomethylation of its promoter. These findings provided a novel insight into the important lipotropic effect of betaine. Due to its fundamental important role as a methyl-donor, betaine is gradually become a new target for the intervention of many chronic metabolic diseases. The hepatic and vascular protective effects of betaine we found provide promising insight into the human nutrition. Our present study has also build up a strong basis for further evaluation of potential mechanisms of betaine in epigenetic studies.
Methods
Chemicals
Choline chloride, Betaine glycine, d9-Betaine (internal standard), d9-choline (internal standard), trichloroacetic acid (TCA), tris (2-carboxylethyl) phosphine (TCEP), ammonium-7- fluorobenzo-2-oxa-1, 3-diazole-4-sulfonic acid (SBD-F), mercaptopropionylglycine (MPG), mineral oil were purchased from Sigma Chemicals. Methanol, chloroform, acetonitrile, formic acid were HPLC grade and obtained from Merck Chemical.
Animals and diet
Male wild-type (WT) C57BL/6 mice (n = 6) and ApoE−/− mice on the C57BL/6 background (n = 12) were obtained from the Jackson Laboratories (Bar Harbor, ME, USA). All mice were acclimated on a standard AIN-93 G diet for one week. The ApoE−/− mice were weight-matched and divided into two groups (n = 6 per group) feeding with the AIN-93 G diets supplemented with 0, 2 g betaine/100 g diet and the WT mice continue to consume the AIN-93 G diet. All mice were housed at a constant temperature, humidity and 12 hR light/dark cycle, and freely accessed to food and water for 7 weeks. Body weight and consumption of diet were recorded weekly. Mice were anesthetized and sacrificed after the experiment, liver were excised and stored at −80°C. This study was approved by the Institutional Animal Care and Use Committee at Sun Yat-Sen University.
Determination of betaine and choline concentration in the animal tissue
One hundred mg frozen liver tissue were homogenized in 600 μl methanol/chloroform (2:1,V/V) using an Ultrasonic Cell Disruptor (PRO scientific Inc. USA). The mixture was vortex-mixed vigorously and left at −20°C overnight. After the extraction, the mixture was centrifuged at 1500 g for 5 min. The supernatants were drawn and transferred to new tubes. The residuals were mixed with 300 μl methanol/chloroform/water (2:1:0.8,V/V/V) and centrifuged at 1500 g for 5 min. The supernatants from the two extractions were pooled. One hundred μl chloroform and 100 μl water were added into the mixture to form two phases. After a centrifugation, the aqueous phase was taken by a syringe, dried by N2 Blowing Concentrator (Organomation Associates Inc, America) and dissolved in 20 μl water. The dissolved water were treated with 800 μl methanol, 100 μl acetonitrile containing 10 μM internal standard and centrifuged at 6000 g for 2 min to precipitate protein. Finally the supernatants were injected into high performance liquid chromatography-tandem mass spectrometry (HPLC-MS) to analysis[27].
Determination of Hcy concentration in the animal tissue
Twenty five mg frozen liver tissue was homogenized in 200 μl cold PBS using an Ultrasonic Cell Disruptor (PRO scientific Inc, USA). The homogenized tissue suspension was treated with 50 μl TCEP (120 mg/ml) and 60 μl MPG (10 μM), vortex-mixed thoroughly, and incubated at 37°C for 30 min. After adding 125 μl TCA (100 g/L with 1 mM EDTA) and vortex-mixing thoroughly, the precipitated proteins were immediately centrifugated at 13000 g for 10 min. One hundred μl supernatant was incubated with 125 μl borate buffer (0.125 M, pH 9.5, with 4 mM EDTA), 10 μl NaOH solution (1.55 M) and 50 μl SBD-F solution (1 g/L in borate buffer) at 60°C for 60 min, placed in the room temperature to cool down, and then samples (10 μl) were injected into HPLC system to analysis[28].
Bisulfite conversion of DNA
Genomic DNA was isolated from the liver tissue with the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China). Treatment of DNA with bisulfite would result in the conversion of unmethylated cytosines to uracils, while methylated cytosines would remain unaltered. Briefly, 5.5 μL of 3 M NaOH was added into 1 ~ 2 μg of DNA (50 μL volume) and incubated at 42°C for 20 min. Following the incubation step, 520 μL of 3.6 M NaHSO3 (pH 5.0) and 30 μL 10 mM hydroquinone were added to each sample (fresh and avoiding exposure to light). All reagents were softly mixed, centrifuged and covered by mineral oil, then incubated at 53°C for 16 h. The bisulfite-converted DNA was purified by Wizard DNA Clean-Up System (Promega, USA). Finally DNA was eluated with 54 μL DDW at 80°C. 6 μL of 3 M NaOH was added into each sample and incubated at 42°C for 15 min. Following the incubation, DNA was precipitated using 6 μl of 3 M sodium acetate and 30 μl 100% cold ethanol at −80°C for 3 h. Each sample was centrifuged 30 min at 12,000 g and the supernatant liquid was poured out. The bisulfite-converted DNA was washed by 70% ethanol, dried and finally resuspended in a total volume of 10 μl.
Real-time quantitative MSP
The chemically modified DNA was subsequently used as a template for a QMSP method to determine the methylation level of the selected CpG dinucleotides in the PPAR α promoter. For the methylation-specific PCR experiment, two pairs of primers were designed to amplify either methylated or unmethylated sequences: one pair was specific for bisulfite-modified methylated DNA (M primers) and the other pair was specific for bisulfite-modified unmethylated DNA (U primers). The sequence was retrieved from NCBI at http://www.ncbi.nlm.nih.gov/gene? with the accession number [Genbank:NM_001113418.1]. Primers were designed via methprimer on line (http://www.urogene.org/methprimer/) and the sequences were shown in Table 1. NIH 3 T3 mouse genomic DNA and fully methylated NIH 3 T3 mouse genomic DNA (New England Biolabs, USA) were separately used as fully unmethylated and methylated positive control. Negative water blanks were also included. For each 20 μl Real-time quantitative PCR, 10 μl 2 × SYBR® Premix Ex Taq TMII, 0.2 μl of 10 μM forward primer, 0.2 μl of 10 μM reverse primer, 0.4 μl of 50 × ROX, 2 μl DNA template and 7.2 μl water were used. All of the regents could be obtained from SYBR® Premix Ex Taq TM II Kit(Takala). For the PPARa M and PPARa U Real-time quantitative MSP, the condition was 94°C for 3 min followed by 40 cycles of 94°C for 30 s, 55°C for 31 s, 72°C for 30 s. Data acquisition at annealing phase were analysed. After amplification, a melt curve was generated to confirm the specificity of PCR product. Serial dilutions of fully unmethylated amplified PCR product and fully methylated amplified PCR product were used to establish calibration curve. Data were collected by the Applied Biosystems 7500 fast real-time PCR system and analysed using 7500 software V2.0.1. The methylation index in a sample and the completeness of bisulfite conversion was estimated as previously described by Y. M. Dennis Lo[29]. Each sample was tested in duplicate. The melt curves in the present study confirmed stable and specific PCR products in both methylated (M) and unmethylated (U) systems. The linearity correlation coefficient of the standard curve of M system was 0.999, which demonstrated a large dynamic range and accuracy of real-time quantitative PCR. Similar results were obtained from U system as well.
RNA isolation and RT-PCR
Total RNA was isolated from the liver tissue with the TRIZOL reagent (Invitrogen, USA) according to the manufacturer’s instruction. The concentration and quality of total RNA was assessed by spectrophotometer, and its integrity was tested on agarose gels. Two μg RNA was reversely transcripted into cDNA with a commercial kit (Invitrogen, USA). Quantitative analysis of gene expression was performed by real-time polymerase chain reaction with an Applied Biosystems 7500 fast real-time PCR system. PCR primer information is summarized in Table 1.
Hepatic biochemical analysis
Total hepatic lipid was extracted by the method presented by Folch and colleagues[30]. Hepatic TG concentrations were determined using commercially available kits (Biosino Bio-Technology and Science Inc, Beijing, China) on a automatic biochemistry analyzer (A25Biosystem, Spain). Hepatic Superoxide dismutase (SOD), glutathione peroxidase (GSH-Px) activities were determined using correspondent kits (Nanjing Jiancheng Bio Inc, Nanjing, China) with ultraviolet spectrophotometry.
Statistic analysis
All results were presented as mean ± standard error (SE) and tested by the analysis of one way ANOVA followed by Bonferroni’s multiple comparison. Linear correlation was conducted to assess the relationship between hepatic betaine concentration and methylation level of PPARα gene. A two-tailed P < 0.05 was considered statistically significant.
Source of funding
This study was supported by grants from National Natural Science Foundation of China (81072302 and 81273050) and Natural Science Foundation of Guangdong Province, People’s Republic of China (10151008901000207).
Abbreviations
- ApoE−/−:
-
ApoE deficient
- WT:
-
Wide type
- TG:
-
Triglyceride
- GSH-Px:
-
Glutathione peroxidase
- SOD:
-
Superoxide dismutase
- Hcy:
-
Homocysteine
- PPARα:
-
Peroxisomal proliferator - activated receptor alpha
- CPT1:
-
Carnitine palmitoyl transferase I
- UCP2:
-
Uncoupling proteins
- ACOX:
-
Acyl-CoA oxidase
- CYP2E1:
-
Cyto-chrome P450 2E1
- FAS:
-
Fatty acid synthase
- ACC:
-
Acetyl-CoA carboxylase
References
Mehta K, Van Thiel DH, Shah N, Mobarhan S: Nonalcoholic fatty liver disease: pathogenesis and the role of antioxidants. Nutr Rev. 2002, 60 (9): 289-293. 10.1301/002966402320387224
Vuppalanchi R, Chalasani N: Nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: selected practical issues in their evaluation and management. Hepatology. 2009, 49 (1): 306-317. 10.1002/hep.22603
Targher G, Bertolini L, Padovani R, Rodella S, Zoppini G, Zenari L, Cigolini M, Falezza G, Arcaro G: Relations between carotid artery wall thickness and liver histology in subjects with nonalcoholic fatty liver disease. Diabetes Care. 2006, 29 (6): 1325-1330. 10.2337/dc06-0135
Kohjima M, Enjoji M, Higuchi N, Kato M, Kotoh K, Yoshimoto T, Fujino T, Yada M, Yada R, Harada N: Re-evaluation of fatty acid metabolism-related gene expression in nonalcoholic fatty liver disease. Int J Mol Med. 2007, 20 (3): 351-358.
Zeisel SH, Mar MH, Howe JC, Holden JM: Concentrations of choline-containing compounds and betaine in common foods. J Nutr. 2003, 133 (5): 1302-1307.
Niculescu MD, Craciunescu CN, Zeisel SH: Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J. 2006, 20 (1): 43-49. 10.1096/fj.05-4707com
Kovacheva VP, Mellott TJ, Davison JM, Wagner N, Lopez-Coviella I, Schnitzler AC, Blusztajn JK: Gestational choline deficiency causes global and Igf2 gene DNA hypermethylation by up-regulation of Dnmt1 expression. J Biol Chem. 2007, 282 (43): 31777-31788. 10.1074/jbc.M705539200
Tryndyak VP, Han T, Muskhelishvili L, Fuscoe JC, Ross SA, Beland FA, Pogribny IP: Coupling global methylation and gene expression profiles reveal key pathophysiological events in liver injury induced by a methyl-deficient diet. Mol Nutr Food Res. 2011, 55 (3): 411-418. 10.1002/mnfr.201000300
Devlin AM, Singh R, Wade RE, Innis SM, Bottiglieri T, Lentz SR: Hypermethylation of Fads2 and altered hepatic fatty acid and phospholipid metabolism in mice with hyperhomocysteinemia. J Biol Chem. 2007, 282 (51): 37082-37090. 10.1074/jbc.M704256200
Craig SA: Betaine in human nutrition. Am J Clin Nutr. 2004, 80 (3): 539-549.
Chang X, Yan H, Fei J, Jiang M, Zhu H, Lu D, Gao X: Berberine reduces methylation of the MTTP promoter and alleviates fatty liver induced by a high-fat diet in rats. J Lipid Res. 2010, 51 (9): 2504-2515. 10.1194/jlr.M001958
Leone TC, Weinheimer CJ, Kelly DP: A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci U S A. 1999, 96 (13): 7473-7478. 10.1073/pnas.96.13.7473
Schwahn BC, Wang XL, Mikael LG, Wu Q, Cohn J, Jiang H, Maclean KN, Rozen R: Betaine supplementation improves the atherogenic risk factor profile in a transgenic mouse model of hyperhomocysteinemia. Atherosclerosis. 2007, 195 (2): e100-e107. 10.1016/j.atherosclerosis.2007.06.030
Abdelmalek MF, Angulo P, Jorgensen RA, Sylvestre PB, Lindor KD: Betaine, a promising new agent for patients with nonalcoholic steatohepatitis: results of a pilot study. Am J Gastroenterol. 2001, 96 (9): 2711-2717. 10.1111/j.1572-0241.2001.04129.x
Abdelmalek MF, Sanderson SO, Angulo P, Soldevila-Pico C, Liu C, Peter J, Keach J, Cave M, Chen T, McClain CJ: Betaine for nonalcoholic fatty liver disease: results of a randomized placebo-controlled trial. Hepatology. 2009, 50 (6): 1818-1826. 10.1002/hep.23239
Xiao HB, Sun ZL, Lu XY, Li DZ, Xu JP, Hu YP: Beneficial effect of 3, 4, 5, 6- tetrahydroxyxanthone on dyslipidemia in apolipoprotein E-deficient mice. Can J Physiol Pharmacol. 2008, 86 (12): 815-826. 10.1139/Y08-091
Lv S, Fan R, Du Y, Hou M, Tang Z, Ling W, Zhu H: Betaine supplementation attenuates atherosclerotic lesion in apolipoprotein E-deficient mice. Eur J Nutr. 2009, 48 (4): 205-212. 10.1007/s00394-009-0003-4
Song Z, Deaciuc I, Zhou Z, Song M, Chen T, Hill D, McClain CJ: Involvement of AMP-activated protein kinase in beneficial effects of betaine on high-sucrose diet-induced hepatic steatosis. Am J Physiol Gastrointest Liver Physiol. 2007, 293 (4): G894-G902. 10.1152/ajpgi.00133.2007
Kathirvel E, Chen P, Morgan K, French SW, Morgan TR: Oxidative stress and regulation of anti-oxidant enzymes in cytochrome P4502E1 transgenic mouse model of non-alcoholic fatty liver. J Gastroenterol Hepatol. 2010, 25 (6): 1136-1143.
Hiltunen MO, Turunen MP, Hakkinen TP, Rutanen J, Hedman M, Makinen K, Turunen AM, Aalto-Setala K, Yla-Herttuala S: DNA hypomethylation and methyltransferase expression in atherosclerotic lesions. Vasc Med. 2002, 7 (1): 5-11. 10.1191/1358863x02vm418oa
Slow S, Garrow TA: Liver choline dehydrogenase and kidney betaine-homocysteine methyltransferase expression are not affected by methionine or choline intake in growing rats. J Nutr. 2006, 136 (9): 2279-2283.
Perrino LA, Pierce SK: Choline dehydrogenase kinetics contribute to glycine betaine regulation differences in chesapeake bay and atlantic oysters. J Exp Zool. 2000, 286 (3): 250-261. 10.1002/(SICI)1097-010X(20000215)286:3<250::AID-JEZ4>3.0.CO;2-X
Chen NC, Yang F, Capecci LM, Gu Z, Schafer AI, Durante W, Yang XF, Wang H: Regulation of homocysteine metabolism and methylation in human and mouse tissues. FASEB J. 2010, 24 (8): 2804-2817. 10.1096/fj.09-143651
Kharbanda KK, Mailliard ME, Baldwin CR, Beckenhauer HC, Sorrell MF, Tuma DJ: Betaine attenuates alcoholic steatosis by restoring phosphatidylcholine generation via the phosphatidylethanolamine methyltransferase pathway. J Hepatol. 2007, 46 (2): 314-321. 10.1016/j.jhep.2006.08.024
Dilger RN, Garrow TA, Baker DH: Betaine can partially spare choline in chicks but only when added to diets containing a minimal level of choline. J Nutr. 2007, 137 (10): 2224-2228.
Mensenkamp AR, Van Luyn MJ, Havinga R, Teusink B, Waterman IJ, Mann CJ, Elzinga BM, Verkade HJ, Zammit VA, Havekes LM: The transport of triglycerides through the secretory pathway of hepatocytes is impaired in apolipoprotein E deficient mice. J Hepatol. 2004, 40 (4): 599-606. 10.1016/j.jhep.2003.12.011
Holm PI, Ueland PM, Kvalheim G, Lien EA: Determination of choline, betaine, and dimethylglycine in plasma by a high-throughput method based on normal-phase chromatography-tandem mass spectrometry. Clin Chem. 2003, 49 (2): 286-294. 10.1373/49.2.286
Nolin TD, McMenamin ME, Himmelfarb J: Simultaneous determination of total homocysteine, cysteine, cysteinylglycine, and glutathione in human plasma by high-performance liquid chromatography: application to studies of oxidative stress. J Chromatogr B Analyt Technol Biomed Life Sci. 2007, 852 (1–2): 554-561.
Lo YM, Wong IH, Zhang J, Tein MS, Ng MH, Hjelm NM: Quantitative analysis of aberrant p16 methylation using real-time quantitative methylation-specific polymerase chain reaction. Cancer Res. 1999, 59 (16): 3899-3903.
Folch J, Lees M, Sloan SGH: A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957, 226: 497-509.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors have no conflict of financial interests to declare.
Authors’ contributions
LW participated in the whole process of the study, including carrying out molecular genetic experiments, data analyses and drafting manuscript. LC participated in the animal feeding experiment. YT and YC were in charge of determining hepatic metabolites by HPLC system. JW was in charge of assessing the levels of hepatic lipids and oxidants activities. TJ participated in the manuscript revising and language modifications. HZ was responsible for designing the research and helped to draft and revise manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Wang, L., Chen, L., Tan, Y. et al. Betaine supplement alleviates hepatic triglyceride accumulation of apolipoprotein E deficient mice via reducing methylation of peroxisomal proliferator-activated receptor alpha promoter. Lipids Health Dis 12, 34 (2013). https://doi.org/10.1186/1476-511X-12-34
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-511X-12-34